Abstract
RNA silencing phenomena were first discovered in plants, yet only the RNA interference pathway in animals has been subject to biochemical analysis. Here, we extend biochemical analysis to plant RNA silencing. We find that standard wheat germ extract contains Dicer-like enzymes that convert double-stranded RNA (dsRNA) into two classes of small interfering RNAs, as well as an RNA-dependent RNA polymerase activity that can convert exogenous single-stranded RNA into dsRNA. In this plant embryo extract, an endogenous microRNA (miRNA) that lacks perfect complementarity to its RNA targets nonetheless acts as a small interfering RNA. The miRNA guides an endonuclease to cleave efficiently wild-type Arabidopsis PHAVOLUTA mRNA, but not a dominant mutant previously shown to perturb leaf development. This finding supports the view that plant miRNAs direct RNAi and that miRNA-specified mRNA destruction is important for proper plant development. Thus, endonuclease complexes guided by small RNAs are a common feature of RNA silencing in both animals and plants.
Keywords: RNAi, PTGS, siRNA, miRNA, RdRP, Dicer
RNA interference (RNAi) in animals and basal eukaryotes, quelling in fungi, and posttranscriptional gene silencing (PTGS) in plants are examples of a broad family of phenomena collectively called RNA silencing (Kooter et al. 1999; Li and Ding 2001; Matzke et al. 2001; Vaucheret et al. 2001; Waterhouse et al. 2001; Hannon 2002; Plasterk 2002). The unifying features of RNA silencing phenomena are the production of small (21–26 nt) RNAs that act as specificity determinants for down-regulating gene expression (Hamilton and Baulcombe 1999; Hammond et al. 2000; Parrish et al. 2000; Zamore et al. 2000; Djikeng et al. 2001; Parrish and Fire 2001; Tijsterman et al. 2002) and the requirement for one or more members of the Argonaute family of proteins (Tabara et al. 1999; Fagard et al. 2000; Hammond et al. 2001; Hutvágner and Zamore 2002; Kennerdell et al. 2002; Martinez et al. 2002a; Pal-Bhadra et al. 2002; Williams and Rubin 2002). We do not yet understand the biochemical function of Argonaute proteins (or PPD proteins, named for their characteristic PAZ and Piwi domains).
Small RNAs are generated in animals by members of the Dicer family of double-stranded RNA (dsRNA)-specific endonucleases (Bernstein et al. 2001; Billy et al. 2001; Grishok et al. 2001; Ketting et al. 2001). Dicer family members are large, multidomain proteins that contain putative RNA helicase, PAZ, two tandem ribonuclease III (RNase III), and one or two dsRNA-binding domains. The tandem RNase III domains are believed to mediate endonucleolytic cleavage of dsRNA into small interfering RNAs (siRNAs), the mediators of RNAi. In Drosophila and mammals, siRNAs, together with one or more Argonaute proteins, form a protein–RNA complex, the RNA-induced silencing complex (RISC), which mediates the cleavage of target RNAs at sequences with extensive complementarity to the siRNA (Hammond et al. 2000, 2001; Zamore et al. 2000; Elbashir et al. 2001a,b,c; Nykänen et al. 2001; Hutvágner and Zamore 2002; Martinez et al. 2002a).
In addition to Dicer and Argonaute proteins, RNA-dependent RNA polymerase (RdRP) genes are required for RNA silencing in Caenorhabditis elegans (Smardon et al. 2000; Sijen et al. 2001), Neurospora crassa (Cogoni and Macino 1999), and Dictyostelium discoideum (Martens et al. 2002), but likely not for RNAi in Drosophila or mammals (Celotto and Graveley 2002; Chiu and Rana 2002; Holen et al. 2002; Martinez et al. 2002b; Schwarz et al. 2002; Roignant et al. 2003). In plants, PTGS initiated by transgenes that overexpress an endogenous mRNA also requires a putative RdRP, SGS2 (SDE1; Dalmay et al. 2000; Mourrain et al. 2000), although transgenes designed to generate dsRNA bypass this requirement (Beclin et al. 2002). Similarly, silencing induced by viruses replicating through a dsRNA intermediate (virus-induced gene silencing, VIGS) does not require SGS2 (Dalmay et al. 2000).
