Abstract
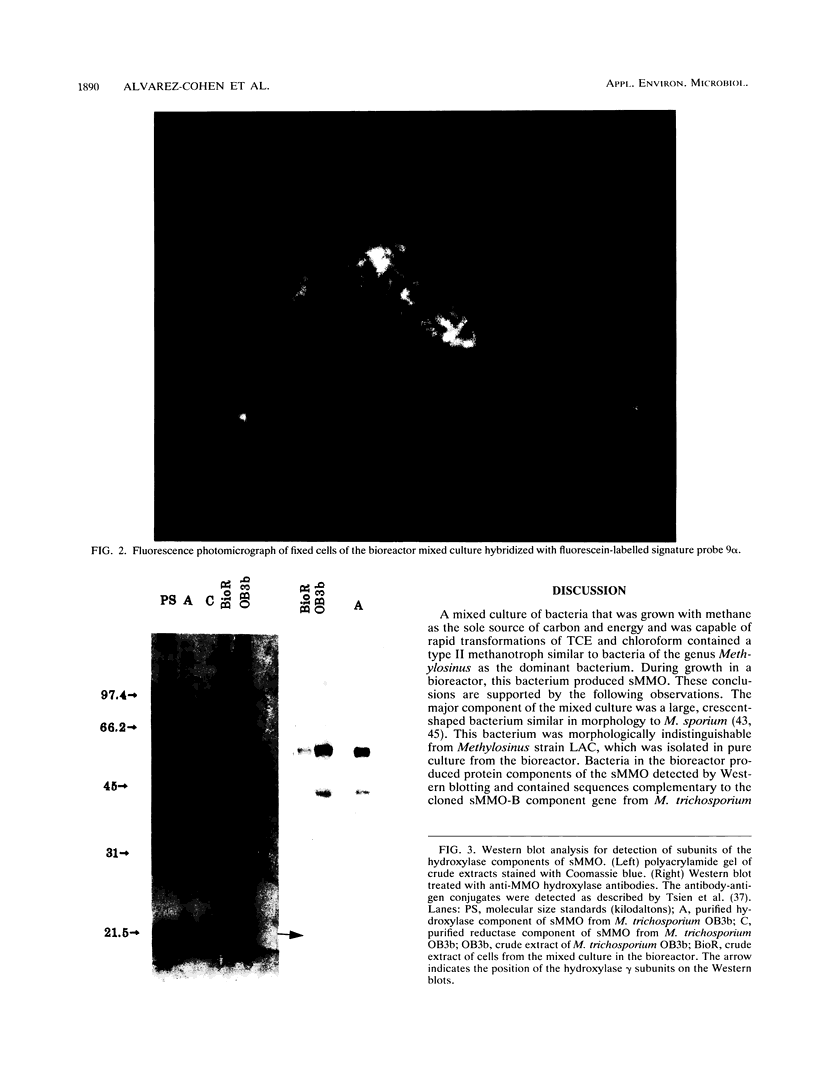
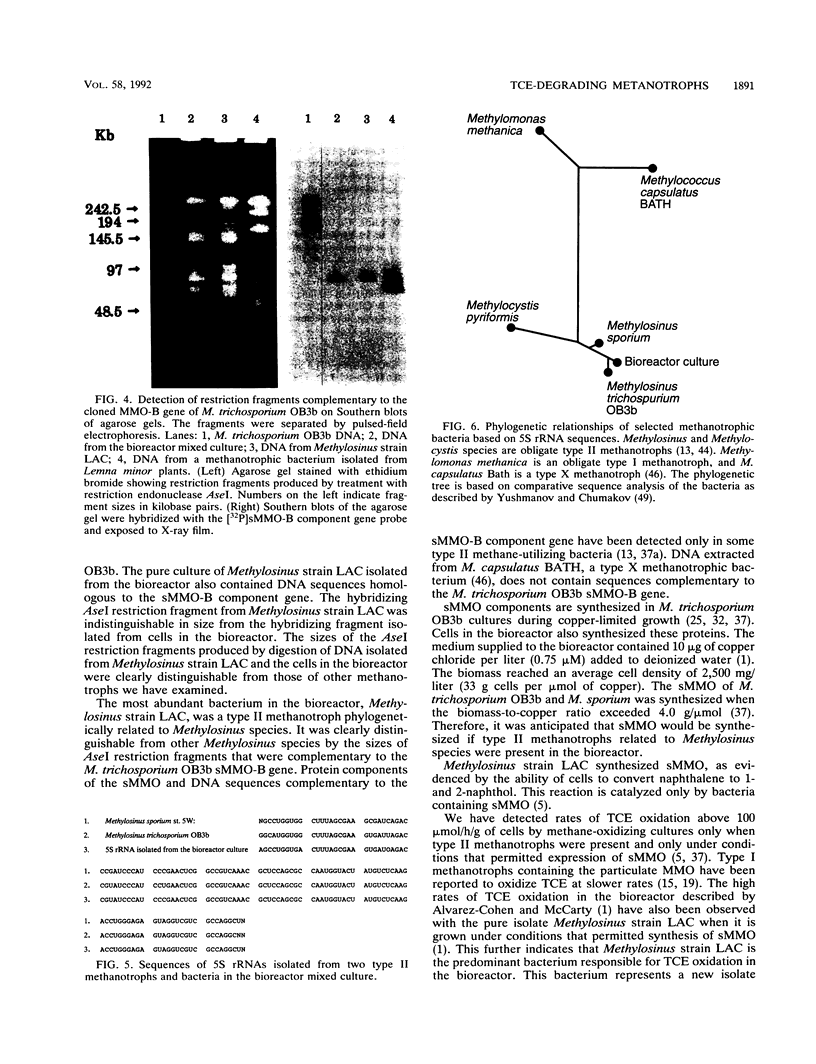
A mixed culture of bacteria grown in a bioreactor with methane as a carbon and energy source rapidly oxidized trichloroethylene and chloroform. The most abundant organism was a crescent-shaped bacterium that bound the fluorescent oligonucleotide signature probes that specifically hybridize to serine pathway methylotrophs. The 5S rRNA from this bacterium was found to be 93.5% homologous to the Methylosinus trichosporium OB3b 5S RNA sequence. A type II methanotrophic bacterium, isolated in pure culture from the bioreactor, synthesized soluble methane monooxygenase during growth in a copper-limited medium and was also capable of rapid trichloroethylene oxidation. The bacterium contained the gene that encodes the soluble methane monooxygenase B component on an AseI restriction fragment identical in size to a restriction fragment present in AseI digests of DNA from bacteria in the mixed culture. The sequence of the 16S rRNA from the pure culture was found to be 92 and 94% homologous to the 16S rRNAs of M. trichosporium OB3b and M. sporium, respectively. Both the pure and mixed cultures oxidized naphthalene to naphthol, indicating the presence of soluble methane monooxygenase. The mixed culture also synthesized soluble methane monooxygenase, as evidenced by the presence of proteins that cross-reacted with antibodies prepared against purified soluble methane monooxygenase components from M. trichosporium OB3b on Western blots (immunoblots). It was concluded that a type II methanotrophic bacterium phylogenetically related to Methylosinus species synthesizes soluble methane monooxygenase and is responsible for trichloroethylene oxidation in the bioreactor.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Alvarez-Cohen L., McCarty P. L. Effects of toxicity, aeration, and reductant supply on trichloroethylene transformation by a mixed methanotrophic culture. Appl Environ Microbiol. 1991 Jan;57(1):228–235. doi: 10.1128/aem.57.1.228-235.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvarez-Cohen L., McCarty P. L. Product toxicity and cometabolic competitive inhibition modeling of chloroform and trichloroethylene transformation by methanotrophic resting cells. Appl Environ Microbiol. 1991 Apr;57(4):1031–1037. doi: 10.1128/aem.57.4.1031-1037.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arciero D., Vannelli T., Logan M., Hooper A. B. Degradation of trichloroethylene by the ammonia-oxidizing bacterium Nitrosomonas europaea. Biochem Biophys Res Commun. 1989 Mar 15;159(2):640–643. doi: 10.1016/0006-291x(89)90042-9. [DOI] [PubMed] [Google Scholar]
