Abstract
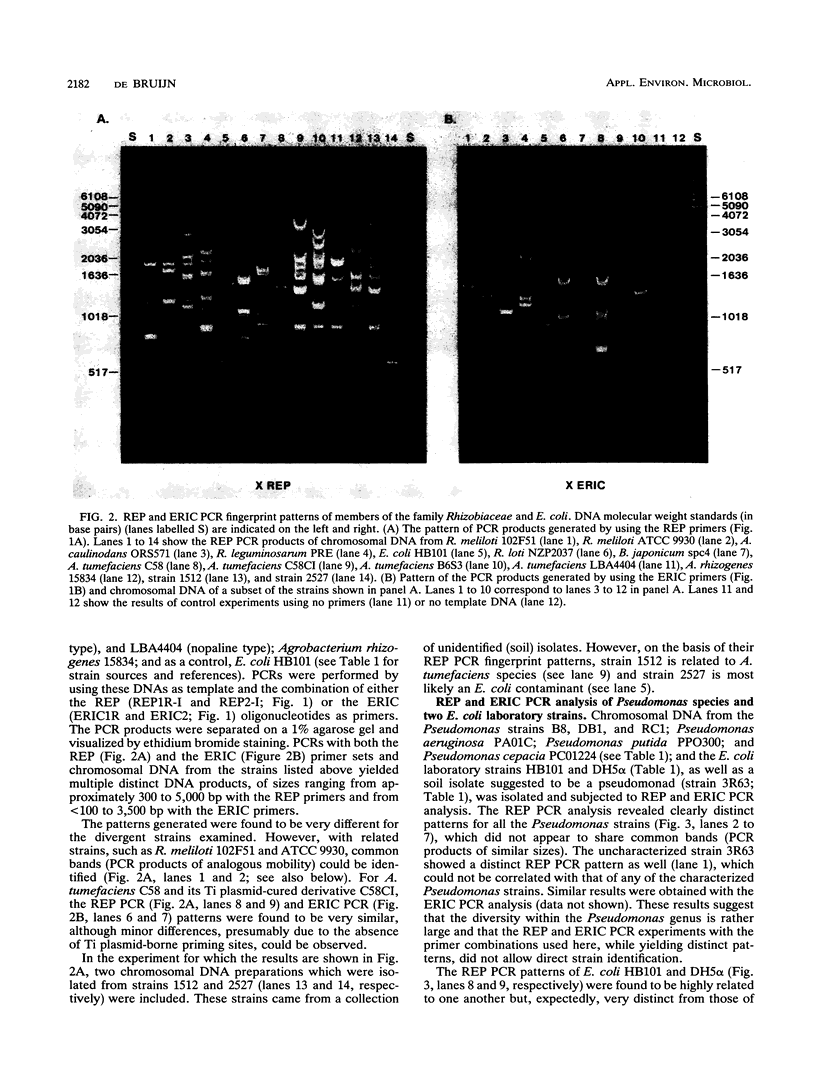
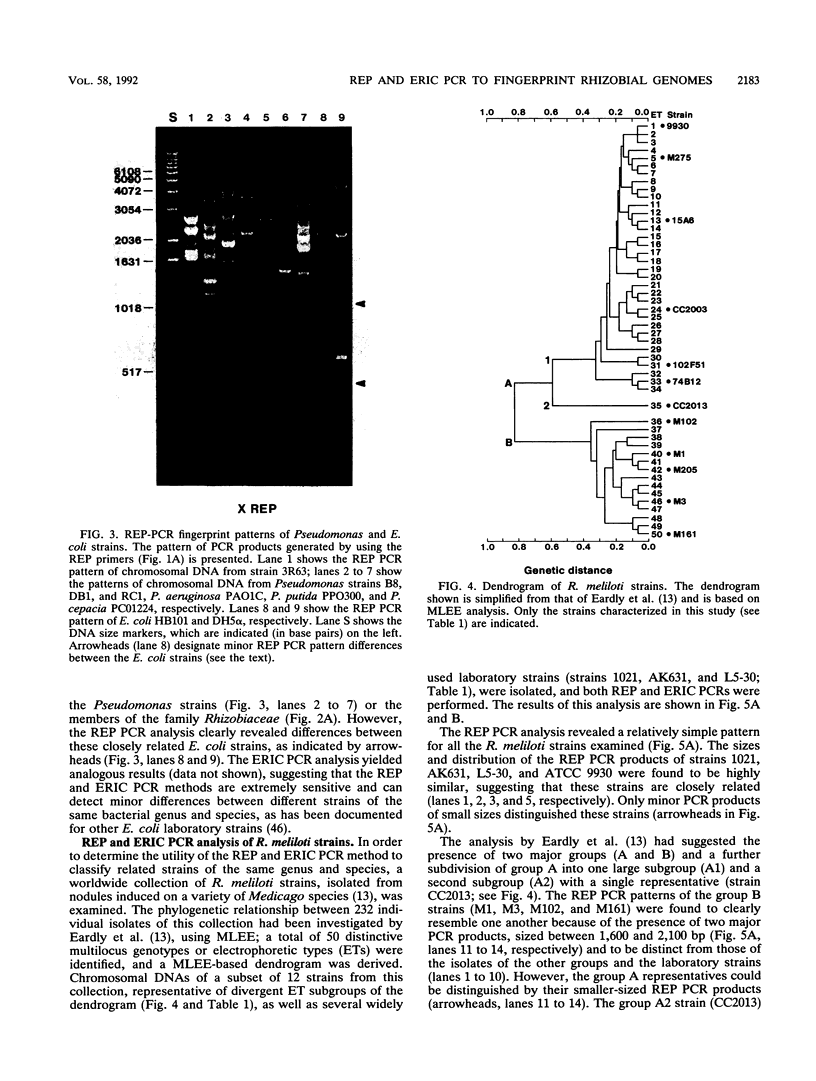
The distribution of dispersed repetitive DNA (repetitive extragenic palindromic [REP] and enterobacterial repetitive intergenic consensus [ERIC]) sequences in the genomes of a number of gram-negative soil bacteria was examined by using conserved primers corresponding to REP and ERIC sequences and the polymerase chain reaction (PCR). The patterns of the resulting PCR products were analyzed on agarose gels and found to be highly specific for each strain. The REP and ERIC PCR patterns of a series of Rhizobium meliloti isolates, previously ordered in a phylogenetic tree based on allelic variations at 14 enzyme loci (B. D. Eardly, L. A. Materon, N. H. Smith, D. A. Johnson, M. D. Rumbaugh, and R. K. Selander, Appl. Environ. Microbiol. 56:187-194), were determined. Isolates which had been postulated to be closely related by multilocus enzyme electrophoresis also revealed similar REP and ERIC PCR patterns, suggesting that the REP and ERIC PCR method is useful for the identification and classification of bacterial strains.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bej A. K., DiCesare J. L., Haff L., Atlas R. M. Detection of Escherichia coli and Shigella spp. in water by using the polymerase chain reaction and gene probes for uid. Appl Environ Microbiol. 1991 Apr;57(4):1013–1017. doi: 10.1128/aem.57.4.1013-1017.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bej A. K., Steffan R. J., DiCesare J., Haff L., Atlas R. M. Detection of coliform bacteria in water by polymerase chain reaction and gene probes. Appl Environ Microbiol. 1990 Feb;56(2):307–314. doi: 10.1128/aem.56.2.307-314.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beringer J. E. R factor transfer in Rhizobium leguminosarum. J Gen Microbiol. 1974 Sep;84(1):188–198. doi: 10.1099/00221287-84-1-188. [DOI] [PubMed] [Google Scholar]
- Boyer H. W., Roulland-Dussoix D. A complementation analysis of the restriction and modification of DNA in Escherichia coli. J Mol Biol. 1969 May 14;41(3):459–472. doi: 10.1016/0022-2836(69)90288-5. [DOI] [PubMed] [Google Scholar]
- Bromfield E. S., Sinha I. B., Wolynetz M. S. Influence of Location, Host Cultivar, and Inoculation on the Composition of Naturalized Populations of Rhizobium meliloti in Medicago sativa Nodules. Appl Environ Microbiol. 1986 May;51(5):1077–1084. doi: 10.1128/aem.51.5.1077-1084.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Greve H., Decraemer H., Seurinck J., Van Montagu M., Schell J. The functional organization of the octopine Agrobacterium tumefaciens plasmid pTiB6s3. Plasmid. 1981 Sep;6(2):235–248. doi: 10.1016/0147-619x(81)90069-x. [DOI] [PubMed] [Google Scholar]