Dicer in animals and CARPEL FACTORY (CAF, a Dicer homolog) in plants also generate microRNAs (miRNAs), 20–24-nt, single-stranded noncoding RNAs thought to regulate endogenous mRNA expression (Lee et al. 1993; Reinhart et al. 2000, 2002; Grishok et al. 2001; Hutvágner et al. 2001; Ketting et al. 2001; Lagos-Quintana et al. 2001, 2002; Lau et al. 2001; Lee and Ambros 2001; Mourelatos et al. 2002; Park et al. 2002). miRNAs are produced by Dicer cleavage of stem–loop precursor RNA transcripts (pre-miRNAs); the miRNA can reside on either the 5′ or 3′ side of the double-stranded stem (Lee et al. 1993; Pasquinelli et al. 2000; Lagos-Quintana et al. 2001; Lau et al. 2001; Lee and Ambros 2001). In animals, pre-miRNAs are transcribed as longer primary transcripts (pri-miRNAs) that are processed in the nucleus into compact, folded structures (pre-miRNAs), then exported to the cytoplasm, where they are cleaved by Dicer to yield mature miRNAs (Lee et al. 2002). Animal miRNAs are only partially complementary to their target mRNAs; this partial complementarity has been proposed to cause miRNAs to repress translation of their targets, rather than direct target cleavage by the RNAi pathway (for review, see Ruvkun 2001; Hutvágner and Zamore 2002). Plant miRNAs have far greater complementarity to cellular mRNAs and have been proposed to mediate target RNA cleavage via an RNAi-like mechanism (Llave et al. 2002b; Rhoades et al. 2002).
Here, we present evidence that extracts of wheat germ, introduced for the study of translation and protein translocation in the 1970s (Roberts and Paterson 1973), recapitulate many of the key features of RNA silencing in plants. Using this in vitro system, we show that in plants, ATP-dependent, Dicer-like enzymes cleave dsRNA into small RNAs that have the structure of siRNAs. Unlike Drosophila embryos or mammalian cells, plants convert dsRNA into two distinct classes of siRNAs, long and short siRNAs. Our inhibitor studies suggest that a different Dicer-like enzyme generates each siRNA class. We further show that a wheat RdRP activity can synthesize dsRNA using exogenous single-stranded RNA as a template without an exogenous primer, and that this dsRNA is preferentially converted into long siRNAs. Finally, we report that wheat germ extracts contain an endogenous RISC programmed with a miRNA. This endogenous miRNA complex can direct efficient cleavage of the wild-type Arabidopsis PHAVOLUTA (PHV) mRNA sequence, but not that of a previously described dominant PHV mutant that perturbs leaf development. This finding supports the view that in plants miRNAs direct RNAi and explains the molecular basis for the dominant PHV mutation in Arabidopsis.
Results
Two distinct classes of small RNAs derived from dsRNA in plant extracts
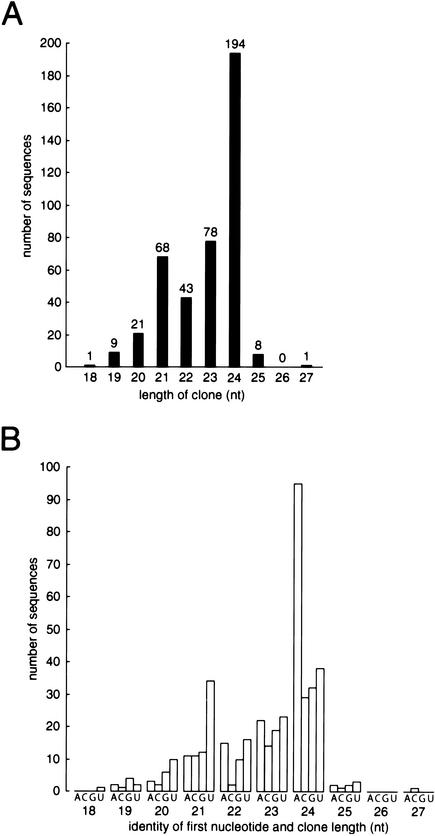
Two distinct classes of small RNAs are produced in transgenic plants bearing silenced transgenes (Hamilton et al. 2002; Mallory et al. 2002). To test if the production of these two classes of small RNAs was a normal feature of plant biology or a specialized response to foreign DNA, we examined the length distribution of a nonredundant set of 423 endogenous small RNAs cloned from Arabidopsis thaliana, including 143 published sequences (Llave et al. 2002a; Reinhart et al. 2002) and an additional 280 sequences not previously published. Excluded from this analysis are cloned fragments of tRNA and rRNA. Included in the set are known and predicted miRNAs, as well as small RNAs of unknown function corresponding to intragenic