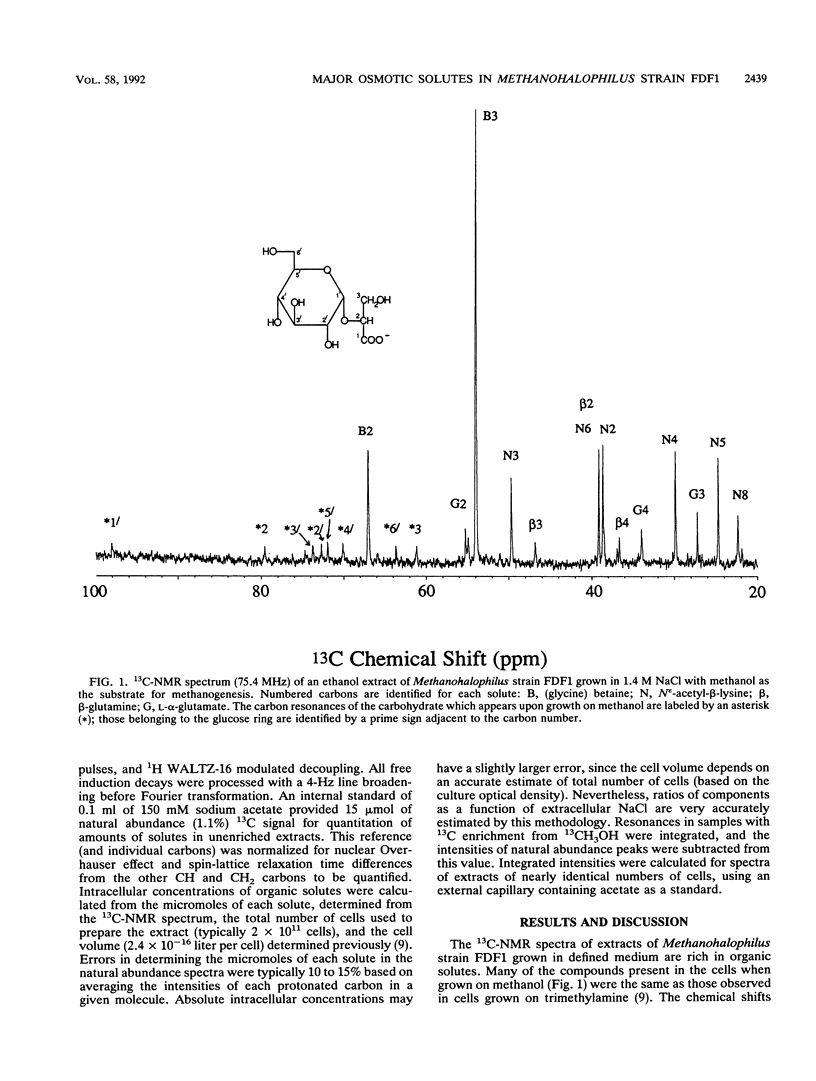
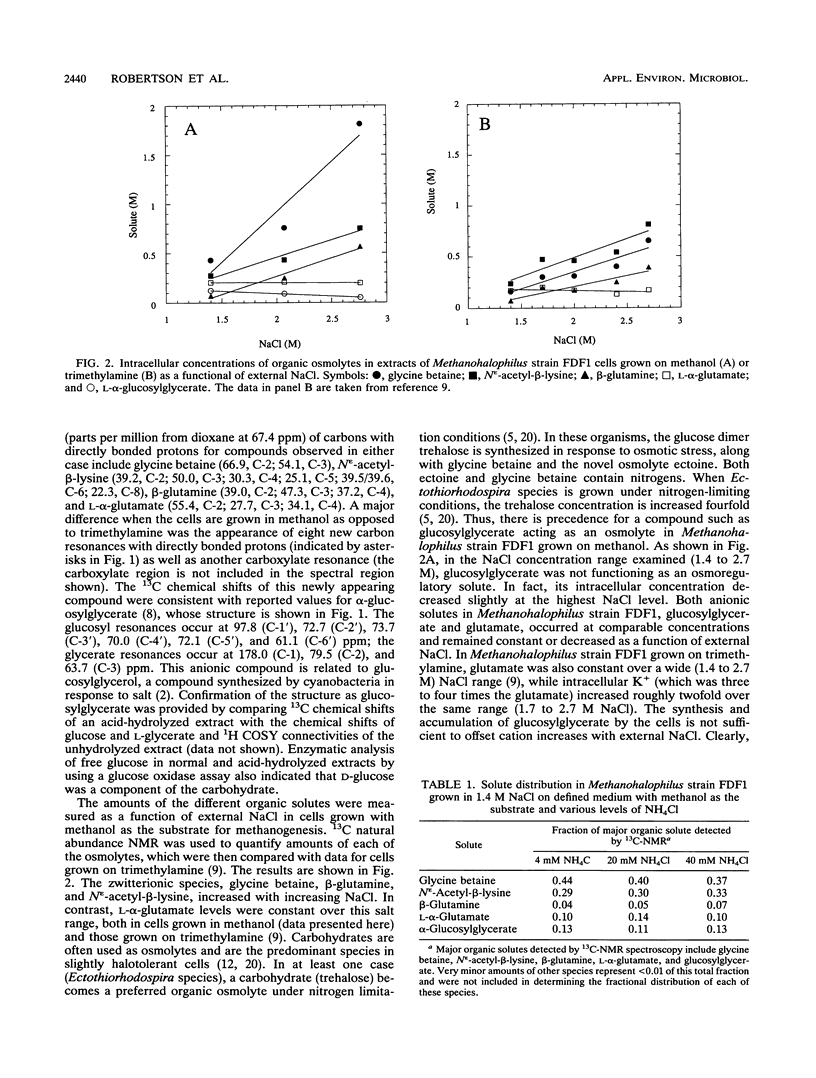
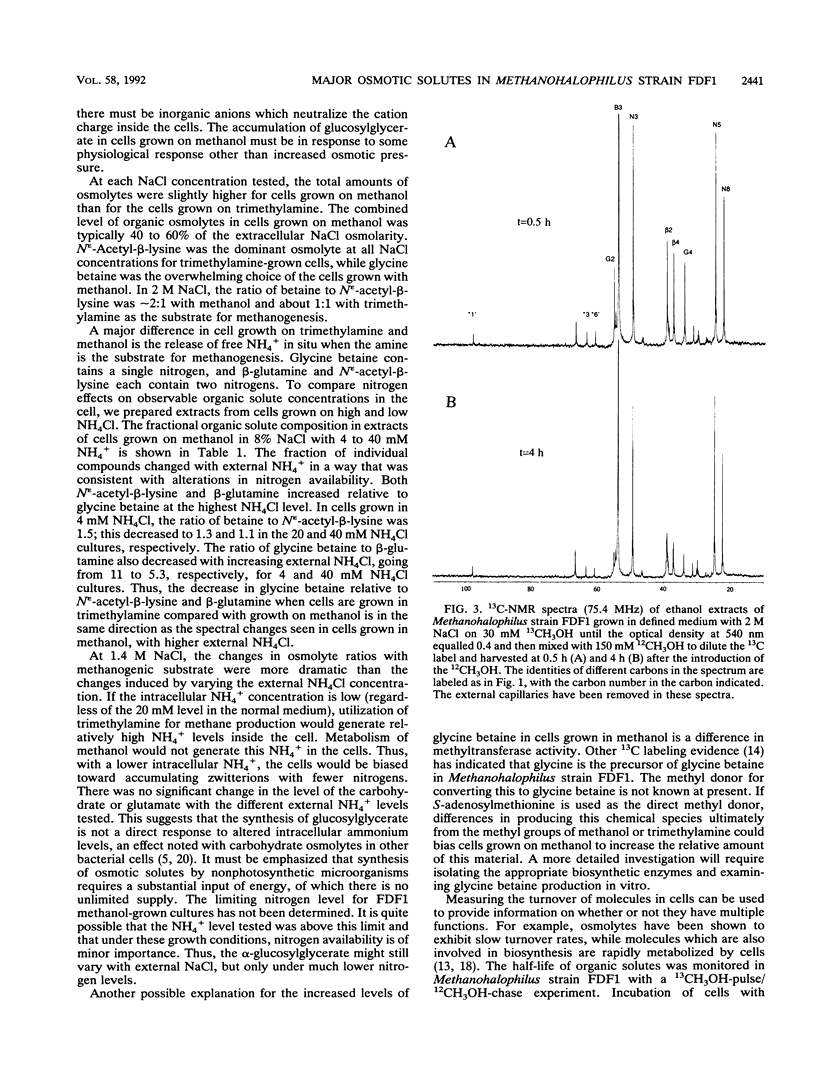
Abstract
Methanohalophilus strain FDF1, a member of the halophilic genus of methanogens, can grow over a range of external NaCl concentrations from 1.2 to 2.9 M and utilize methanol, trimethylamine, and dimethyl sulfide as substrates for methanogenesis. It produces the osmolytes glycine betaine, β-glutamine, and Nε-acetyl-β-lysine with increasing external NaCl, but the relative ratio of these zwitterions depends primarily on the methanogenic substrate and less on the external osmolarity. When the cells are grown on methanol in defined medium, accumulation of glycine betaine predominates over the other zwitterionic solutes. The cells also synthesized a carbohydrate which was not detected in cells grown on trimethylamine. This negatively charged compound, identified as α-glucosylglycerate from the 13C and 1H chemical shifts, does not act as an osmoregulatory solute in the salt range 1.4 to 2.7 M in this methanogen as evidenced by its invariant intracellular concentration. 13CH3OH-pulse/12CH3OH-chase experiments were used to determine half-lifes for these organic solute pools in the cells. l-α-Glutamate showed a rapid loss of heavy isotope, indicating that l-α-glutamate functions as a biosynthetic intermediate in these cells. Measurable turnover rates for both β-glutamine, which acts as an osmolyte, and α-glucosylglycerate suggest that they function as metabolic intermediates as well. Molecules which function solely as osmolytes (glycine betaine and Nε-acetyl-β-lysine) showed a slower turnover consistent with their roles as osmotic solutes in Methanohalophilus strain FDF1.
