Abstract
DNA polymerase zeta (Polζ) promotes the mutagenic bypass of DNA lesions in eukaryotes. Genetic studies in Saccharomyces cerevisiae have indicated that relative to the contribution of other pathways, Polζ makes only a modest contribution to lesion bypass. Intriguingly, however, disruption of the REV3 gene, which encodes the catalytic subunit of Polζ, causes early embryonic lethality in mice. Here, we present genetic and biochemical evidence for the requirement of yeast Polζ for predominantly error-free replication past thymine glycol (Tg), a DNA lesion formed frequently by free radical attack. These results raise the possibility that, as in yeast, in higher eukaryotes also, Polζ makes a major contribution to the replicative bypass of Tgs as well as other lesions that block synthesis by replicative DNA polymerases. Such a preeminent role of Polζ in lesion bypass would ensure that rapid cell divisions continue unabated during early embryonic development, thereby minimizing the generation of DNA strand breaks, chromosome aberrations, and the ensuing apoptotic response.
Keywords: DNA polymerase ζ, thymine glycol, translesion DNA synthesis, Polζ as an extender, error-free translesion DNA synthesis by Polζ, yeast
Genetic studies in the yeast Saccharomyces cerevisiae have indicated the requirement of Rad6–Rad18-dependent processes in promoting replication of damaged DNA. For example, the replicative bypass of UV-induced DNA lesions is carried out by three different Rad6–Rad18-controlled pathways (Torres-Ramos et al. 2002) in which Rad5, an SWI/SNF DNA-dependent ATPase (Johnson et al. 1992, 1994), plays an important role in promoting error-free replication through UV-induced DNA lesions. The other two pathways utilize the RAD30-encoded DNA polymerase eta (Polη; Johnson et al. 1999), and the REV3, REV7-encoded DNA polymerase zeta (Polζ; Nelson et al. 1996), which primarily function in the error-free and mutagenic modes, respectively, of translesion DNA synthesis (TLS) through UV lesions. The mechanism by which Rad5 affects lesion bypass is not known; however, it has been suggested to promote a copy choice-type of DNA synthesis (Torres-Ramos et al. 2002). Polη is very efficient at replicating through a cis-syn thymine–thymine (T–T) dimer, and steady-state kinetic studies have shown that it replicates through the two Ts of this DNA lesion with the same efficiency and accuracy as it replicates through undamaged Ts (Johnson et al. 2000b; Washington et al. 2000). Genetic studies in yeast have additionally indicated a role for Polη in the error-free bypass of cyclobutane pyrimidine dimers (CPDs) formed at TC and CC sites (Yu et al. 2001).
Polζ differs from Polη in its inability to replicate through CPDs, and that is because Polζ is very inefficient at inserting nucleotides opposite the 3′T of a T–T dimer. Polζ, however, promotes the mutagenic bypass of DNA lesions by extending from nucleotides inserted opposite the lesion by another DNA polymerase (Johnson et al. 2000a). For example, Polζ efficiently extends from a G inserted opposite the 3′T of either a T–T dimer or a (6–4) T–T photoproduct (Johnson et al. 2000a, 2001), and it efficiently extends from an A opposite an abasic site (Haracska et al. 2001a). However, as inferred from genetic studies in yeast, Polζ plays a relatively minor role in the bypass of UV lesions or of abasic sites. Thus, a rev3Δ mutant displays only a modest increase in UV sensitivity in yeast (Johnson et al. 1992), and the discontinuities that are formed in DNA strands synthesized from UV-damaged templates become fully repaired on subsequent incubation in a strain lacking Polζ (Torres-Ramos et al. 2002). In addition, introduction of the rev3Δ mutation into the yeast apn1Δ apn2Δ strain, defective in the removal of abasic sites, causes no significant increase in sensitivity to the alkylating agent methyl methane sulfonate (Johnson et al. 1998).
In the absence of any extraneous DNA damage, the rev3Δ mutation in yeast confers no obvious adverse effect on cell viability or growth. Disruption of the REV3 gene in mice, however, causes early embryonic lethality (Bemark et al. 2000; Esposito et al. 2000; Wittschieben et al. 2000). This observation raised the possibility that Polζ plays a more critical role in TLS through DNA lesions than had been envisaged based on the genetic studies carried out in yeast thus far.
Free radical-induced DNA damage makes a substantial contribution to the spontaneous mutational burden, and consequently, it has been implicated in carcinogenesis and aging (Ames et al. 1993). Hydroxyl radicals, the principal DNA-damaging species formed from aerobic respiration or from exposure to chemical oxidizing agents or ionizing radiation, interact with the thymine or cytosine residues, principally at the 5,6 double bond, leading to the conversion of thymine to thymine glycol (5,6-dihyroxy-5,6-dihydrothymine; Tg; Fig. 1A) and of cytosine to cytosine glycol (Cg). Tg is a major stable product of thymine modification in vitro and in vivo (Teoule et al. 1974; Cathcart et al. 1984), whereas Cg is unstable and deaminates to uracil glycol (Ug; Wagner et al. 1992). Cg can also lead to the formation of two other stable products, 5-hydroxycytosine (5-OHC; Dizdaroglu and Simic 1984; Teoule 1987; Wagner et al. 1992), and 5-hydroxyuracil (5-OHU; Wagner et al. 1992).
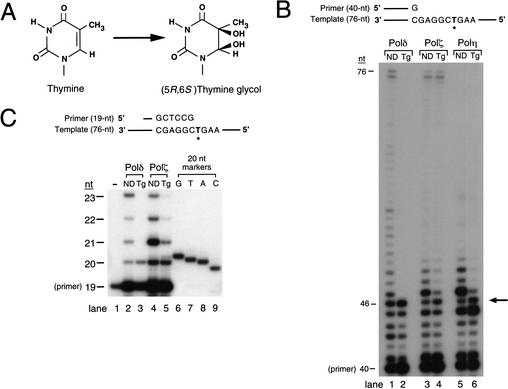
Figure 1.
Replicative bypass of Tg by yeast DNA polymerases. (A) Structure of Tg formed upon oxidation of thymine. (B) DNA synthesis by yeast Polδ (1 nM), Polζ (1.5 nM), and Polη (1 nM) on undamaged and Tg-containing DNA substrates. (Lanes 1,2) Synthesis by Polδ on undamaged DNA (ND) and on DNA containing a Tg, respectively. (Lanes 3,4) Synthesis by Polζ on undamaged DNA and on DNA containing a Tg, respectively. (Lanes 5,6) Synthesis by Polη on undamaged DNA and on DNA containing a Tg, respectively. DNA length in nucleotides (nt) is given on the left. The position of the Tg is indicated by the arrow on the right. (C) Identification of the nucleotide inserted opposite Tg by Polδ (1 nM) and Polζ (1.5 nM). Synthesis was assayed on a DNA substrate containing a 19-nt primer annealed to either the undamaged or Tg-containing template. (Lane 1) A 19-nt primer. (Lanes 2,3) Synthesis by Polδ on undamaged DNA (ND) and on DNA containing a Tg, respectively. (Lanes 4,5) Synthesis by Polζ on undamaged DNA and on DNA containing a Tg, respectively. Lanes 6–9 show 20-nt markers containing the 19-nt primer sequence with an additional G, T, A, or C residue, respectively. For both B and C, reactions were carried out using the standard DNA polymerase assay containing 100 μM of each dNTP for 10 min at 30°C. The * in B and C indicates the position of the undamaged thymine or Tg in the template.