regions or to mRNA sequences in either the sense or antisense orientation. The distribution of lengths within this set was bimodal, with peaks at 21 and 24 nt (Fig. 1A). In contrast, the length distribution of cloned small RNAs from C. elegans forms a single broad peak (Lau et al. 2001). The two classes of green fluorescent protein (GFP)-derived small RNAs were proposed to be siRNAs with distinct RNA silencing functions: the ∼21-mers to direct posttranscriptional silencing via mRNA degradation and the ∼24-mers to trigger systemic silencing and the methylation of homologous DNA (Hamilton et al. 2002). Our analysis of the two classes of endogenous small RNAs indicates that each class has a distinct sequence bias, with a 5′-uridine predominating in the shorter class and a 5′-adenosine in the longer class (Fig. 1B). The 5′ sequence bias of the short class is produced by the inclusion in our data set of miRNAs, which in plants and animals typically begin with uridine (Lagos-Quintana et al. 2001, 2002; Lau et al. 2001; Lee and Ambros 2001; Reinhart et al. 2002). Thus, the non-miRNA small RNAs in the shorter class display no 5′ sequence bias, whereas a 5′-adenosine is overrepresented in the longer class. The two classes are either generated by different enzymes, function in separate effector complexes, or both.
Figure 1.
Arabidopsis thaliana small RNAs form two distinct size classes. (A) Size distribution of small RNA clones. (B) Sequence composition of the 5′ ends of cloned small RNA as a function of length.
Plant small RNAs are bona fide siRNAs
Although the small RNAs that correlate with the posttranscriptional silencing of homologous target mRNAs were first discovered in plants (Hamilton and Baulcombe 1999), they have not yet been shown to be the direct products of endonucleolytic cleavage of long dsRNA. To begin to test if small RNAs are, in fact, siRNAs, we prepared plant extracts and monitored them for Dicer-like activity. When uniformly 32P-radiolabeled dsRNA was incubated in wheat germ extract, it was efficiently cleaved into small RNAs (Fig. 2A). As reported previously for extracts of Drosophila (Zamore et al. 2000) and for purified Drosophila (Bernstein et al. 2001) and human Dicer (Billy et al. 2001), no intermediate products were detected in the conversion of dsRNA into small RNAs. Unlike the fly and human Dicer reactions, two discrete size classes, one ∼21-nt and the other 24–25-nt long, were produced from the dsRNA upon incubation in wheat germ extract (Fig. 2B). The ratio of wheat 24–25-mers to ∼21-mers in 14 separate reactions was 4 ± 1.7, similar to the roughly 2.5-fold excess of longer small RNA sequences cloned from Arabidopsis. (The 2.5-fold excess of long to short, cloned endogenous small RNAs underestimates the ratio, because it includes miRNAs, which are predominantly short.) Silencing-related small RNAs have thus far only been demonstrated in vivo for dicots, and wheat is a monocot. Extracts of the dicot cauliflower, a member of the mustard family like Arabidopsis, also converted dsRNA into two discrete sizes of small RNAs, ∼21 and ∼24 nt (Fig. 2C). In both Drosophila and C. elegans, Dicer requires ATP for efficient production of both siRNAs (Zamore et al. 2000; Bernstein et al. 2001; Nykänen et al. 2001) and miRNAs (Hutvágner et al. 2001; Ketting et al. 2001). Consistent with the idea that both classes of small RNAs are produced by plant orthologs of Dicer, efficient production of both the ∼21-nt and the ∼24-nt small RNAs in wheat germ extract required ATP (Fig. 2D).