- Brusseau G. A., Tsien H. C., Hanson R. S., Wackett L. P. Optimization of trichloroethylene oxidation by methanotrophs and the use of a colorimetric assay to detect soluble methane monooxygenase activity. Biodegradation. 1990;1(1):19–29. doi: 10.1007/BF00117048. [DOI] [PubMed] [Google Scholar]
- England T. E., Uhlenbeck O. C. 3'-terminal labelling of RNA with T4 RNA ligase. Nature. 1978 Oct 12;275(5680):560–561. doi: 10.1038/275560a0. [DOI] [PubMed] [Google Scholar]
- Fogel M. M., Taddeo A. R., Fogel S. Biodegradation of chlorinated ethenes by a methane-utilizing mixed culture. Appl Environ Microbiol. 1986 Apr;51(4):720–724. doi: 10.1128/aem.51.4.720-724.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox B. G., Borneman J. G., Wackett L. P., Lipscomb J. D. Haloalkene oxidation by the soluble methane monooxygenase from Methylosinus trichosporium OB3b: mechanistic and environmental implications. Biochemistry. 1990 Jul 10;29(27):6419–6427. doi: 10.1021/bi00479a013. [DOI] [PubMed] [Google Scholar]
- Fox B. G., Froland W. A., Dege J. E., Lipscomb J. D. Methane monooxygenase from Methylosinus trichosporium OB3b. Purification and properties of a three-component system with high specific activity from a type II methanotroph. J Biol Chem. 1989 Jun 15;264(17):10023–10033. [PubMed] [Google Scholar]
- Harder W., Attwood M. M. Biology, physiology and biochemistry of hyphomicrobia. Adv Microb Physiol. 1978;17:303–359. doi: 10.1016/s0065-2911(08)60060-0. [DOI] [PubMed] [Google Scholar]
- Henry S. M., Grbić-Galić D. Influence of endogenous and exogenous electron donors and trichloroethylene oxidation toxicity on trichloroethylene oxidation by methanotrophic cultures from a groundwater aquifer. Appl Environ Microbiol. 1991 Jan;57(1):236–244. doi: 10.1128/aem.57.1.236-244.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Little C. D., Palumbo A. V., Herbes S. E., Lidstrom M. E., Tyndall R. L., Gilmer P. J. Trichloroethylene biodegradation by a methane-oxidizing bacterium. Appl Environ Microbiol. 1988 Apr;54(4):951–956. doi: 10.1128/aem.54.4.951-956.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarty P. L. Bioengineering issues related to in situ remediation of contaminated soils and groundwater. Basic Life Sci. 1988;45:143–162. doi: 10.1007/978-1-4899-0824-7_9. [DOI] [PubMed] [Google Scholar]