The oxidized pyrimidines are removed in yeast by two DNA glycosylases, Ntg1 and Ntg2 (Senturker et al. 1998), that are related to Escherichia coli endonuclease III (Wallace 1997), which also functions in the removal of these lesions. Nucleotide excision repair (NER) is a versatile repair system that functions in the removal of a diversity of DNA lesions, including UV-induced CPDs and (6–4) dipyrimidine photoproducts, intrastrand and interstrand DNA cross-links, abasic sites, and oxidized purines and pyrimidines (Huang et al. 1994; Sancar 1996; Reardon et al. 1997; Prakash and Prakash 2000; Torres-Ramos et al. 2000). If the oxidized base is not repaired by base excision repair (BER) or NER, it can be a block to replicative DNA polymerases and can be potentially lethal, unless the progression of the replication fork is rescued by lesion bypass processes. Alternatively, the lesion would be mutagenic if it is bypassed by the replicative DNA polymerases by mispairing.
Of the oxidized pyrimidine bases that have been studied to date, only Tg presents a strong block to DNA polymerases in vitro. For example, synthesis by E. coli DNA polymerase I Klenow fragment and T4 DNA polymerase is arrested at the site of the lesion; both polymerases incorporate the correct nucleotide A opposite Tg, but no extension from the inserted nucleotide occurs (Clark and Beardsley 1987). The other oxidized pyrimidines (Ug, 5-OHC, and 5-OHU) do not block DNA polymerases and are readily bypassed, but they are mutagenic because of the incorporation of an A opposite them (Purmal et al. 1994, 1998).
Here we examine the mechanism of Tg bypass in S. cerevisiae. Tg presents a block to the yeast replicative DNA polymerase delta (Polδ). However, it can efficiently insert an A opposite this lesion, but it is unable to carry out the subsequent extension reaction. Polζ, on the other hand, is able to replicate through this DNA lesion, and it does so by inserting an A opposite it and by efficiently extending from the inserted nucleotide. Although Polη can replicate through the Tg lesion, it is quite inefficient at both the insertion and extension steps. Importantly, and quite unexpectedly, we found that of the three Rad6–Rad18-dependent lesion bypass pathways, only the Polζ pathway is necessary for Tg bypass, whereas the Polη- and Rad5-dependent pathways are dispensable. Moreover, Polζ predominantly promotes the error-free bypass of this DNA lesion. These findings indicate that rather than merely playing a minor role in the mutagenic bypass of DNA lesions, for certain DNA lesions, Polζ provides the primary if not the sole means of their bypass. Consequently, efficient error-free TLS by Polζ could be crucial for maintaining the speed of rapid cell divisions during early embryonic development, thereby accounting for the indispensability of this polymerase in higher eukaryotes.
Results
Replication of DNA containing a Tg by yeast DNA polymerases δ, ζ, and η
To examine the effect of a Tg on the DNA synthetic activity of yeast DNA polymerases δ, ζ, and η, we incorporated a site-specific Tg into a 76-nucleotide (nt) template, and annealed it to a 40-nt 5′ 32P-labeled primer. DNA synthesis was assayed in a running start reaction, which allows for the synthesis of 5 nucleotides before the Tg lesion is encountered (Fig. 1B). Although the replicative polymerase Polδ is able to incorporate a nucleotide opposite the Tg lesion, it is unable to carry out DNA synthesis beyond it (Fig. 1B, lane 2). In comparison to undamaged DNA, wherein Polδ synthesized past ∼40% of the unmodified T residue (Fig. 1B, lane 1), there was no observed bypass of the Tg (Fig. 1B, lane 2). The absence of any significant stall site just before the lesion indicates that Polδ can efficiently insert a nucleotide opposite the Tg lesion and that the inserted nucleotide is not subject to its 3′→5′ exonuclease activity.
Although Polζ is required for the mutagenic bypass of DNA lesions, we previously showed that Polζ is highly inefficient at inserting nucleotides opposite a variety of DNA lesions; rather, it promotes the bypass of lesions by extending from the nucleotides inserted opposite the lesion site by another polymerase. On the Tg-containing substrate, however, Polζ is able to carry out unassisted lesion bypass by inserting a nucleotide opposite the lesion and subsequently extending from it (Fig. 1B, lane 4).
Yeast Polη replicates through UV-induced CPDs efficiently and accurately. On the Tg-containing substrate, however, Polη appears to be blocked both at the nucleotide incorporation step and at the subsequent extension step opposite from the lesion site (Fig. 1B, lane 6), and this intimation was confirmed by steady-state kinetic analyses described below.
Nucleotide incorporated opposite Tg by yeast DNA polymerases δ and ζ
To determine the identity of the nucleotide(s) inserted by Polδ and Polζ opposite the Tg residue, we used a standing-start 19-nt primer which anneals to the template one nucleotide before the Tg residue, and the reaction products were distinguished on a 20% polyacrylamide gel. In the presence of 100 μM of each of the four dNTPs, Polδ inserted an A across from the Tg residue (Fig. 1C, cf. lanes 3 and 8), and again, there was no observable bypass of the Tg lesion by Polδ. Polζ also primarily inserted an A opposite the Tg, but some very weak incorporation of G was also observed (Fig. 1C, cf. lanes 5 and 6,8).
Steady-state kinetic analyses of nucleotide incorporation and extension reactions opposite Tg by yeast DNA polymerase ζ
To quantify the efficiency of nucleotide insertion opposite the Tg by Polζ, we carried out steady-state kinetic analyses, and the results are shown in Table 1. Polζ can efficiently insert an A residue opposite the Tg, as it does so only about eightfold less efficiently than it inserts an A opposite an undamaged T. The other nucleotides G, T, and C are also incorporated opposite the Tg, but with greatly reduced efficiencies.
Table 1.
Steady-state kinetic parameters for nucleotide incorporation opposite undamaged thymine (T) and thymine glycol (Tg) by yeast Polζ
| Template nucleotide
|
Incoming residue
|
Vmax (nM/min)
|
Km (μM)
|
Vmax/Km
|
finc
|
Relative efficiencya
|
|---|---|---|---|---|---|---|
| T | G | 0.1 ± 0.005 | 213 ± 30 | 4.7 × 10−4 | 2.2 × 10−3 | — |
| A | 0.08 ± 0.01 | 0.39 ± 0.1 | 0.21 | 1 | — | |
| T | 0.04 ± 0.09 | 143 ± 97 | 2.8 × 10−4 | 1.3 × 10−3 | — | |
| C | 0.14 ± 0.05 | 453 ± 67 | 3.1 × 10−4 | 1.5 × 10−3 | — | |
| Tg | G | 0.11 ± 0.01 | 189 ± 60 | 5.8 × 10−4 | 2.1 × 10−2 | 1:362 |
| A | 0.06 ± 0.01 | 2.2 ± 0.2 | 2.7 × 10−2 | 1 | 1:8 | |
| T | 0.08 ± 0.01 | 444 ± 151 | 1.8 × 10−4 | 6.7 × 10−3 | 1:1166 | |
| C | 0.07 ± 0.01 | 319 ± 203 | 2.2 × 10−4 | 8.1 × 10−3 | 1:955 |
Indicates the efficiency of incorporation of different nucleotides opposite Tg relative to the efficiency of incorporation of an A opposite an undamaged T.