Full text
PDF


Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Balch W. E., Fox G. E., Magrum L. J., Woese C. R., Wolfe R. S. Methanogens: reevaluation of a unique biological group. Microbiol Rev. 1979 Jun;43(2):260–296. doi: 10.1128/mr.43.2.260-296.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borowitzka L. J., Demmerle S., Mackay M. A., Norton R. S. Carbon-13 nuclear magnetic resonance study of osmoregulation in a blue-green alga. Science. 1980 Nov 7;210(4470):650–651. doi: 10.1126/science.210.4470.650. [DOI] [PubMed] [Google Scholar]
- Daniels L., Sparling R., Sprott G. D. The bioenergetics of methanogenesis. Biochim Biophys Acta. 1984 Sep 6;768(2):113–163. doi: 10.1016/0304-4173(84)90002-8. [DOI] [PubMed] [Google Scholar]
- Ekiel I., Smith I. C., Sprott G. D. Biosynthetic pathways in Methanospirillum hungatei as determined by 13C nuclear magnetic resonance. J Bacteriol. 1983 Oct;156(1):316–326. doi: 10.1128/jb.156.1.316-326.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones W. J., Nagle D. P., Jr, Whitman W. B. Methanogens and the diversity of archaebacteria. Microbiol Rev. 1987 Mar;51(1):135–177. doi: 10.1128/mr.51.1.135-177.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai M. C., Sowers K. R., Robertson D. E., Roberts M. F., Gunsalus R. P. Distribution of compatible solutes in the halophilic methanogenic archaebacteria. J Bacteriol. 1991 Sep;173(17):5352–5358. doi: 10.1128/jb.173.17.5352-5358.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathrani I. M., Boone D. R. Isolation and characterization of a moderately halophilic methanogen from a solar saltern. Appl Environ Microbiol. 1985 Jul;50(1):140–143. doi: 10.1128/aem.50.1.140-143.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts M. F., Choi B. S., Robertson D. E., Lesage S. Free amino acid turnover in methanogens measured by 15N NMR spectroscopy. J Biol Chem. 1990 Oct 25;265(30):18207–18212. [PubMed] [Google Scholar]
- Robertson D. E., Lesage S., Roberts M. F. Beta-aminoglutaric acid is a major soluble component of Methanococcus thermolithotrophicus. Biochim Biophys Acta. 1989 Sep 15;992(3):320–326. doi: 10.1016/0304-4165(89)90091-3. [DOI] [PubMed] [Google Scholar]
- Robertson D. E., Noll D., Roberts M. F., Menaia J. A., Boone D. R. Detection of the osmoregulator betaine in methanogens. Appl Environ Microbiol. 1990 Feb;56(2):563–565. doi: 10.1128/aem.56.2.563-565.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson D. E., Roberts M. F., Belay N., Stetter K. O., Boone D. R. Occurrence of beta-glutamate, a novel osmolyte, in marine methanogenic bacteria. Appl Environ Microbiol. 1990 May;56(5):1504–1508. doi: 10.1128/aem.56.5.1504-1508.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson D. E., Roberts M. F. Organic osmolytes in methanogenic archaebacteria. Biofactors. 1991 Jan;3(1):1–9. [PubMed] [Google Scholar]
- Sowers K. R., Robertson D. E., Noll D., Gunsalus R. P., Roberts M. F. N epsilon-acetyl-beta-lysine: an osmolyte synthesized by methanogenic archaebacteria. Proc Natl Acad Sci U S A. 1990 Dec;87(23):9083–9087. doi: 10.1073/pnas.87.23.9083. [DOI] [PMC free article] [PubMed] [Google Scholar]