Figure 2.
dsRNA is cleaved into two discrete classes of bona fide siRNAs in plant extracts. (A) Upon incubation in wheat germ extract, 32P-dsRNA was cleaved into small RNAs in a highly processive reaction, as in fly embryo lysate. (B) 32P-dsRNA was cleaved in wheat germ extract into two sizes of small RNAs, ∼21-nt and 24–25-nt long, relative to synthetic 5′-32P-radiolabeled RNA markers. (C) 32P-dsRNA was cleaved in cauliflower extract into two sizes of small RNAs. (D) Efficient production of small RNAs in wheat germ extract required ATP. ATP, creatine phosphate, and creatine kinase were included (+ATP) or omitted (−ATP) from the reaction. (E) Small RNAs produced in vitro in wheat germ extract are double-stranded. 32P-dsRNA was incubated in wheat germ extract or Drosophila embryo lysate, deproteinized at room temperature without organic extraction, then analyzed by gel filtration on a Superdex 200 HR column. The peak positions of double- and single-stranded synthetic siRNA standards are indicated. (F) Scheme for detecting 3′ overhanging ends on small RNAs by nuclease protection. (G) Small RNAs produced by incubation of 32P-dsRNA in wheat germ extract have ∼2-nt 3′ overhanging ends and a central double-stranded body, characteristics of the products of Dicer cleavage. Brackets indicate the nuclease digestion products. The positions of 5′-32P-radiolabeled size markers are indicated at left. 3′-phosphorylated markers were generated by reacting synthetic RNAs 1 base longer than indicated with periodate, followed by β-elimination, yielding an RNA 1 base shorter, but bearing a 3′-phosphate in place of a hydroxyl.
Although small, silencing-associated RNAs in plants are commonly called siRNAs, and synthetic siRNA duplexes initiate plant RNA silencing (Klahre et al. 2002), plant small RNAs have not been demonstrated to be double-stranded RNAs with 2-nt, 3′ overhanging ends and 3′-hydroxyl termini. Such attributes reflect the unique production of siRNAs by members of the Dicer family of ribonuclease III enzymes. To determine if the small RNAs generated from dsRNA in wheat germ extracts were bona fide siRNAs, we analyzed their structure. Uniformly 32P-radiolabeled dsRNA was incubated in wheat germ extract, deproteinized, and fractionated by gel filtration to resolve single-stranded from double-stranded siRNA (Nykänen et al. 2001). Both classes of small RNA products of the in vitro wheat germ reaction comigrated with a synthetic siRNA duplex and with Drosophila siRNA duplexes generated by processing dsRNA in Drosophila embryo lysate (Fig. 2E). Therefore, the small RNAs generated by incubating dsRNA in wheat germ extract are double-stranded.
Next, we examined the end structure of the small RNAs. Treatment of 5′-32P-radiolabeled, synthetic siRNA duplexes with the single-stranded RNA-specific nucleases T1 and RNase A removes the 2-nt, 3′ overhanging ends typical of siRNAs, generating 1-nt and 2-nt shorter RNAs. In a denaturing polyacrylamide gel, such nuclease products of siRNAs migrate faster, because they contain 3′-phosphates (diagramed in Fig. 2F). When synthetic 25-nt duplexes with 2-nt, 3′ overhangs were digested with T1 and RNase A, the expected 24-nt and 23-nt, 3′ phosphorylated products were generated (Fig. 2G). The small RNAs produced by incubation of dsRNA in the wheat germ extract are a mixture of ∼21-nt and 24–25-nt species. Digestion of this mixture with single-stranded nucleases produced a faster-migrating population of RNA species whose length distribution is consistent with the original mixture having the single-stranded overhangs and double-stranded body characteristic of siRNAs (Fig. 2G). Both size classes of small RNAs produced upon incubation of dsRNA in wheat germ extract have 2‘,3′-hydroxyl and 5′ monophosphate termini (data not shown). In sum, the small RNAs have all the hallmarks of the products of Dicer-mediated cleavage of dsRNA. We conclude that they are bona fide siRNAs.