Because Polζ can replicate through the Tg lesion, we next measured the efficiency with which it is able to extend from the primer terminus opposite the Tg lesion. As shown in Table 2, compared to extension from the normal A:T primer-terminal base pair, Polζ is able to extend from an A paired with a Tg with an efficiency that is reduced by only ∼50%. On the undamaged template, Polζ extends primers that contain a 3′-terminal G or T opposite undamaged T quite efficiently; compared to the extension from an A:T base pair, Polζ extends from a G or T opposite an undamaged T with an ∼20-fold reduced efficiency. Compared to the extension from an A opposite Tg, extension from a G or a T opposite this lesion by Polζ was reduced ∼50-fold. Thus, Polζ is very proficient at extending from an A opposite Tg, and its proficiency in extending from a G or a T opposite a Tg lesion remains about the same as opposite an undamaged T.
Table 2.
Steady-state kinetic parameters for primer extension from nucleotides incorporated opposite thymine (T) and thymine glycol (Tg) by yeast Polζ
| Base pair at primer terminusa
|
Incoming residueb
|
Vmax (nM/min)
|
Km (μM)
|
Vmax/Km
|
foext
|
Efficiency relative to normal T:A
|
|---|---|---|---|---|---|---|
| T · G | C | 0.33 ± 0.023 | 90 ± 15 | 3.6 × 10−3 | 0.05 | — |
| T · A | C | 0.18 ± 0.02 | 2.6 ± 0.8 | 7 × 10−2 | 1 | — |
| T · T | C | 0.24 ± 0.01 | 66 ± 11 | 3.6 × 10−3 | 0.05 | — |
| T · C | C | N.D.c | >125 | 3.0 × 10−4,d | 0.004 | — |
| Tg · G | C | 0.08 ± 0.002 | 107 ± 7 | 7.5 × 10−4 | 0.02 | 1:93 |
| Tg · A | C | 0.08 ± 0.004 | 2.3 ± 0.4 | 3.5 × 10−2 | 1 | 1:2 |
| Tg · T | C | 0.09 ± 0.008 | 135 ± 15 | 6.7 × 10−4 | 0.02 | 1:104 |
| Tg · C | C | — | — | — | N.D.c | — |
The first nucleotide in the base pair listed is the template base, and the second nucleotide is at the 3′ end of the primer paired opposite the template base.
Reactions were carried out in the presence of the next correct nucleotide, dCTP.
N.D., not determined.
Since the nucleotide incorporation rate remained linear throughout the nucleotide concentration range used, the Vmax/Km value was obtained from the slope of the line.
Steady-state kinetics of nucleotide insertion and extension reactions opposite Tg by yeast DNA polymerase η
Because Polη could replicate through the Tg lesion, we examined its ability to insert nucleotides opposite the Tg lesion and to extend past it, using steady-state kinetics. In comparison to the insertion of an A opposite an undamaged T residue, Polη is ∼60-fold less efficient at inserting an A opposite the Tg lesion. The other nucleotides were also inserted opposite Tg, and their incorporation was only a fewfold less efficient than the incorporation of an A (Table 3). The steady-state kinetic parameters for extension past the Tg lesion by Polη are shown in Table 4. In comparison to extension from a normal A:T base pair, Polη is ∼300-fold less efficient at extending from an A opposite the Tg lesion. Although extension from a G opposite Tg is somewhat better than the extension from an A opposite Tg, it is still ∼200-fold less efficient than extension from the normal A:T base pair.
Table 3.
Steady-state kinetic parameters for nucleotide incorporation opposite thymine (T) and thymine glycol (Tg) by yeast Polη
| Template nucleotide
|
Incoming residue
|
Vmax (nM/min)
|
Km (μM)
|
Vmax/Km
|
finc
|
Relative efficiencya
|
|---|---|---|---|---|---|---|
| T | G | 0.17 ± 0.02 | 95 ± 38 | 1.8 × 10−3 | 1.0 × 10−2 | — |
| A | 0.33 ± 0.03 | 1.9 ± 0.5 | 0.17 | 1 | — | |
| T | 0.12 ± 0.008 | 200 ± 38 | 6.0 × 10−4 | 3.5 × 10−3 | — | |
| C | 0.17 ± 0.01 | 69 ± 24 | 2.5 × 10−3 | 1.5 × 10−2 | — | |
| Tg | G | 0.17 ± 0.02 | 316 ± 84 | 5.4 × 10−4 | 1.8 × 10−1 | 1:315 |
| A | 0.53 ± 0.03 | 187 ± 44 | 3 × 10−3 | 1 | 1:57 | |
| T | 0.06 ± 0.003 | 391 ± 53 | 1.5 × 10−4 | 5.0 × 10−2 | 1:1133 | |
| C | 0.15 ± 0.01 | 171 ± 44 | 8.8 × 10−4 | 3.1 × 10−1 | 1:193 |
aIndicates the efficiency of incorporation of different nucleotides opposite Tg relative to the efficiency of incorporation of an A opposite an undamaged T.
Table 4.
Steady-state kinetic parameters for primer extension from nucleotides opposite thymine (T) and thymine glycol (Tg) by yeast Polη
| Base pair at primer terminusa
|
Incoming residueb
|
Vmax (nM/min)
|
Km (μM)
|
Vmax/Km
|
foext
|
Relative to normal T:A
|
|---|---|---|---|---|---|---|
| T · A | C | 0.8 ± 0.06 | 1.3 ± 0.4 | 0.6 | 1 | — |
| Tg · G | C | 0.17 ± 0.001 | 50 ± 12 | 3 × 10−3 | 1.5 | 1:200 |
| Tg · A | C | 0.25 ± 0.02 | 151 ± 33 | 2 × 10−3 | 1 | 1:300 |
| Tg · T | C | 0.07 ± 0.006 | 59 ± 2 | 1 × 10−3 | 0.5 | 1:600 |
| Tg · C | C | N.D.c | ≥500 | 4.6 × 10−5(d) | 0.023 | 1:13,000 |
The first nucleotide in the base pair listed is the template base, and the second nucleotide is at the 3′ end of the primer paired opposite the template base.
Reactions were carried out in the presence of the next correct nucleotide, dCTP.
N.D., not determined.
Since the nucleotide incorporation rate remained linear throughout the nucleotide concentration range used, the Vmax/Km value was obtained from the slope of the line.