- Demezas D. H., Reardon T. B., Watson J. M., Gibson A. H. Genetic Diversity among Rhizobium leguminosarum bv. Trifolii Strains Revealed by Allozyme and Restriction Fragment Length Polymorphism Analyses. Appl Environ Microbiol. 1991 Dec;57(12):3489–3495. doi: 10.1128/aem.57.12.3489-3495.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eardly B. D., Materon L. A., Smith N. H., Johnson D. A., Rumbaugh M. D., Selander R. K. Genetic structure of natural populations of the nitrogen-fixing bacterium Rhizobium meliloti. Appl Environ Microbiol. 1990 Jan;56(1):187–194. doi: 10.1128/aem.56.1.187-194.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilson E., Clément J. M., Brutlag D., Hofnung M. A family of dispersed repetitive extragenic palindromic DNA sequences in E. coli. EMBO J. 1984 Jun;3(6):1417–1421. doi: 10.1002/j.1460-2075.1984.tb01986.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilson E., Perrin D., Hofnung M. DNA polymerase I and a protein complex bind specifically to E. coli palindromic unit highly repetitive DNA: implications for bacterial chromosome organization. Nucleic Acids Res. 1990 Jul 11;18(13):3941–3952. doi: 10.1093/nar/18.13.3941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant S. G., Jessee J., Bloom F. R., Hanahan D. Differential plasmid rescue from transgenic mouse DNAs into Escherichia coli methylation-restriction mutants. Proc Natl Acad Sci U S A. 1990 Jun;87(12):4645–4649. doi: 10.1073/pnas.87.12.4645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgins C. F., Ames G. F., Barnes W. M., Clement J. M., Hofnung M. A novel intercistronic regulatory element of prokaryotic operons. Nature. 1982 Aug 19;298(5876):760–762. doi: 10.1038/298760a0. [DOI] [PubMed] [Google Scholar]
- Higgins C. F., McLaren R. S., Newbury S. F. Repetitive extragenic palindromic sequences, mRNA stability and gene expression: evolution by gene conversion? A review. Gene. 1988 Dec 10;72(1-2):3–14. doi: 10.1016/0378-1119(88)90122-9. [DOI] [PubMed] [Google Scholar]
- Holsters M., Silva B., Van Vliet F., Genetello C., De Block M., Dhaese P., Depicker A., Inzé D., Engler G., Villarroel R. The functional organization of the nopaline A. tumefaciens plasmid pTiC58. Plasmid. 1980 Mar;3(2):212–230. doi: 10.1016/0147-619x(80)90110-9. [DOI] [PubMed] [Google Scholar]
