Abstract
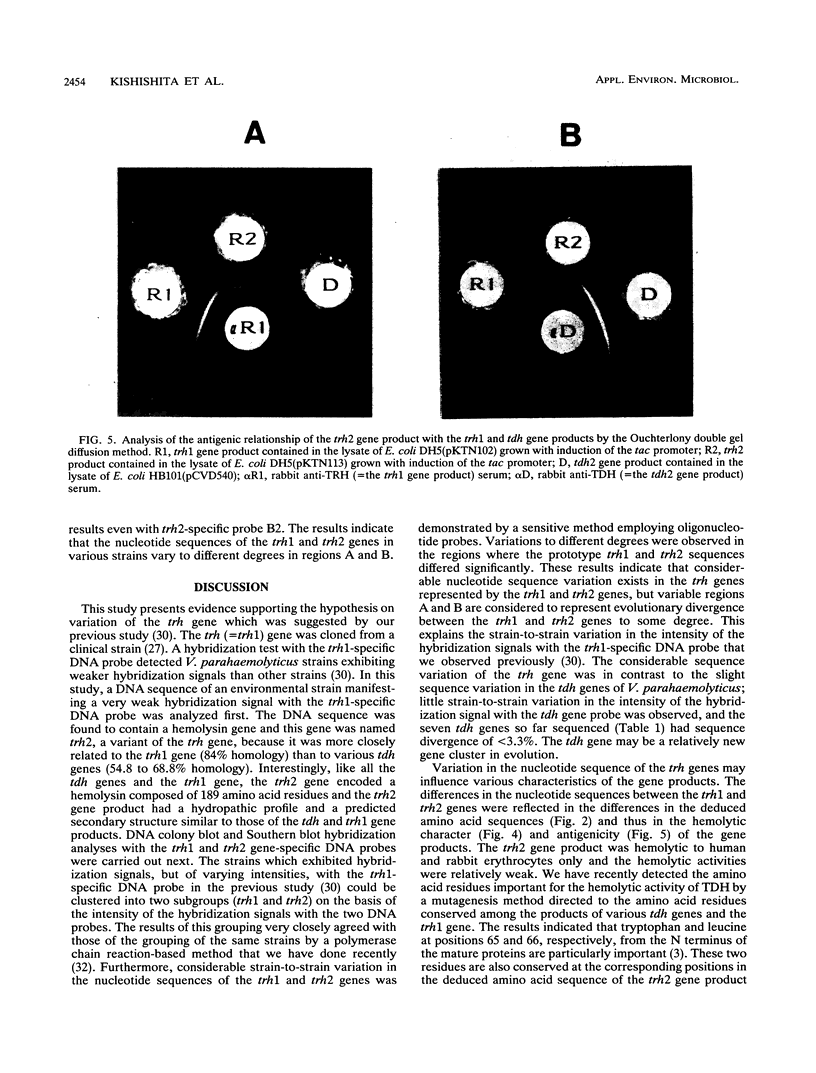
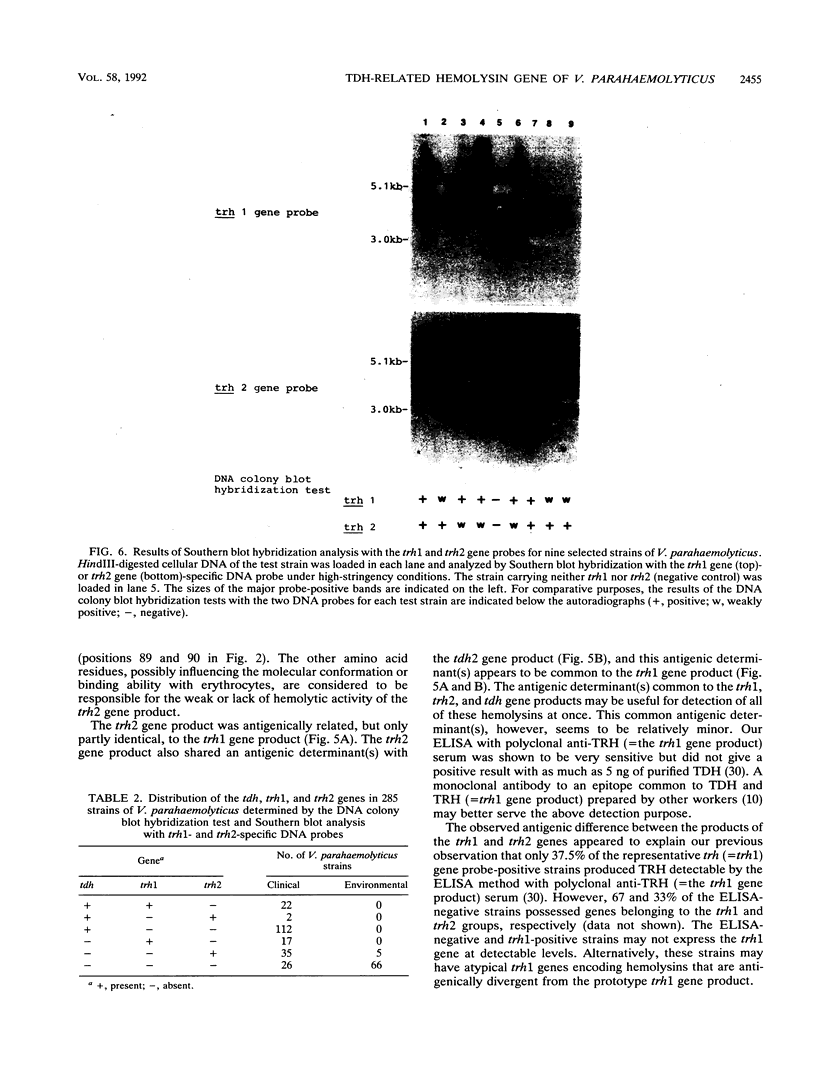
Our previous molecular epidemiologic study with gene probes (H. Shirai, H. Ito, T. Hirayama, Y. Nakamoto, N. Nakabayashi, K. Kumagai, Y. Takeda, and M. Nishibuchi, Infect. Immun. 58:3568-3573, 1990) demonstrated that the gene (trh) encoding a thermostable direct hemolysin-related hemolysin was strongly associated with clinical strains of Vibrio parahaemolyticus. Strain-to-strain variation in the intensities of the hybridization signals observed in the above study also suggested that the trh genes in different strains may have significantly divergent nucleotide sequences. To assess the public health significance of the rare environmental strains which exhibited very weak hybridization signals with the trh gene-specific DNA probe, the trh-like sequence was cloned from one of the environmental strains and the nucleotide sequence was determined in this study. A hemolysin gene (trh2) which was 84% homologous to the trh gene (newly named trh1) and 54.8 to 68.8% homologous to the genes (tdh) encoding thermostable direct hemolysins was detected in the cloned sequence. The trh2 gene product showed a profile of hemolytic activities against various animal erythrocytes different from that of the trh1 gene product. The trh2 gene product was antigenically related (partially identical) to the trh1 and tdh gene products. DNA colony blot and Southern blot hybridization analyses with trh1- and trh2-specific DNA probes showed that the trh1 probe-positive strains exhibiting hybridization signals with varying intensities could be clustered into trh1 and trh2 subgroups. In addition, hybridization analysis with oligonucleotide probes demonstrated significant strain-to-strain variation in the trh1 and trh2 gene sequences.(ABSTRACT TRUNCATED AT 250 WORDS)
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Baba K., Shirai H., Terai A., Kumagai K., Takeda Y., Nishibuchi M. Similarity of the tdh gene-bearing plasmids of Vibrio cholerae non-O1 and Vibrio parahaemolyticus. Microb Pathog. 1991 Jan;10(1):61–70. doi: 10.1016/0882-4010(91)90066-j. [DOI] [PubMed] [Google Scholar]
- Baba K., Shirai H., Terai A., Takeda Y., Nishibuchi M. Analysis of the tdh gene cloned from a tdh gene- and trh gene-positive strain of Vibrio parahaemolyticus. Microbiol Immunol. 1991;35(3):253–258. doi: 10.1111/j.1348-0421.1991.tb01554.x. [DOI] [PubMed] [Google Scholar]