Requirement of DNA polymerase ζ for error-free bypass of Tg
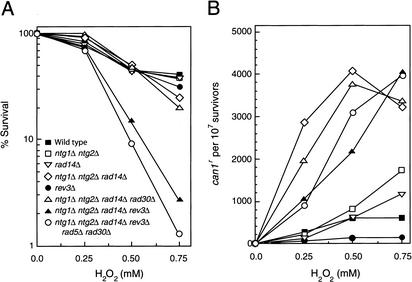
To determine whether Polζ is required for the bypass of Tgs in vivo, we examined the effect of a rev3Δ mutation on survival and mutagenesis following treatment with hydrogen peroxide. Although the rev3Δ strain shows no significant increase in sensitivity to H2O2 over wild type, the frequency of H2O2-induced mutations was greatly reduced in this strain (Fig. 2). However, since H2O2 generates a variety of DNA lesions in addition to the formation of Tg and other oxidized pyrimidines, to better observe the specific effect of Tg and other oxidized pyrimidines in the absence of REV3, we examined H2O2 sensitivity and mutagenesis in yeast cells in which the NTG1, NTG2, and RAD14 genes had been deleted. The yeast Ntg1 and Ntg2 DNA glycosylases are both able to remove Tg as well as other oxidized pyrimidines from DNA, and Rad14 is a component of the nucleotide excision repair machinery which would also contribute to the removal of these oxidized pyrimidines. As shown in Figure 2, neither the ntg1Δ ntg2Δ nor the rad14Δ mutations cause any increase in H2O2 sensitivity, but they confer some increase in the frequency of H2O2-induced can1r mutations. However, whereas only a small increase in H2O2 sensitivity occurs in the ntg1Δ ntg2Δ rad14Δ strain, there is a dramatic rise in the frequency of H2O2-induced can1r mutations in this strain. Gellon et al. (2001) reported a similar increase in H2O2 sensitivity and H2O2-induced mutagenesis in the ntg1Δ ntg2Δ rad14Δ strain. Importantly, we found that introduction of the rev3Δ mutation into the ntg1Δ ntg2Δ rad14Δ strain confers a synergistic enhancement in H2O2 sensitivity, but the frequency of H2O2-induced can1r mutations declines only somewhat in the ntg1Δ ntg2Δ rad14Δ rev3Δ mutant strain compared to that in the ntg1Δ ntg2Δ rad14Δ strain. To examine the in vivo role of Polη in Tg bypass, we introduced a rad30Δ mutation into the ntg1Δ ntg2Δ rad14Δ strain and examined its sensitivity and its ability to mutate in response to H2O2 treatment. However, the rad30Δ mutation had no significant effect on survival or mutagenesis in this genetic background (Fig. 2). Furthermore, introduction of both the rad5Δ and rad30Δ mutations into the ntg1Δ ntg2Δ rad14Δ rev3Δ strain did not cause any significant increase in H2O2 sensitivity, and the frequency of H2O2-induced can1r mutations was not affected (Fig. 2). We elaborate on the implications of these genetic results in the Discussion, and from those considerations, we infer a preeminent role for Polζ in Tg bypass, whereas Polη and Rad5 make no significant contribution to the bypass of this DNA lesion.
Figure 2.
Requirement of REV3 for the replicative bypass of Tg. (A) Synergistic enhancement of H2O2 sensitivity by the rev3Δ mutation in yeast strains lacking the NTG1, NTG2, and RAD14 genes. (B) H2O2-induced CAN1S-to-can1r forward mutations in various yeast strains. Symbols for various strains are as shown in A. The sensitivity to H2O2 and the frequency of H2O2-induced can1r mutations were the same in the ntg1Δ ntg2Δ rad14Δ and ntg1Δ ntg2Δ rad14Δ rad5Δ strains.
Discussion
Here we show that Tg is a block to replication by Polδ, but it can efficiently insert an A opposite this lesion. On the other hand, Polζ is able to replicate through this DNA lesion by inserting an A and then extending from it. Using steady-state kinetic analyses, we found that compared to the incorporation of an A opposite undamaged T, Polζ is about eightfold reduced in its efficiency to incorporate an A opposite Tg, but it extends from the A nucleotide opposite Tg almost as proficiently as from an A opposite undamaged T. Although Polη can replicate through the Tg lesion by inserting an A, its efficiency of incorporating the nucleotide is reduced by ∼60-fold, and it shows an ∼300-fold reduction in the efficiency of extending from the inserted nucleotide.
NMR studies of duplex DNA containing a site-specific Tg have indicated that this lesion induces a significant and localized alteration in the structure of DNA, and that the Tg residue in DNA is approximately half extrahelical (Kao et al. 1993; Kung and Bolton 1997). The structural perturbations caused by Tg are likely to be quite different from that found in DNA containing an abasic site, and we found that all three yeast DNA polymerases examined here are more efficient at inserting a nucleotide opposite the Tg lesion than opposite an abasic site. Thus, although Polδ preferentially inserts an A opposite both DNA lesions, it displays a strong stall site just before the abasic site (Haracska et al. 2001a), whereas no such stall site is evident for the Tg lesion, indicating that Tg is not a significant block to nucleotide incorporation by Polδ, but an abasic site is. In addition, compared to the highly inefficient incorporation of nucleotides opposite an abasic site by Polζ (Haracska et al. 2001a) and Polη (Haracska et al. 2001b), both these polymerases incorporate an A opposite Tg much more efficiently.
We previously showed that Polζ functions in the mutagenic bypass of a cis-syn T–T dimer or a (6–4) T–T photoproduct by extending from the nucleotide inserted opposite the 3′T of these lesions by another DNA polymerase (Johnson et al. 2000a, 2001). A cis-syn T–T dimer has only a modest effect on the DNA structure, and it does not affect the ability of the two Ts in the dimer to pair with As (Ciarrocchi and Pedrini 1982; Kim et al. 1995). Although a (6–4) T–T photoproduct, wherein the 3′T is oriented perpendicular to the 5′T, induces a large structural distortion, the ability of the 5′T to form normal hydrogen bonds with the A in the complementary strand is not affected (Kim et al. 1995; Lee et al. 1999). Polζ also extends efficiently from nucleotides, particularly an A, incorporated opposite an abasic site by another DNA polymerase (Haracska et al. 2001a). An abasic site also is not expected to significantly affect the ability of the next 5′ template base to pair with its cognate base. Thus, although the template-primer ends at the lesion site will incur some degree of geometric distortion in all of these cases, the presence of any of these lesions in the template strand opposite the primer end will not significantly affect the formation of the next base pair. In contrast, modeling studies with DNA containing a Tg, by energy minimization (Clark et al. 1987) and by molecular dynamics simulations (Miaskiewicz et al. 1995), have indicated that this damaged base destabilizes the stacking interactions with the 5′ base pair, and that is due, at least in part, to an unfavorable steric overlap between the pseudoaxial methyl group in Tg and the 5′ template base. In these studies, Clark et al. (1987) inferred a large increase in the tilt angle of the template base immediately 5′ to Tg, and Miaskiewicz et al. (1995) found a dramatic increase in the rise parameter between the lesion and the 5′ adjacent base, and this change was accompanied by the weakening of Watson-Crick hydrogen bonds in the next base pair on the 5′ side of the lesion. Thus, both of these studies indicate that the 5′ side of the lesion is the site of major structural distortions to DNA, and these observations explain well the inability of most DNA polymerases to extend from the base opposite Tg. The proficient ability of Polζ to extend from the A nucleotide opposite Tg shows that this polymerase is uniquely adapted to carry out the extension reaction even in the presence of such steric interference with the 5′ template base.
The oxidized pyrimidines, Tg, Ug, 5-OHC, and 5-OHU will be removed in yeast by the Ntg1 and Ntg2 DNA glycosylases and by NER. However, although the simultaneous inactivation of all of these pathways elicits only a small increase in H2O2 sensitivity, there is a large increase in the frequency of H2O2-induced can1r mutations. These observations suggest that yeast cells can efficiently replicate through Tg and other oxidized pyrimidines, and replication through some of these damaged bases, presumably Cg, 5-OHC, and 5-OHU, is mutagenic. To determine the contribution of the three Rad6–Rad18-dependent pathways to the replication of oxidized pyrimidines, we examined the H2O2 sensitivity and H2O2-induced can1r mutagenesis in the ntg1Δ ntg2Δ rad14Δ strain from which the REV3, RAD30, or RAD5 gene had been deleted. Quite unexpectedly, we found that only the deletion of the REV3 gene confers a large increase in H2O2 sensitivity, whereas even deletion of both the RAD5 and RAD30 genes in the ntg1Δ ntg2Δ rad14Δ rev3Δ strain causes no further increase in H2O2 sensitivity. The frequency of H2O2-induced can1r mutations was reduced somewhat in the ntg1Δ ntg2Δ rad14Δ strain upon the introduction of the rev3Δ mutation, but the additional introduction of the rad5Δ and rad30Δ mutations into such a strain caused no further change in mutation frequencies.