- Miller R. E., Guengerich F. P. Metabolism of trichloroethylene in isolated hepatocytes, microsomes, and reconstituted enzyme systems containing cytochrome P-450. Cancer Res. 1983 Mar;43(3):1145–1152. [PubMed] [Google Scholar]
- Nelson M. J., Montgomery S. O., Mahaffey W. R., Pritchard P. H. Biodegradation of trichloroethylene and involvement of an aromatic biodegradative pathway. Appl Environ Microbiol. 1987 May;53(5):949–954. doi: 10.1128/aem.53.5.949-954.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oldenhuis R., Vink R. L., Janssen D. B., Witholt B. Degradation of chlorinated aliphatic hydrocarbons by Methylosinus trichosporium OB3b expressing soluble methane monooxygenase. Appl Environ Microbiol. 1989 Nov;55(11):2819–2826. doi: 10.1128/aem.55.11.2819-2826.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel R. N., Savas J. C. Purification and properties of the hydroxylase component of methane monooxygenase. J Bacteriol. 1987 May;169(5):2313–2317. doi: 10.1128/jb.169.5.2313-2317.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patt T. E., Cole G. C., Bland J., Hanson R. S. Isolation and characterization of bacteria that grow on methane and organic compounds as sole sources of carbon and energy. J Bacteriol. 1974 Nov;120(2):955–964. doi: 10.1128/jb.120.2.955-964.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peattie D. A. Direct chemical method for sequencing RNA. Proc Natl Acad Sci U S A. 1979 Apr;76(4):1760–1764. doi: 10.1073/pnas.76.4.1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stainthorpe A. C., Lees V., Salmond G. P., Dalton H., Murrell J. C. The methane monooxygenase gene cluster of Methylococcus capsulatus (Bath). Gene. 1990 Jul 2;91(1):27–34. doi: 10.1016/0378-1119(90)90158-n. [DOI] [PubMed] [Google Scholar]