- Bolivar F., Rodriguez R. L., Greene P. J., Betlach M. C., Heyneker H. L., Boyer H. W., Crosa J. H., Falkow S. Construction and characterization of new cloning vehicles. II. A multipurpose cloning system. Gene. 1977;2(2):95–113. [PubMed] [Google Scholar]
- Chou P. Y., Fasman G. D. Empirical predictions of protein conformation. Annu Rev Biochem. 1978;47:251–276. doi: 10.1146/annurev.bi.47.070178.001343. [DOI] [PubMed] [Google Scholar]
- Feinberg A. P., Vogelstein B. A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity. Anal Biochem. 1983 Jul 1;132(1):6–13. doi: 10.1016/0003-2697(83)90418-9. [DOI] [PubMed] [Google Scholar]
- Henikoff S. Unidirectional digestion with exonuclease III creates targeted breakpoints for DNA sequencing. Gene. 1984 Jun;28(3):351–359. doi: 10.1016/0378-1119(84)90153-7. [DOI] [PubMed] [Google Scholar]
- Honda T., Ni Y. X., Miwatani T. Purification and characterization of a hemolysin produced by a clinical isolate of Kanagawa phenomenon-negative Vibrio parahaemolyticus and related to the thermostable direct hemolysin. Infect Immun. 1988 Apr;56(4):961–965. doi: 10.1128/iai.56.4.961-965.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honda T., Ni Y., Yoh M., Miwatani T. Production of monoclonal antibodies against thermostable direct hemolysin of Vibrio parahaemolyticus and application of the antibodies for enzyme-linked immunosorbent assay. Med Microbiol Immunol. 1989;178(5):245–253. doi: 10.1007/BF00191059. [DOI] [PubMed] [Google Scholar]
- Iida T., Yamamoto K. Cloning and expression of two genes encoding highly homologous hemolysins from a Kanagawa phenomenon-positive Vibrio parahaemolyticus T4750 strain. Gene. 1990 Sep 1;93(1):9–15. doi: 10.1016/0378-1119(90)90129-f. [DOI] [PubMed] [Google Scholar]
- Joseph S. W., Colwell R. R., Kaper J. B. Vibrio parahaemolyticus and related halophilic Vibrios. Crit Rev Microbiol. 1982;10(1):77–124. doi: 10.3109/10408418209113506. [DOI] [PubMed] [Google Scholar]
- Kyte J., Doolittle R. F. A simple method for displaying the hydropathic character of a protein. J Mol Biol. 1982 May 5;157(1):105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- Marsh P. Ptac-85, an E. coli vector for expression of non-fusion proteins. Nucleic Acids Res. 1986 Apr 25;14(8):3603–3603. doi: 10.1093/nar/14.8.3603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyamoto Y., Kato T., Obara Y., Akiyama S., Takizawa K., Yamai S. In vitro hemolytic characteristic of Vibrio parahaemolyticus: its close correlation with human pathogenicity. J Bacteriol. 1969 Nov;100(2):1147–1149. doi: 10.1128/jb.100.2.1147-1149.