From these genetic results, we infer that of the three Rad6–Rad18-dependent pathways, only the Polζ pathway makes a predominant contribution to the replicative bypass of Tg. Because Polδ is unable to replicate through a Tg, this lesion would be lethal if not bypassed. The requirement of Polζ for extension past Tg could then account for the large increase in H2O2 sensitivity that occurs in the ntg1Δ ntg2Δ rad14Δ strain upon the introduction of the rev3Δ mutation. As the Polζ-mediated bypass of Tg is expected to be predominantly error-free (see below; Fig. 3), the reduction in the frequency of H2O2-induced can1r mutations that is seen in the ntg1Δ ntg2Δ rad14Δ rev3Δ strain compared to that in the ntg1Δ ntg2Δ rad14Δ strain may arise from the role of Polζ in the mutagenic bypass of H2O2-induced DNA lesions other than oxidized pyrimidines. Because the oxidized pyrimidines Ug, 5-OHC, and 5-OHU are not a block to DNA polymerases and can be bypassed by mispairing, the high incidence of H2O2-induced can1r mutations that accrues in the ntg1Δ ntg2Δ rad14Δ strain in either the absence or presence of the rev3Δ mutation is likely due to the mutagenic bypass of such oxidized pyrimidines by Polδ.
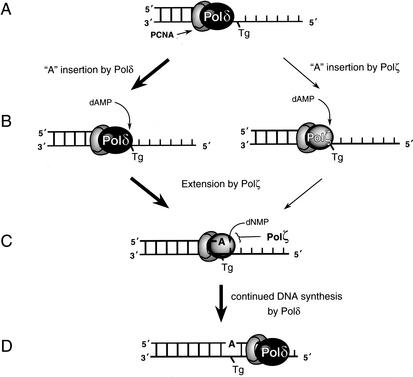
Figure 3.
Error-free Tg bypass in yeast by the coordinated action of Polδ and Polζ. (A) Polδ encounters a Tg during replication. (B) Although Polζ could contribute to the insertion of an A opposite Tg, because of the efficient nucleotide insertion by Polδ, and because of its proximity to the lesion site, we expect Polδ to primarily carry out the insertion reaction. (C) Polζ extends from the A nucleotide inserted opposite Tg. (D) Polδ resumes normal DNA synthesis.
In summary, the large increase in H2O2 sensitivity and the persistence of high levels of H2O2-induced mutagenesis in the ntg1Δ ntg2Δ rad14Δ rev3Δ strain, plus the absence of any significant effect on H2O2 sensitivity and induced mutagenesis when the rad5Δ and rad30Δ mutations are introduced into this strain, are all consistent with the requirement of Polζ for the error-free bypass of Tg lesions, whereas Rad5 and Polη make no significant contribution to the bypass of this DNA lesion.
Although Polζ could on its own replicate through a Tg lesion by carrying out both the nucleotide incorporation and subsequent extension reactions, we consider it more likely that replication through a Tg lesion in yeast occurs by the coordinated action of Polδ and Polζ (Fig. 3), in which Polδ acts at the nucleotide incorporation step and Polζ functions at the extension step. We think that Polδ would have precedence over Polζ at the nucleotide incorporation step, because Polδ would be the first polymerase to encounter the Tg lesion, and because this lesion imposes no block on nucleotide incorporation by it. We suggest that the reason the Polζ-dependent bypass pathway prevails over the other Rad6–Rad18-dependent bypass pathways is because of the proficient ability of Polδ for A incorporation opposite the lesion site from which Polζ efficiently extends (Fig. 3). As a corollary, for DNA lesions where Polδ stops before the lesion site, pathways other than Polζ could come into play. Thus, for example, the stalling of Polδ just before the 3′T of a cis-syn T–T dimer (Johnson et al. 1999), or other UV-induced DNA lesions such as (6–4) dipyrimidine photoproducts, could present Rad5 or Polη the opportunity to gain entry into the replicational ensemble before the lesion site and to carry out the bypass reaction. Consequently, although all three Rad6–Rad18-dependent pathways, namely Rad5, Polη, and Polζ, effect replication through UV-induced DNA lesions, the Rad5 pathway, and to a lesser extent, Polη, play a more prominent role in the bypass of UV lesions than does Polζ (Torres-Ramos et al. 2002). In the absence of any stalling of Polδ before the lesion site, as is the case for a Tg lesion, lesion bypass would become highly dependent upon a polymerase able to efficiently elongate from the nucleotide inserted opposite the lesion site by Polδ, thereby accounting for the requirement of Polζ for Tg bypass.
As inferred here from genetic studies in yeast, in higher eukaryotes also, Polζ could be a major contributor to Tg bypass. The ability of human Polη and Polκ to replicate through a Tg lesion was recently examined. Although human Polη is more efficient at the nucleotide incorporation step than is yeast Polη, Tg is a significant block for extension by human Polη, as evidenced from the inhibition of synthesis that occurs right after the incorporation of a nucleotide opposite the lesion, as well as after the incorporation of the next nucleotide (Kusumoto et al. 2002). Human Polκ is inhibited by Tg at both the nucleotide incorporation step and the subsequent extension step, with inhibition at the extension step being more severe (Fischhaber et al. 2002). The proficient ability of Polζ to extend from the A nucleotide inserted opposite Tg by Polδ reported here for yeast would suggest that in humans too, this represents the primary pathway for Tg bypass.
A preeminent role of Polζ in Tg bypass in higher eukaryotes could explain the embryonic lethality of Rev3−/− mice. In addition, we consider it very likely that Polζ similarly promotes the error-free bypass of many other frequently formed spontaneous DNA lesions. Furthermore, proficient replication through a variety of frequently formed DNA lesions would ensure that rapid cell divisions continue unhindered during early embryonic development. Hence, in the absence of Polζ, replication forks would stall at spontaneously arising DNA lesions, leading to the formation of double-stranded DNA breaks and chromatid and chromosome aberrations that are seen in Rev3−/− embryos, which would then cause the apoptotic response and embryonic lethality (Van Sloun et al. 2002). Thus, by promoting efficient and error-free TLS through DNA lesions, Polζ could make a critical contribution to early embryonic development in higher eukaryotes.
Materials and methods
Enzyme purification
Yeast DNA polymerase δ was kindly provided by Peter Burgers (Washington University School of Medicine, St. Louis, MO), and DNA polymerase η from which the GST portion had been cleaved was purified as described (Johnson et al. 2001). To purify yeast Polζ, the yeast REV3 gene was cloned in frame with the glutathione S-transferase (GST) gene containing the Prescission protease recognition sequence (LEVLFQGP), under control of the galactose-inducible phosphoglycerate kinase promoter in plasmid pBJ842, generating plasmid pREV3.30. The native Rev7 protein was expressed from the alcohol dehydrogenase (ADH) promotor in plasmid pREV7.27. Yeast strain BJ5464 harboring both pREV3.30 and pREV7.27 was grown in synthetic complete media lacking leucine and tryptophan (SC–leu,-trp), induced with 2% galactose, and harvested as described (Johnson et al. 2000b). Frozen yeast cells were resuspended in 2.5 mL of cell breakage buffer (50 mM Tris-HCl at pH 7.5, 10% sucrose, 300 mM NaCl, 1 mM EDTA, 0.5 mM benzamidine/phenylmethylsulfonyl fluoride, 10 mM β-mercaptoethanol) per gram of yeast and lysed in a French press before centrifugation at 100,000 g. The extract was then passed over a 100 μL glutathione sepharose 4B column (Pharmacia) at 4°C, and the column was washed with 10 volumes of cell breakage buffer containing 1M NaCl. The column was equilibrated in Prescission cleavage buffer (50 mM Tris-HCl at pH 7.5, 150 mM NaCl, 10% glycerol, 1 mM dithiolthreitol, 0.01% NP-40), and Polζ (Rev3/Rev7 complex) without the GST tag was batch-eluted from the matrix by treatment with 2 units of Prescission protease (Amersham Pharmacia) overnight at 4°C. Prescission protease cleaves between the glutamine (Q) and glycine (G) residues of the recognition sequence (see above) and leaves a 7-amino-acid leader peptide on the N terminus of the Rev3 protein. Polζ aliquots were stored at −70°C.