Different Dicer-like enzymes produce each class of siRNA
We can imagine at least two mechanisms by which long dsRNA could be converted in plants into distinct size classes of small RNAs. Local dsRNA sequence might determine siRNA length, irrespective of which Dicer ortholog cleaves the dsRNA. In this case, we anticipate that the two classes of small RNAs would have distinct sequence compositions. Instead, only the 5′ ends of the two classes show sequence bias (Fig. 1B). An alternative explanation is that different Dicer orthologs produce each class. Both the Arabidopsis and rice genomes encode at least four different Dicer-like proteins, including the Arabidopsis protein CARPEL FACTORY/SHORT INTEGUMENTS-1 (CAF). The number of wheat Dicer orthologs is presently unknown, because the hexaploid wheat genome remains to be sequenced.
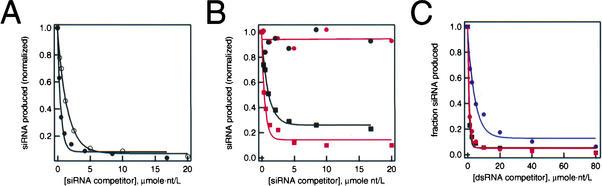
Drosophila Dicer binds tightly to siRNAs (P.D. Zamore and B. Haley, unpubl.). Therefore, we reasoned that different Dicer orthologs might be differentially inhibited by their products, siRNAs. We tested the ability of 21-nt and 25-nt synthetic siRNA duplexes to inhibit the production of siRNAs in Drosophila embryo lysates and the production of the two distinct classes of siRNA in wheat germ extract. Drosophila Dicer produces siRNAs 21–22 nt long. Drosophila Dicer was inhibited more strongly by a 21-nt siRNA duplex than by a 25-mer (Fig. 3A). Conversely, production of 24–25-nt siRNAs by wheat germ extract was inhibited more strongly by an ∼25-nt synthetic siRNA duplex competitor than a 21-mer (Fig. 3B). These results are consistent with the idea that the authentic siRNA product of Dicer should bind more strongly to its active site than an siRNA of an inauthentic length. Surprisingly, production of the ∼21-nt siRNAs was completely refractory to inhibition by either 21-nt or 25-nt synthetic siRNA duplexes, at siRNA concentrations as high as 800 nM (Fig. 3B). The simplest explanation for these data is that a different Dicer-like enzyme generates each class of siRNA and that the enzyme responsible for producing the 24–25-nt siRNAs is strongly inhibited by its siRNA product, whereas the enzyme that produces the ∼21-nt siRNAs is not inhibited by siRNA product at the concentrations tested. An alternative explanation is that the concentration of the enzyme that produces the ∼21-mers is higher than the highest concentration of inhibitor we tested, 800 nM. For this to be true, the enzyme would need to be present at micromolar concentration in the extract, which seems unlikely, as it would then correspond to ∼1% of total protein. The finding that production of both classes of siRNAs were equally and strongly inhibited by long dsRNA competitor (Fig. 3C) also supports an argument against this view. If the enzyme that generates the 21-mers were present in the extract at very high concentration, its activity should not have been competed by the same concentrations of long dsRNA competitor that saturate the enzyme that produces the 24–25-nt products. We conclude that each class of siRNA is produced by the ATP- dependent, endonucleolytic cleavage of dsRNA by a different Dicer ortholog.
Figure 3.
The two classes of plant siRNAs are produced by different enzymes. (A,B) 32P-dsRNA was incubated in either Drosophila embryo lysate or wheat germ extract for 3 h in the presence of increasing concentrations of 21-nt or 25-nt siRNA duplexes, then analyzed by denaturing gel electrophoresis and quantified. The siRNA concentration is presented in micromoles nucleotide per liter to permit comparison of the different lengths of siRNA duplex used. The relative efficiency of the reactions was determined by fitting the data to a single exponential and comparing the rate constant. (A) 21-nt siRNA duplexes (filled circles) are more efficient inhibitors of Drosophila Dicer than 25-nt siRNA duplexes (open circles). (B) Production of 25-nt siRNAs in wheat germ extract (squares) was inhibited more efficiently by a 25-nt synthetic siRNA duplex (red symbols) than by a 21-nt siRNA duplex (black symbols), but production of the 21-nt siRNAs (circles) was not inhibited by either synthetic siRNA duplex. (C) dsRNA competitor inhibited the production of both 25-nt (black squares) and 21-nt (red circles) siRNAs in wheat germ extract. Production of siRNAs in Drosophila embryo lysate (blue circles) was also inhibited by dsRNA competitor, but to a lower extent, perhaps reflecting a higher concentration of Dicer in Drosophila embryo lysate than in wheat germ extract.