- Hulton C. S., Higgins C. F., Sharp P. M. ERIC sequences: a novel family of repetitive elements in the genomes of Escherichia coli, Salmonella typhimurium and other enterobacteria. Mol Microbiol. 1991 Apr;5(4):825–834. doi: 10.1111/j.1365-2958.1991.tb00755.x. [DOI] [PubMed] [Google Scholar]
- Martínez-Romero E., Segovia L., Mercante F. M., Franco A. A., Graham P., Pardo M. A. Rhizobium tropici, a novel species nodulating Phaseolus vulgaris L. beans and Leucaena sp. trees. Int J Syst Bacteriol. 1991 Jul;41(3):417–426. doi: 10.1099/00207713-41-3-417. [DOI] [PubMed] [Google Scholar]
- McClelland M., Jones R., Patel Y., Nelson M. Restriction endonucleases for pulsed field mapping of bacterial genomes. Nucleic Acids Res. 1987 Aug 11;15(15):5985–6005. doi: 10.1093/nar/15.15.5985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meade H. M., Long S. R., Ruvkun G. B., Brown S. E., Ausubel F. M. Physical and genetic characterization of symbiotic and auxotrophic mutants of Rhizobium meliloti induced by transposon Tn5 mutagenesis. J Bacteriol. 1982 Jan;149(1):114–122. doi: 10.1128/jb.149.1.114-122.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullis K. B., Faloona F. A. Specific synthesis of DNA in vitro via a polymerase-catalyzed chain reaction. Methods Enzymol. 1987;155:335–350. doi: 10.1016/0076-6879(87)55023-6. [DOI] [PubMed] [Google Scholar]
- Murphy P. J., Heycke N., Banfalvi Z., Tate M. E., de Bruijn F., Kondorosi A., Tempé J., Schell J. Genes for the catabolism and synthesis of an opine-like compound in Rhizobium meliloti are closely linked and on the Sym plasmid. Proc Natl Acad Sci U S A. 1987 Jan;84(2):493–497. doi: 10.1073/pnas.84.2.493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newbury S. F., Smith N. H., Robinson E. C., Hiles I. D., Higgins C. F. Stabilization of translationally active mRNA by prokaryotic REP sequences. Cell. 1987 Jan 30;48(2):297–310. doi: 10.1016/0092-8674(87)90433-8. [DOI] [PubMed] [Google Scholar]
- Ooms G., Hooykaas P. J., Moolenaar G., Schilperoort R. A. Grown gall plant tumors of abnormal morphology, induced by Agrobacterium tumefaciens carrying mutated octopine Ti plasmids; analysis of T-DNA functions. Gene. 1981 Jun-Jul;14(1-2):33–50. doi: 10.1016/0378-1119(81)90146-3. [DOI] [PubMed] [Google Scholar]