Tg substrate
The site-specific Tg was generated as described (McNulty et al. 1998). The 11-nt oligomer 5′-GCAAGTCGGAG-3′ (OD50 Abs260), containing a single thymine residue at the underlined position, was treated with 1 mM KMnO4 for 5 min at 4°C. The oligonucleotide was then purified by reverse-phase HPLC using a Waters Delta Pak C-18 analytical column. The identity of the Tg containing 11-mer oligonucleotide was verified by mass spectrometry. The unmodified and the Tg containing 11-nt oligomers were each incorporated into a 76-nt template DNA (5′-AGCAAGTCACCAATGTCTAAGAGTTGCAAGTCGGAG CTACACTGGAGTACCGGAGCATCGTCGTGACTGGGAA AAC-3′) by ligation to flanking 25-nt and 40-nt oligomers, where the underlined T was either undamaged or Tg. The full-length 76-nt template DNAs were separated from unligated oligonucleotides using 12% polyacrylamide gels containing 8 M urea, cut from the gels, and subsequently isolated by electroelution. Primer oligonucleotides were 5′ 32P-labeled using γ-32P ATP (Amersham-Pharmacia) and T4 kinase (Roche). For running start DNA synthesis reactions, the 40-nt primer N4264 (5′-GTTTTCCCAGTCACGACGATGCTCCGGTACTCCAGT GTAG-3′) was annealed to either the nondamaged or Tg-containing template. For determining the identity of the deoxynucleotide incorporated opposite the unmodified T or the Tg, the 19-nt primer (5′-GTACTCCAGTGTAGCTCCG-3′) was used. The 20-nt marker oligonucleotides were identical in sequence to the 19-nt primer, except that they contained an additional G, A, T, or C residue at the 3′ end. For steady-state kinetic nucleotide incorporation assays, the 45-nt primer N8282 (5′-GTTTTCCCAGTCACGACGATGCTCCGGTACT CCAGTGTAGCTCCG-3′) was used. To analyze the steady-state kinetic parameters of primer extension by Polζ from a G, A, T, or C residue paired with either a T or a Tg in the template, oligomers identical to N8282 but containing an additional G, A, T, or C nucleotide at the 3′ end were used, respectively.
DNA polymerase assays
The standard DNA polymerase reaction (5 μL) contained 25 mM Tris-HCl at pH 7.5, 5 mM MgCl2, 1 mM dithiolthreitol, 100 μg/mL bovine serum albumin, 10% glycerol, 100 μM of each deoxynucleotide (dGTP, dATP, dTTP, and/or dCTP), and 10 nM DNA substrate consisting of a 5′ 32P-labeled primer annealed to a DNA template. Enzyme concentrations used for examining TLS through a Tg are indicated in the legend for Figure 1. Reactions were carried out at 30°C for 10 min and terminated by the addition of 4 volumes of loading buffer (95% formamide, 0.05% cyanol blue, 0.05% bromophenol blue, and 20 mM EDTA) before resolving on 10% or 20% polyacrylamide gels containing 8M urea. Gels were dried before autoradiography at −70°C.
Steady-state kinetic analysis
Analyses of kinetic parameters for deoxynucleotide incorporation opposite the Tg lesion and for primer extension from nucleotides opposite this lesion were done as described (Creighton et al. 1995). For examining the kinetics of nucleotide incorporation, yeast Polζ (0.5 nM) or yeast Polη (1 nM) were assayed for 5–10 min at 30°C using the standard DNA polymerase assay except that only a single deoxynucleotide was included. For yeast Polζ, the correct deoxynucleotide (dATP) concentration was varied from 0.05 to 1 μM and from 0.25 to 10 μM, in assays containing nondamaged or Tg-containing DNA, respectively. Incorrect nucleotides were varied from 50 to 5000 μM. For yeast Polη, the correct deoxynucleotide (dATP) concentration was varied from 0.5 to 250 μM, and incorrect nucleotide concentrations ranged from 10 to 3000 μM. Gel band intensities of the substrate and products of the deoxynucleotide incorporation reactions were quantified using a Storm phosphorimager and ImageQuant software (Molecular Dynamics). The observed rate of deoxynucleotide incorporation, vobs, was determined by dividing the amount of product formed by the reaction time. We plotted vobs as a function of the deoxynucleotide concentration and fit these data to the Michaelis-Menten equation describing a hyperbola: vobs = (Vmax × [dNTP])/(Km + [dNTP]).
From the best fit curve, the apparent Km and Vmax steady-state kinetics parameters were obtained for the incorporation of both incorrect and correct deoxynucleotides. These parameters were also used to calculate the frequency of deoxynucleotide misincorporation, finc, using the following equation: finc = (Vmax/Km)incorrect/(Vmax/Km)correct.
To examine the kinetics of primer extension from a G, A, T, or C residue paired opposite a normal thymine or opposite a Tg, the standard DNA polymerase assay conditions were used as described above, except that the concentration of dCTP, the next correct nucleotide, was varied. For assays containing yeast Polζ, the dCTP concentration ranged from 0.25 to 50 μM for extension from an A residue paired opposite a normal thymine or a Tg, and ranged from 5 to 500 μM for the mispaired primer termini. For yeast Polη, the dCTP concentration ranged from 0.1 to 500 μM for extension from an A residue paired opposite a normal thymine or a Tg, and ranged from 10 to 3000 μM for all other primer:template combinations. The intrinsic efficiency of mismatch extension, f°ext, a constant that represents the efficiency of extending mismatched termini in competition with matched termini at equal DNA concentrations in the limit of zero next nucleotide, was calculated using the equation: f°ext = (Vmax/Km)mismatch/(Vmax/Km)matched.
Generation of yeast null mutant strains
Genomic deletion mutations of the REV3 and RAD14 genes were generated using plasmids pPM292 and pR14.4, respectively. To create the ntg1Δ-generating plasmid, a 2.9-kb region of yeast genomic DNA encompassing the entire S. cerevisiae NTG1 open reading frame (ORF) plus 800 bp and 420 bp of 5′ and 3′ flanking DNA, respectively, was amplified by PCR and cloned into pUC19. A BamHI linker was then inserted at the EcoRV site at position +144 of the NTG1 ORF, and subsequently, the 1.6-kb 3′ DNA of NTG1 from nt +144 to +420 bp downstream of the termination codon was replaced with a 467-bp PCR product containing only nt +1153 to +420 bp downstream of the termination codon. The resulting plasmid contains PCR products of 930 bp and 470 bp, corresponding to the 5′ and 3′ flanking region of NTG1, respectively, separated by a BamHI restriction endonuclease site. The URA3 gene blaster fragment was then cloned into the BamHI site, generating the ntg1Δ-generating plasmid pSKSL58. This plasmid, when digested with the restriction endonucleases EcoRI and SphI, results in a 5.7-kb DNA fragment that when transformed into yeast, deletes the NTG1 ORF from position +144 to +1153.