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizusawa S., Nishimura S., Seela F. Improvement of the dideoxy chain termination method of DNA sequencing by use of deoxy-7-deazaguanosine triphosphate in place of dGTP. Nucleic Acids Res. 1986 Feb 11;14(3):1319–1324. doi: 10.1093/nar/14.3.1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishibuchi M., Hill W. E., Zon G., Payne W. L., Kaper J. B. Synthetic oligodeoxyribonucleotide probes to detect Kanagawa phenomenon-positive Vibrio parahaemolyticus. J Clin Microbiol. 1986 Jun;23(6):1091–1095. doi: 10.1128/jcm.23.6.1091-1095.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishibuchi M., Ishibashi M., Takeda Y., Kaper J. B. Detection of the thermostable direct hemolysin gene and related DNA sequences in Vibrio parahaemolyticus and other vibrio species by the DNA colony hybridization test. Infect Immun. 1985 Sep;49(3):481–486. doi: 10.1128/iai.49.3.481-486.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishibuchi M., Kaper J. B. Duplication and variation of the thermostable direct haemolysin (tdh) gene in Vibrio parahaemolyticus. Mol Microbiol. 1990 Jan;4(1):87–99. doi: 10.1111/j.1365-2958.1990.tb02017.x. [DOI] [PubMed] [Google Scholar]
- Nishibuchi M., Kaper J. B. Nucleotide sequence of the thermostable direct hemolysin gene of Vibrio parahaemolyticus. J Bacteriol. 1985 May;162(2):558–564. doi: 10.1128/jb.162.2.558-564.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishibuchi M., Kumagai K., Kaper J. B. Contribution of the tdh1 gene of Kanagawa phenomenon-positive Vibrio parahaemolyticus to production of extracellular thermostable direct hemolysin. Microb Pathog. 1991 Dec;11(6):453–460. doi: 10.1016/0882-4010(91)90042-9. [DOI] [PubMed] [Google Scholar]
- Nishibuchi M., Murakami A., Arita M., Jikuya H., Takano J., Honda T., Miwatani T. Detection with synthetic oligonucleotide probes of nucleotide sequence variations in the genes encoding enterotoxins of Escherichia coli. J Clin Microbiol. 1989 Oct;27(10):2272–2276. doi: 10.1128/jcm.27.10.2272-2276.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishibuchi M., Taniguchi T., Misawa T., Khaeomanee-Iam V., Honda T., Miwatani T. Cloning and nucleotide sequence of the gene (trh) encoding the hemolysin related to the thermostable direct hemolysin of Vibrio parahaemolyticus. Infect Immun. 1989 Sep;57(9):2691–2697. doi: 10.1128/iai.57.9.2691-2697.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oku Y., Uesaka Y., Hirayama T., Takeda Y. Development of a highly sensitive bead-ELISA to detect bacterial protein toxins. Microbiol Immunol. 1988;32(8):807–816. doi: 10.1111/j.1348-0421.1988.tb01442.x. [DOI] [PubMed] [Google Scholar]