An RNA-dependent RNA polymerase activity in wheat germ extracts
Genetic evidence implicates an RNA-dependent RNA polymerase (RdRP) in PTGS triggered by transgenes expressing sense mRNA (S-PTGS; Dalmay et al. 2000; Mourrain et al. 2000). Plant RdRPs have been proposed to generate dsRNA from aberrantly expressed single-stranded RNA, thereby leading to the production of siRNAs that silence that RNA (Vaucheret et al. 2001). No direct biochemical evidence has yet been presented demonstrating that such a pathway is plausible.
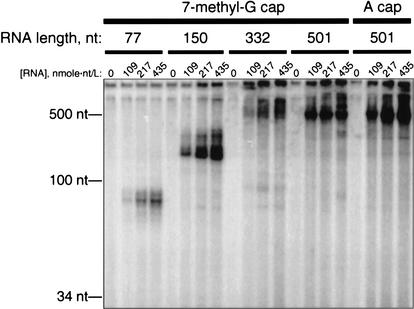
Wheat germ extracts contain an RdRP activity (Fig. 4). We incubated increasing concentrations of single-stranded RNA with the extract and ribonucleotide triphosphates, including α-32P-UTP. Single-stranded RNA ranging from 77 to 501 nt, either bearing a 7-methyl-G(5′)ppp(5′)G or an A(5′)ppp(5′) cap structure, all led to the incorporation of 32P into RNA with approximately the same length as the exogenous, nonradioactive single-stranded RNA (Fig. 4). These radioactive RNAs correspond to bona fide complementary RNA (cRNA) generated by an RdRP that copied the single-stranded RNA by initiating RNA synthesis at the extreme 3′ end of the exogenous template RNA (data not shown). In theory, these newly radioactive RNAs could have arisen by transfer of radiolabel to the input RNA itself. We observed this type of label transfer when similar experiments were performed using Drosophila embryo lysates, but not with wheat germ extract. Instead, the 32P-RNA represents newly synthesized cRNA produced by a wheat enzyme using exogenous single-stranded RNA as a template in the absence of an exogenous nucleic acid primer.
Figure 4.
Wheat germ extract contains an RdRP activity. Single-stranded RNA of the indicated size and cap structure was incubated in wheat germ extract for 3 h in the presence of ATP, CTP, GTP, and α-32P-UTP. The products of the reaction were analyzed by denaturing polyacrylamide gel electrophoresis.
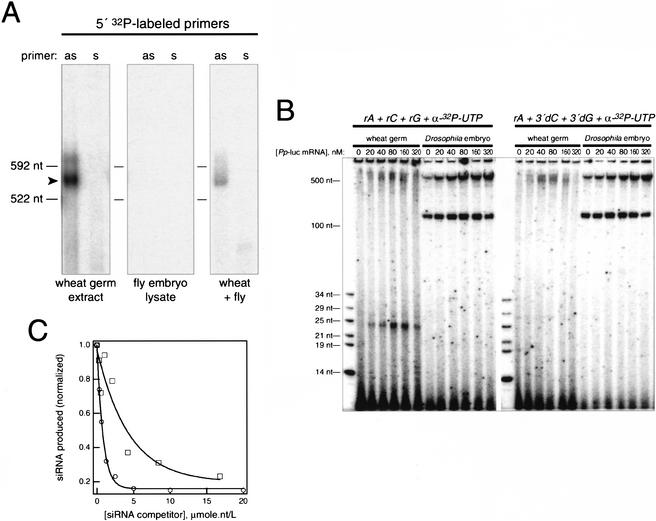
In addition to copying single-stranded RNA into approximately full-length cRNA, RdRPs have also been reported to extend primers, using single-stranded RNA as a template (e.g., Schiebel et al. 1998). We similarly find that the RdRP activity or activities in wheat germ extract could extend a 32P-radiolabeled primer (Fig. 5A), but only when the RNA primer was complementary (antisense) to the template RNA. Under identical conditions, we can detect no such primer-extension activity in lysates of syncitial blastoderm Drosophila embryos, despite earlier reports to the contrary (Lipardi et al. 2001). We can, however, detect the RNA-dependent, RNA-primer-extension activity when wheat and fly extracts are mixed (Fig. 5A). In neither Drosophila embryo lysate nor wheat germ extract can we detect primer extension of a single-stranded RNA template using a 21-nt siRNA duplex rather than a 21-nt antisense primer.