To construct the ntg2Δ-generating plasmid, a 3.3-kb DNA fragment containing the entire NTG2 ORF, and 1.2 kb and 0.95 kb of 5′ and 3′ flanking DNA, respectively, was amplified by PCR and cloned into pUC19. The internal region of the ORF from nucleotide +50 to +958 was removed and replaced by a BamHI linker. The URA3 gene blaster fragment was subsequently inserted into this BamHI site, resulting in the ntg2Δ-generating plasmid pPM982. This plasmid, when digested with the restriction endonucleases Asp718 and SphI, releases a 6.2-kb DNA fragment that when transformed into yeast deletes the 1140 bp NTG2 ORF from nucleotides +50 to +958.
Yeast strain EMY74.7 (MATahis3Δ, leu2-3,112, ura3-52 trp1Δ) and its derivatives were transformed with the deletion-generating plasmids using the lithium acetate method. The presence of each deletion was confirmed by PCR analysis of yeast genomic DNA and by back-crossing to known yeast deletion mutants.
H2O2 sensitivity and induced mutagenesis
Yeast strains grown to mid-log phase at 30°C in YPD medium were treated with H2O2 (Fluka) at the concentrations indicated in the legend for Figure 2. Cells were then incubated for an additional h at 30° C with vigorous shaking, harvested by filtration, and resuspended in sterile glass-distilled H2O. Appropriate dilutions were then plated on YPD for survival analysis or on synthetic complete medium lacking arginine but containing canavanine to measure the frequency of CAN1S to can1r forward mutations. Plates were incubated at 30°C for 3–4 d for survival and for 5–6 d for mutagenesis before counting colonies.
Acknowledgments
The Tg-containing DNA substrate was constructed in the Synthetic Organic Chemistry Core Laboratory, supported by NIEHS Center Grant P30-ES06676, and we thank Richard Hodge for providing this DNA. This work was supported by NIH grants GM19261 and CA41261.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL l.prakash@utmb.edu; FAX (409) 747-8608.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.1048303.
References
- Ames BN, Shigenaga MK, Hagen TM. Oxidants, antioxidants, and the dengerative diseases of aging. Proc Nat Acad Sci. 1993;90:7915–7922. doi: 10.1073/pnas.90.17.7915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bemark M, Khamlichi AA, Davies SL, Neuberger MS. Disruption of mouse polymerase ζ (Rev3) leads to embryonic lethality and impairs blastocyst development in vitro. Curr Biol. 2000;10:1213–1216. doi: 10.1016/s0960-9822(00)00724-7. [DOI] [PubMed] [Google Scholar]
- Cathcart R, Schwiers E, Saul RL, Ames BN. Thymine glycol and thymidine glycol in human and rat urine: A possible assay for oxidative DNA damage. Proc Nat Acad Sci. 1984;81:5633–5637. doi: 10.1073/pnas.81.18.5633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciarrocchi G, Pedrini AM. Determination of pyrimidine dimer unwinding angle by measurement of DNA electrophoretic mobility. J Mol Biol. 1982;155:177–183. doi: 10.1016/0022-2836(82)90445-4. [DOI] [PubMed] [Google Scholar]
- Clark JM, Beardsley GP. Functional effects of cis- thymine glycol lesions on DNA synthesis in vitro. Biochem. 1987;26:5398–5403. doi: 10.1021/bi00391a027. [DOI] [PubMed] [Google Scholar]
- Clark JM, Pattabiraman N, Jarvis W, Beardsley GP. Modeling and molecular mechanical studies of the cis- thymine glycol radiation damage lesion in DNA. Biochem. 1987;26:5404–5409. doi: 10.1021/bi00391a028. [DOI] [PubMed] [Google Scholar]
- Creighton S, Bloom LB, Goodman MF. Gel fidelity assay measuring nucleotide misinsertion, exonucleolytic proofreading, and lesion bypass efficiencies. Methods Enzymol. 1995;262:232–256. doi: 10.1016/0076-6879(95)62021-4. [DOI] [PubMed] [Google Scholar]
- Dizdaroglu M, Simic MG. Radiation-induced crosslinking of cytosine. Radiat Res. 1984;100:41–46. [PubMed] [Google Scholar]
- Esposito G, Godin I, Klein U, Yaspo M-L, Cumano A, Rajewsky K. Disruption of the Rev31- encoded catalytic subunit of polymerase ζ in mice results in early embryonic lethality. Curr Biol. 2000;10:1221–1224. doi: 10.1016/s0960-9822(00)00726-0. [DOI] [PubMed] [Google Scholar]
- Fischhaber PL, Gerlach VL, Feaver WJ, Hatahet Z, Wallace SS, Friedberg EC. Human DNA polymerase κ bypasses and extends beyond thymine glycols during translesion synthesis in vitro, preferentially incorporating correct nucleotides. J Biol Chem. 2002;277:37604–37611. doi: 10.1074/jbc.M206027200. [DOI] [PubMed] [Google Scholar]
- Gellon L, Barbey R, Auffret van der Kemp P, Thomas D, Boiteux S. Synergism between base excision repair, mediated by the DNA glycosylases Ntg1 and Ntg2, and nucleotide excision repair in the removal of oxidatively damaged DNA bases in Saccharomyces cerevisiae. Mol Genet Genomics. 2001;265:1087–1096. doi: 10.1007/s004380100507. [DOI] [PubMed] [Google Scholar]
- Haracska L, Unk I, Johnson RE, Johansson E, Burgers PMJ, Prakash S, Prakash L. Roles of yeast DNA polymerases δ and ζ and of Rev1 in the bypass of abasic sites. Genes & Dev. 2001a;15:945–954. doi: 10.1101/gad.882301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haracska L, Washington MT, Prakash S, Prakash L. Inefficient bypass of an abasic site by DNA polymerase η. J Biol Chem. 2001b;276:6861–6866. doi: 10.1074/jbc.M008021200. [DOI] [PubMed] [Google Scholar]
- Huang J-C, Hsu DS, Kazantsev A, Sancar A. Substrate spectrum of human excinuclease: Repair of abasic sites, methylated bases, mismatches, and bulky adducts. Proc Nat Acad Sci. 1994;91:12213–12217. doi: 10.1073/pnas.91.25.12213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson RE, Henderson ST, Petes TD, Prakash S, Bankmann M, Prakash L. Saccharomyces cerevisiae RAD5-encoded DNA repair protein contains DNA helicase and zinc-binding sequence motifs and affects the stability of simple repetitive sequences in the genome. Mol Cell Biol. 1992;12:3807–3818. doi: 10.1128/mcb.12.9.3807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson RE, Prakash S, Prakash L. Yeast DNA repair protein RAD5 that promotes instability of simple repetitive sequences is a DNA-dependent ATPase. J Biol Chem. 1994;269:28259–28262. [PubMed] [Google Scholar]
- Johnson RE, Torres-Ramos CA, Izumi T, Mitra S, Prakash S, Prakash L. Identification of APN2, the Saccharomyces cerevisiae homolog of the major human AP endonuclease HAP1, and its role in the repair of abasic sites. Genes & Dev. 1998;12:3137–3143. doi: 10.1101/gad.12.19.3137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson RE, Prakash S, Prakash L. Efficient bypass of a thymine-thymine dimer by yeast DNA polymerase, Polη. Science. 1999;283:1001–1004. doi: 10.1126/science.283.5404.1001. [DOI] [PubMed] [Google Scholar]