- Strand S. E., Shippert L. Oxidation of chloroform in an aerobic soil exposed to natural gas. Appl Environ Microbiol. 1986 Jul;52(1):203–205. doi: 10.1128/aem.52.1.203-205.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsien H. C., Bratina B. J., Tsuji K., Hanson R. S. Use of oligodeoxynucleotide signature probes for identification of physiological groups of methylotrophic bacteria. Appl Environ Microbiol. 1990 Sep;56(9):2858–2865. doi: 10.1128/aem.56.9.2858-2865.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsien H. C., Brusseau G. A., Hanson R. S., Waclett L. P. Biodegradation of trichloroethylene by Methylosinus trichosporium OB3b. Appl Environ Microbiol. 1989 Dec;55(12):3155–3161. doi: 10.1128/aem.55.12.3155-3161.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuji K., Tsien H. C., Hanson R. S., DePalma S. R., Scholtz R., LaRoche S. 16S ribosomal RNA sequence analysis for determination of phylogenetic relationship among methylotrophs. J Gen Microbiol. 1990 Jan;136(1):1–10. doi: 10.1099/00221287-136-1-1. [DOI] [PubMed] [Google Scholar]
- Vannelli T., Logan M., Arciero D. M., Hooper A. B. Degradation of halogenated aliphatic compounds by the ammonia- oxidizing bacterium Nitrosomonas europaea. Appl Environ Microbiol. 1990 Apr;56(4):1169–1171. doi: 10.1128/aem.56.4.1169-1171.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel T. M., McCarty P. L. Biotransformation of tetrachloroethylene to trichloroethylene, dichloroethylene, vinyl chloride, and carbon dioxide under methanogenic conditions. Appl Environ Microbiol. 1985 May;49(5):1080–1083. doi: 10.1128/aem.49.5.1080-1083.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wackett L. P., Brusseau G. A., Householder S. R., Hanson R. S. Survey of microbial oxygenases: trichloroethylene degradation by propane-oxidizing bacteria. Appl Environ Microbiol. 1989 Nov;55(11):2960–2964. doi: 10.1128/aem.55.11.2960-2964.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whittenbury R., Phillips K. C., Wilkinson J. F. Enrichment, isolation and some properties of methane-utilizing bacteria. J Gen Microbiol. 1970 May;61(2):205–218. doi: 10.1099/00221287-61-2-205. [DOI] [PubMed] [Google Scholar]
- Wilson J. T., Wilson B. H. Biotransformation of trichloroethylene in soil. Appl Environ Microbiol. 1985 Jan;49(1):242–243. doi: 10.1128/aem.49.1.242-243.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolters J., Erdmann V. A. Compilation of 5S rRNA and 5S rRNA gene sequences. Nucleic Acids Res. 1988;16 (Suppl):r1–70. doi: 10.1093/nar/16.suppl.r1. [DOI] [PMC free article] [PubMed] [Google Scholar]