Figure 5.
Characterization of the wheat RdRP activity. (A) Wheat germ extract, but not Drosophila embryo lysate, contains an RdRP activity that can extend a primer. The arrowhead indicates the primer extension product produced when an antisense 21-nt RNA primer, but not a sense primer, was incubated in the wheat germ extract with a 592-nt single-stranded RNA. The primers correspond to nucleotides 511–532 of the RNA template. (B) RdRP-dependent production of small RNAs in wheat germ extract. Increasing concentrations of a 2.7-kb Photinus pyralis (Pp) luciferase mRNA triggered production of 32P-radiolabeled small RNAs in wheat germ extract when ribonucleotide triphosphates (including α-32P-UTP), but not when 3′-deoxy GTP and 3′-deoxy CTP were included in the reaction. (C) Production of newly synthesized small RNAs was more efficiently inhibited by a 25-nt synthetic siRNA duplex (open circles) than by a 21-nt synthetic siRNA duplex (open squares).
We envision that aberrant single-stranded RNA triggers silencing in plants when it serves as a template for the production of cRNA, generating dsRNA, which can then be cleaved by Dicer into siRNA duplexes. Our data suggest that such copying does not require primers, but is triggered merely by an exceptionally high concentration of single-stranded RNA. To test if high concentrations of single-stranded mRNA could lead to the production of siRNAs, we repeated the RdRP reactions using a 2.7-kb single-stranded firefly luciferase mRNA. Increasing concentrations of the mRNA were incubated in either wheat germ extract or Drosophila embryo lysate in the presence of ATP, CTP, GTP, and α-32P-UTP, and examined for the production of 21–25-nt radioactive RNAs. Figure 5B (left) shows that when the incubations were performed in wheat germ lysates, a single class of small RNA, ∼24 nt long, was produced with increasing concentrations of the exogenous, single-stranded template RNA. No such radioactive product was observed in Drosophila embryo lysates, but we note that these lysates contain endogenous UTP, which may preclude detection of 32P small RNAs. To test if the radiolabeled ∼24-nt products were generated by the de novo synthesis of RNA, we repeated the experiment, replacing CTP and GTP with 3′-deoxy CTP and 3′-deoxy GTP, inhibitors of RNA synthesis. In the presence of these inhibitors, no radioactive small RNAs were observed in the wheat reaction (Fig. 5B, right). Thus, single-stranded RNA can trigger in wheat germ extract the de novo synthesis of ∼24-nt small RNAs.
We do not detect the production of 21-nt RNAs in this assay. The assay should have detected such 21-nt small RNAs if they were present at 1/10 the concentration of the ∼24-mers, but we would be unlikely to detect them far below this threshold. Experiments with double-stranded RNA suggest that the 21-mers are produced in wheat at about 1/4 the rate of the 24–25-nt small RNAs (Fig. 2). Thus, the production of dsRNA by the RdRP activity may be coupled to the production of the longer class of small RNAs. We note that such coupling does not imply that production of ∼24-nt siRNAs from exogenous dsRNA requires the participation of an RdRP. We envision that dsRNA generated by RdRP copying of single-stranded RNA is preferentially processed by a wheat Dicer ortholog that produces long siRNAs, perhaps because the two proteins are physically linked.