- Pinero D., Martinez E., Selander R. K. Genetic diversity and relationships among isolates of Rhizobium leguminosarum biovar phaseoli. Appl Environ Microbiol. 1988 Nov;54(11):2825–2832. doi: 10.1128/aem.54.11.2825-2832.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Putnoky P., Petrovics G., Kereszt A., Grosskopf E., Ha D. T., Bánfalvi Z., Kondorosi A. Rhizobium meliloti lipopolysaccharide and exopolysaccharide can have the same function in the plant-bacterium interaction. J Bacteriol. 1990 Sep;172(9):5450–5458. doi: 10.1128/jb.172.9.5450-5458.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Regensburger B., Hennecke H. RNA polymerase from Rhizobium japonicum. Arch Microbiol. 1983 Aug;135(2):103–109. doi: 10.1007/BF00408017. [DOI] [PubMed] [Google Scholar]
- Roberts G. P., Leps W. T., Silver L. E., Brill W. J. Use of two-dimensional polyacrylamide gel electrophoresis to identify and classify Rhizobium strains. Appl Environ Microbiol. 1980 Feb;39(2):414–422. doi: 10.1128/aem.39.2.414-422.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sayler G. S., Layton A. C. Environmental application of nucleic acid hybridization. Annu Rev Microbiol. 1990;44:625–648. doi: 10.1146/annurev.mi.44.100190.003205. [DOI] [PubMed] [Google Scholar]
- Selander R. K., Caugant D. A., Ochman H., Musser J. M., Gilmour M. N., Whittam T. S. Methods of multilocus enzyme electrophoresis for bacterial population genetics and systematics. Appl Environ Microbiol. 1986 May;51(5):873–884. doi: 10.1128/aem.51.5.873-884.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharples G. J., Lloyd R. G. A novel repeated DNA sequence located in the intergenic regions of bacterial chromosomes. Nucleic Acids Res. 1990 Nov 25;18(22):6503–6508. doi: 10.1093/nar/18.22.6503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shyamala V., Schneider E., Ames G. F. Tandem chromosomal duplications: role of REP sequences in the recombination event at the join-point. EMBO J. 1990 Mar;9(3):939–946. doi: 10.1002/j.1460-2075.1990.tb08192.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sobral B. W., Honeycutt R. J., Atherly A. G., McClelland M. Electrophoretic separation of the three Rhizobium meliloti replicons. J Bacteriol. 1991 Aug;173(16):5173–5180. doi: 10.1128/jb.173.16.5173-5180.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steffan R. J., Atlas R. M. Polymerase chain reaction: applications in environmental microbiology. Annu Rev Microbiol. 1991;45:137–161. doi: 10.1146/annurev.mi.45.100191.001033. [DOI] [PubMed] [Google Scholar]
- Stern M. J., Ames G. F., Smith N. H., Robinson E. C., Higgins C. F. Repetitive extragenic palindromic sequences: a major component of the bacterial genome. Cell. 1984 Jul;37(3):1015–1026. doi: 10.1016/0092-8674(84)90436-7. [DOI] [PubMed] [Google Scholar]
