Abstract
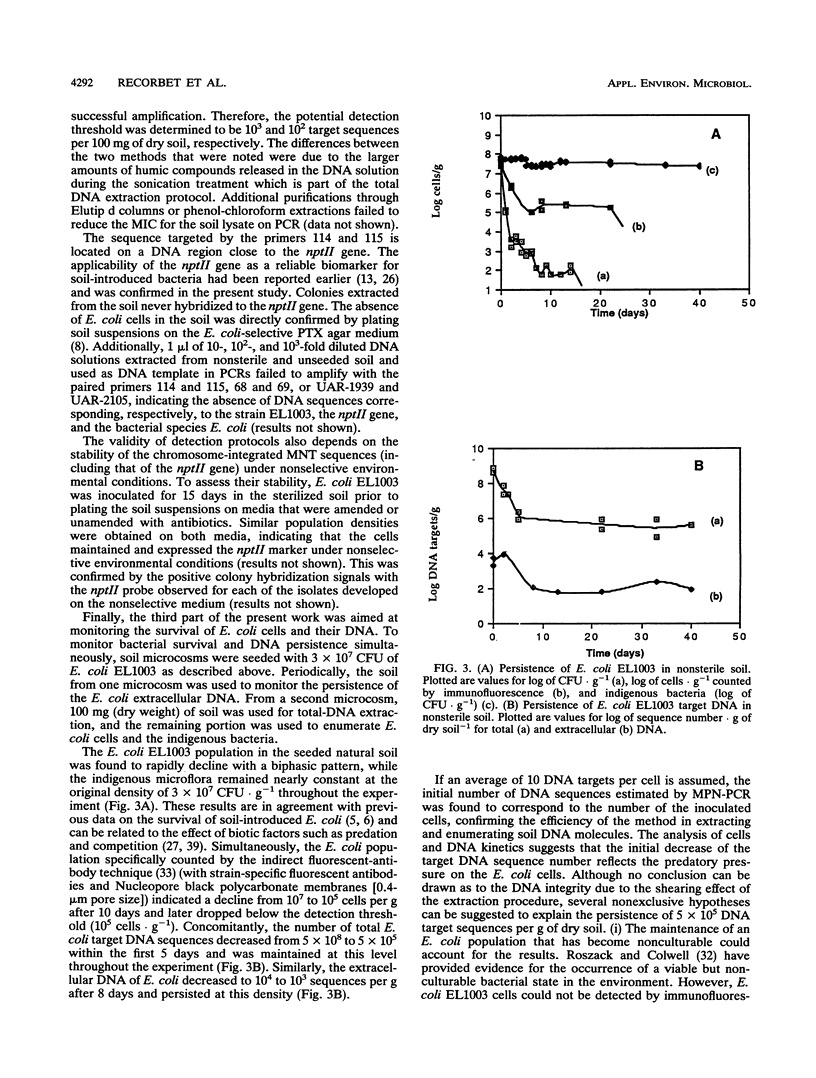
Investigations to quantify bacterial survival and DNA persistence of a genetically engineered population of Escherichia coli introduced into soil microcosms were carried out. The survival of E. coli was monitored by plate counting and immunofluorescence methods, whereas the persistence of the DNA was evaluated by using a most-probable-number-polymerase chain reaction method. Whereas the E. coli population density declined below the plate-counting-technique detection threshold (10(2) CFU.g-1) after 15 days, 10(3) extracellular and 5 x 10(5) total DNA target sequences were still detected after 40 days. Additionally, the E. coli cell counts fell below the detection limit of the immunofluorescence method (10(5) cells.g-1) before the end of the experiment. Colony hybridizations did not reveal gene transfer to the indigenous microflora. These results confirm the persistence of residual E. coli target sequences that could not be detected by the classical cell counting method and offer promising applications for the environmental detection of microorganisms, either engineered, pathogenic, or released for beneficial effects.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Aardema B. W., Lorenz M. G., Krumbein W. E. Protection of sediment-adsorbed transforming DNA against enzymatic inactivation. Appl Environ Microbiol. 1983 Aug;46(2):417–420. doi: 10.1128/aem.46.2.417-420.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bej A. K., DiCesare J. L., Haff L., Atlas R. M. Detection of Escherichia coli and Shigella spp. in water by using the polymerase chain reaction and gene probes for uid. Appl Environ Microbiol. 1991 Apr;57(4):1013–1017. doi: 10.1128/aem.57.4.1013-1017.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunel B., Cleyet-Marel J. C., Normand P., Bardin R. Stability of Bradyrhizobium japonicum Inoculants after Introduction into Soil. Appl Environ Microbiol. 1988 Nov;54(11):2636–2642. doi: 10.1128/aem.54.11.2636-2642.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- COCHRAN W. G. Estimation of bacterial densities by means of the "most probable number". Biometrics. 1950 Jun;6(2):105–116. [PubMed] [Google Scholar]
- Fredrickson J. K., Bezdicek D. F., Brockman F. J., Li S. W. Enumeration of Tn5 mutant bacteria in soil by using a most- probable-number-DNA hybridization procedure and antibiotic resistance. Appl Environ Microbiol. 1988 Feb;54(2):446–453. doi: 10.1128/aem.54.2.446-453.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henschke R. B., Henschke E. J., Schmidt F. R. Monitoring survival and gene transfer in soil microcosms of recombinant Escherichia coli designed to represent an industrial production strain. Appl Microbiol Biotechnol. 1991 May;35(2):247–252. doi: 10.1007/BF00184696. [DOI] [PubMed] [Google Scholar]
- Holben William E., Jansson Janet K., Chelm Barry K., Tiedje James M. DNA Probe Method for the Detection of Specific Microorganisms in the Soil Bacterial Community. Appl Environ Microbiol. 1988 Mar;54(3):703–711. doi: 10.1128/aem.54.3.703-711.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khanna M., Stotzky G. Transformation of Bacillus subtilis by DNA bound on montmorillonite and effect of DNase on the transforming ability of bound DNA. Appl Environ Microbiol. 1992 Jun;58(6):1930–1939. doi: 10.1128/aem.58.6.1930-1939.