- Sakazaki R., Tamura K., Kato T., Obara Y., Yamai S. Studies on the enteropathogenic, facultatively halophilic bacterium, Vibrio parahaemolyticus. 3. Enteropathogenicity. Jpn J Med Sci Biol. 1968 Oct;21(5):325–331. doi: 10.7883/yoken1952.21.325. [DOI] [PubMed] [Google Scholar]
- Shirai H., Ito H., Hirayama T., Nakamoto Y., Nakabayashi N., Kumagai K., Takeda Y., Nishibuchi M. Molecular epidemiologic evidence for association of thermostable direct hemolysin (TDH) and TDH-related hemolysin of Vibrio parahaemolyticus with gastroenteritis. Infect Immun. 1990 Nov;58(11):3568–3573. doi: 10.1128/iai.58.11.3568-3573.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirai H., Nishibuchi M., Ramamurthy T., Bhattacharya S. K., Pal S. C., Takeda Y. Polymerase chain reaction for detection of the cholera enterotoxin operon of Vibrio cholerae. J Clin Microbiol. 1991 Nov;29(11):2517–2521. doi: 10.1128/jcm.29.11.2517-2521.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taniguchi H., Hirano H., Kubomura S., Higashi K., Mizuguchi Y. Comparison of the nucleotide sequences of the genes for the thermostable direct hemolysin and the thermolabile hemolysin from Vibrio parahaemolyticus. Microb Pathog. 1986 Oct;1(5):425–432. doi: 10.1016/0882-4010(86)90004-5. [DOI] [PubMed] [Google Scholar]
- Terai A., Shirai H., Yoshida O., Takeda Y., Nishibuchi M. Nucleotide sequence of the thermostable direct hemolysin gene (tdh gene) of Vibrio mimicus and its evolutionary relationship with the tdh genes of Vibrio parahaemolyticus. FEMS Microbiol Lett. 1990 Sep 15;59(3):319–323. doi: 10.1016/0378-1097(90)90241-h. [DOI] [PubMed] [Google Scholar]
- Vieira J., Messing J. Production of single-stranded plasmid DNA. Methods Enzymol. 1987;153:3–11. doi: 10.1016/0076-6879(87)53044-0. [DOI] [PubMed] [Google Scholar]
- Vieira J., Messing J. The pUC plasmids, an M13mp7-derived system for insertion mutagenesis and sequencing with synthetic universal primers. Gene. 1982 Oct;19(3):259–268. doi: 10.1016/0378-1119(82)90015-4. [DOI] [PubMed] [Google Scholar]
- Yamasaki S., Shirai H., Takeda Y., Nishibuchi M. Analysis of the gene of Vibrio hollisae encoding the hemolysin similar to the thermostable direct hemolysin of Vibrio parahaemolyticus. FEMS Microbiol Lett. 1991 May 15;64(2-3):259–263. doi: 10.1016/0378-1097(91)90606-b. [DOI] [PubMed] [Google Scholar]