- Johnson RE, Washington MT, Haracska L, Prakash S, Prakash L. Eukaryotic polymerases ι and ζ act sequentially to bypass DNA lesions. Nature. 2000a;406:1015–1019. doi: 10.1038/35023030. [DOI] [PubMed] [Google Scholar]
- Johnson RE, Washington MT, Prakash S, Prakash L. Fidelity of human DNA polymerase η. J Biol Chem. 2000b;275:7447–7450. doi: 10.1074/jbc.275.11.7447. [DOI] [PubMed] [Google Scholar]
- Johnson RE, Haracska L, Prakash S, Prakash L. Role of DNA polymerase η in the bypass of a (6–4) TT photoproduct. Mol Cell Biol. 2001;21:3558–3563. doi: 10.1128/MCB.21.10.3558-3563.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kao JY, Goljer I, Phan TA, Bolton PH. Characterization of the effects of a thymine glycol residue on the structure, dynamics, and stability of duplex DNA by NMR. J Biol Chem. 1993;268:17787–17793. [PubMed] [Google Scholar]
- Kim J-K, Patel D, Choi B-S. Contrasting structural impacts induced by cis-syn cyclobutane dimer and (6–4) adduct in DNA duplex decamers: Implication in mutagenesis and repair activity. Photochem Photobiol. 1995;62:44–50. doi: 10.1111/j.1751-1097.1995.tb05236.x. [DOI] [PubMed] [Google Scholar]
- Kung HC, Bolton PH. Structure of a duplex DNA containing a thymine glycol residue in solution. J Biol Chem. 1997;272:9227–9236. doi: 10.1074/jbc.272.14.9227. [DOI] [PubMed] [Google Scholar]
- Kusumoto R, Masutani C, Iwai S, Hanaoka F. Translesion synthesis by human DNA polymerase η across thymine glycol lesions. Biochem. 2002;41:6090–6099. doi: 10.1021/bi025549k. [DOI] [PubMed] [Google Scholar]
- Lee J-H, Hwang G-S, Choi B-S. Solution structure of a DNA decamer duplex containing the stable 3′ T.G base pair of the pyrimidine(6–4)pyrimidone photoproduct [(6–4) adduct]: Implications for the highly specific 3′ T → C transition of the (6–4) adduct. Proc Nat Acad Sci. 1999;96:6632–6636. doi: 10.1073/pnas.96.12.6632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNulty JM, Jerkovic B, Bolton PH, Basu AK. Replication inhibition and miscoding properties of DNA templates containing a site-specific cis-thymine glycol or urea rsidue. Chem Res Toxicol. 1998;11:666–673. doi: 10.1021/tx970225w. [DOI] [PubMed] [Google Scholar]
- Miaskiewicz K, Miller J, Ornstein R, Osman R. Molecular dynamics simulations of the effects of ring-saturated thymine lesions on DNA structure. Biopolymers. 1995;35:113–124. doi: 10.1002/bip.360350112. [DOI] [PubMed] [Google Scholar]
- Nelson JR, Lawrence CW, Hinkle DC. Thymine thymine dimer bypass by yeast DNA polymerase ζ. Science. 1996;272:1646–1649. doi: 10.1126/science.272.5268.1646. [DOI] [PubMed] [Google Scholar]
- Prakash S, Prakash L. Nucleotide excision repair in yeast. Mutat Res. 2000;451:13–24. doi: 10.1016/s0027-5107(00)00037-3. [DOI] [PubMed] [Google Scholar]
- Purmal AA, Kow YW, Wallace SS. Major oxidative products of cytosine, 5-hydroxycytosine and 5-hydroxyuracil, exhibit sequence context-dependent mispairing in vitro. Nucleic Acids Res. 1994;22:72–78. doi: 10.1093/nar/22.1.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purmal AA, Lampman GW, Bond JP, Hatahet Z, Wallace SS. Enzymatic processing of uracil glycol, a major oxidative product of DNA cytosine. J Biol Chem. 1998;273:10026–10035. doi: 10.1074/jbc.273.16.10026. [DOI] [PubMed] [Google Scholar]
- Reardon JT, Bessho T, Kung HC, Bolton PH, Sancar A. In vitro repair of oxidative DNA damage by human nucleotide excision repair system: Possible explanation for neurodegeneration in xeroderma pigmentosum patients. Proc Nat Acad Sci. 1997;94:9463–9468. doi: 10.1073/pnas.94.17.9463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sancar A. DNA excision repair. Ann Rev Biochem. 1996;65:43–81. doi: 10.1146/annurev.bi.65.070196.000355. [DOI] [PubMed] [Google Scholar]
- Senturker S, van der Kemp PA, You HJ, Doetsch PW, Dizdaroglu M, Boiteux S. Substrate specificities of the Ntg1 and Ntg2 proteins of Saccharomyces cerevisiae for oxidized DNA bases are not identical. Nucleic Acids Res. 1998;26:5270–5276. doi: 10.1093/nar/26.23.5270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teoule R. Radiation-induced DNA damage and its repair. Int J of Radiat Biol. 1987;51:573–589. doi: 10.1080/09553008414552111. [DOI] [PubMed] [Google Scholar]
- Teoule R, Bonicel A, Bert C, Cadet J, Polverelli M. Identification of radioproducts resulting from the breakage of thymine moiety by γ irradiation of E. coli DNA in an aerated aqueous solution. Radiat Res. 1974;57:46–58. [PubMed] [Google Scholar]
- Torres-Ramos CA, Johnson RE, Prakash L, Prakash S. Evidence for the involvement of nucleotide excision repair in the removal of abasic sites in yeast. Mol Cell Biol. 2000;20:3522–3528. doi: 10.1128/mcb.20.10.3522-3528.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres-Ramos C, Prakash S, Prakash L. Requirement of RAD5 and MMS2 for post-replication repair of UV-damaged DNA in Saccharomyces cerevisiae. Mol Cell Biol. 2002;22:2419–2426. doi: 10.1128/MCB.22.7.2419-2426.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Sloun PPH, Varlet I, Sonneveld E, Boei JJWA, Romeijn RJ, Eeken JCJ, De Wind N. Involvement of mouse Rev3 in tolerance of endogenous and exogenous DNA damage. Mol Cell Biol. 2002;22:2159–2169. doi: 10.1128/MCB.22.7.2159-2169.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner JR, Hu C-C, Ames BN. Endogenous oxidative damage of deoxycytidine in DNA. Proc Nat Acad Sci. 1992;89:3380–3384. doi: 10.1073/pnas.89.8.3380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace S. Oxidative stress defenses. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1997. Oxidative damage to DNA and its repair; pp. 49–90. [Google Scholar]
- Washington MT, Johnson RE, Prakash S, Prakash L. Accuracy of thymine-thymine dimer bypass by Saccharomyces cerevisiae DNA polymerase η. Proc Nat Acad Sci. 2000;97:3094–3099. doi: 10.1073/pnas.050491997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittschieben J, Shivji MKK, Lalani E, Jacobs MS, Marini F, Gearhart PM, Rosewell I, Stamp G, Wood RD. Disruption of the developmentally regulated Rev31 gene causes embryonic lethality. Curr Biol. 2000;10:1217–1220. doi: 10.1016/s0960-9822(00)00725-9. [DOI] [PubMed] [Google Scholar]
- Yu S-L, Johnson RE, Prakash S, Prakash L. Requirement of DNA polymerase η for error-free bypass of UV-induced CC and TC photoproducts. Mol Cell Biol. 2001;21:185–188. doi: 10.1128/MCB.21.1.185-188.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]