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenz M. G., Wackernagel W. Adsorption of DNA to sand and variable degradation rates of adsorbed DNA. Appl Environ Microbiol. 1987 Dec;53(12):2948–2952. doi: 10.1128/aem.53.12.2948-2952.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picard C., Ponsonnet C., Paget E., Nesme X., Simonet P. Detection and enumeration of bacteria in soil by direct DNA extraction and polymerase chain reaction. Appl Environ Microbiol. 1992 Sep;58(9):2717–2722. doi: 10.1128/aem.58.9.2717-2722.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pillai S. D., Josephson K. L., Bailey R. L., Gerba C. P., Pepper I. L. Rapid method for processing soil samples for polymerase chain reaction amplification of specific gene sequences. Appl Environ Microbiol. 1991 Aug;57(8):2283–2286. doi: 10.1128/aem.57.8.2283-2286.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romanowski G., Lorenz M. G., Sayler G., Wackernagel W. Persistence of free plasmid DNA in soil monitored by various methods, including a transformation assay. Appl Environ Microbiol. 1992 Sep;58(9):3012–3019. doi: 10.1128/aem.58.9.3012-3019.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romanowski G., Lorenz M. G., Wackernagel W. Adsorption of plasmid DNA to mineral surfaces and protection against DNase I. Appl Environ Microbiol. 1991 Apr;57(4):1057–1061. doi: 10.1128/aem.57.4.1057-1061.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roszak D. B., Colwell R. R. Survival strategies of bacteria in the natural environment. Microbiol Rev. 1987 Sep;51(3):365–379. doi: 10.1128/mr.51.3.365-379.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selenska S., Klingmüller W. DNA recovery and direct detection of Tn5 sequences from soil. Lett Appl Microbiol. 1991 Jul;13(1):21–24. doi: 10.1111/j.1472-765x.1991.tb00559.x. [DOI] [PubMed] [Google Scholar]
- Smit E., van Elsas J. D., van Veen J. A., de Vos W. M. Detection of Plasmid Transfer from Pseudomonas fluorescens to Indigenous Bacteria in Soil by Using Bacteriophage phiR2f for Donor Counterselection. Appl Environ Microbiol. 1991 Dec;57(12):3482–3488. doi: 10.1128/aem.57.12.3482-3488.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steffan R. J., Atlas R. M. DNA amplification to enhance detection of genetically engineered bacteria in environmental samples. Appl Environ Microbiol. 1988 Sep;54(9):2185–2191. doi: 10.1128/aem.54.9.2185-2191.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steffan R. J., Goksøyr J., Bej A. K., Atlas R. M. Recovery of DNA from soils and sediments. Appl Environ Microbiol. 1988 Dec;54(12):2908–2915. doi: 10.1128/aem.54.12.2908-2915.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tate R. L., 3rd Cultural and environmental factors affecting the longevity of Escherichia coli in Histosols. Appl Environ Microbiol. 1978 May;35(5):925–929. doi: 10.1128/aem.35.5.925-929.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai Y. L., Olson B. H. Detection of low numbers of bacterial cells in soils and sediments by polymerase chain reaction. Appl Environ Microbiol. 1992 Feb;58(2):754–757. doi: 10.1128/aem.58.2.754-757.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai Y. L., Olson B. H. Rapid method for direct extraction of DNA from soil and sediments. Appl Environ Microbiol. 1991 Apr;57(4):1070–1074. doi: 10.1128/aem.57.4.1070-1074.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]