- Stern M. J., Prossnitz E., Ames G. F. Role of the intercistronic region in post-transcriptional control of gene expression in the histidine transport operon of Salmonella typhimurium: involvement of REP sequences. Mol Microbiol. 1988 Jan;2(1):141–152. doi: 10.1111/j.1365-2958.1988.tb00015.x. [DOI] [PubMed] [Google Scholar]
- Subramanian P. S., Versalovic J., McCabe E. R., Lupski J. R. Rapid mapping of Escherichia coli::Tn5 insertion mutations by REP-Tn5 PCR. PCR Methods Appl. 1992 Feb;1(3):187–192. doi: 10.1101/gr.1.3.187. [DOI] [PubMed] [Google Scholar]
- Versalovic J., Koeuth T., Lupski J. R. Distribution of repetitive DNA sequences in eubacteria and application to fingerprinting of bacterial genomes. Nucleic Acids Res. 1991 Dec 25;19(24):6823–6831. doi: 10.1093/nar/19.24.6823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welsh J., McClelland M. Fingerprinting genomes using PCR with arbitrary primers. Nucleic Acids Res. 1990 Dec 25;18(24):7213–7218. doi: 10.1093/nar/18.24.7213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welsh J., McClelland M. Genomic fingerprints produced by PCR with consensus tRNA gene primers. Nucleic Acids Res. 1991 Feb 25;19(4):861–866. doi: 10.1093/nar/19.4.861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White F. F., Nester E. W. Hairy root: plasmid encodes virulence traits in Agrobacterium rhizogenes. J Bacteriol. 1980 Mar;141(3):1134–1141. doi: 10.1128/jb.141.3.1134-1141.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams J. G., Kubelik A. R., Livak K. J., Rafalski J. A., Tingey S. V. DNA polymorphisms amplified by arbitrary primers are useful as genetic markers. Nucleic Acids Res. 1990 Nov 25;18(22):6531–6535. doi: 10.1093/nar/18.22.6531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woese C. R. Bacterial evolution. Microbiol Rev. 1987 Jun;51(2):221–271. doi: 10.1128/mr.51.2.221-271.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolk C. P., Cai Y., Panoff J. M. Use of a transposon with luciferase as a reporter to identify environmentally responsive genes in a cyanobacterium. Proc Natl Acad Sci U S A. 1991 Jun 15;88(12):5355–5359. doi: 10.1073/pnas.88.12.5355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y., Ames G. F. DNA gyrase binds to the family of prokaryotic repetitive extragenic palindromic sequences. Proc Natl Acad Sci U S A. 1988 Dec;85(23):8850–8854. doi: 10.1073/pnas.85.23.8850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Bruijn F. J. Transposon Tn5 mutagenesis to map genes. Methods Enzymol. 1987;154:175–196. doi: 10.1016/0076-6879(87)54077-0. [DOI] [PubMed] [Google Scholar]