Are the ∼24-nt RNAs that are synthesized in the RdRP reactions really the products of Dicer cleavage of dsRNA? Production of wheat 24–25-nt siRNAs from 32P-radiolabeled dsRNA is efficiently inhibited by synthetic siRNA duplexes; 25-nt synthetic siRNA duplexes are more potent inhibitors than 21-nt duplexes (Fig. 3B). We asked if production of the ∼24-nt small RNAs in the RdRP reactions was similarly inhibited by synthetic siRNA duplexes. Figure 5C shows that the production of ∼24-nt small RNAs in the RdRP reactions programmed with a 2.7-kb single-stranded RNA template was inhibited by synthetic siRNA duplexes. Like the production of 24–25-nt siRNAs from exogenous dsRNA, production of the de novo synthesized ∼24-mers was inhibited to a greater extent by 25-nt synthetic siRNA duplexes than by 21-nt duplexes (Fig. 5C). Half-maximal inhibition of small RNA production in the RdRP-dependent reactions occurred at roughly the same concentration of synthetic siRNA duplex as inhibition of the processing of 32P dsRNA (cf. Figs. 5C and 3B). We conclude that in wheat germ extract, exogenous single-stranded RNA provides the template for the synthesis of cRNA by an RdRP and that the resulting template-RNA:cRNA hybrid is then preferentially cleaved into ∼24-nt siRNAs by a Dicer-like enzyme.
miRNAs act as siRNAs in plants
In addition to siRNAs, another class of small RNAs, microRNAs (miRNAs), has been detected in plants (Llave et al. 2002a; Park et al. 2002; Reinhart et al. 2002). Like their animal counterparts, plant miRNAs are generated by a Dicer family member, CAF. miRNAs are encoded in stem–loop precursor RNAs that are cleaved by CAF into 21–24-nt single-stranded small RNAs (Park et al. 2002; Reinhart et al. 2002). Exogenous miRNA precursors were not faithfully processed into mature miRNAs in wheat germ extract (data not shown). Instead, in vitro transcribed pre-miRNAs were cleaved into small RNAs too long to correspond to authentic, mature miRNAs. Perhaps the Dicer ortholog responsible for miRNA maturation in wheat—presumably wheat CAF—is absent from wheat germ extracts. In Arabidopsis, CAF transcripts that encode a protein with a nuclear localization signal have been reported, suggesting that CAF protein may be nuclear (Jacobsen et al. 1999). Because wheat germ extracts are essentially cytoplasm, nuclear CAF might not be present in the extract.
Plant miRNAs differ from animal miRNAs in that there are corresponding mRNA sequences in the Arabidopsis and rice genomes with significant complementarity to miRNA sequences (Llave et al. 2002a,b; Reinhart et al. 2002; Rhoades et al. 2002). The high degree of complementarity between 14 recently analyzed plant miRNAs and specific families of developmentally important plant mRNAs led to the proposal that plant miRNAs direct developmentally controlled mRNA destruction (Rhoades et al. 2002). That is, after the plant miRNAs are generated by the cleavage of pre-miRNAs by CAF, they enter the RNAi pathway and function as siRNAs. In contrast, animal miRNAs are thought to act as translational repressors (for review, see Ruvkun 2001). An untested feature of this proposal is that an RNAi-like pathway in plants tolerates the three to four mismatches sometimes observed between an miRNA and its predicted mRNA target.
If plant miRNAs are endogenous mediators of RNAi, we reasoned that wheat germ extracts should contain miRNA-programmed complexes that specify endonucleolytic cleavage of corresponding target RNAs. In particular, miR165 has been proposed to down-regulate PHV and PHABULOSA (PHB) mRNA expression in Arabidopsis by an RNAi-like mechanism (Rhoades et al. 2002). PHV and PHB encode homeodomain-leucine zipper transcription factors implicated in the perception of radial position in the shoot tissues that give rise to leaves (McConnell and Barton 1998; McConnell et al. 2001). Dominant phv and phb mutations alter a single amino acid (glycine → glutamic acid) in the sterol/lipid-binding domain of the proteins, suggesting that the mutant phenotype results from a change in the function of PHV and PHB (McConnell and Barton 1998; McConnell et al. 2001). However, the discovery of plant miRNAs complementary to this site in PHV led to the suggestion that the molecular basis of the dominance is the persistence of PHV and PHB expression at developmental stages when these mRNAs are normally destroyed (Rhoades et al. 2002). This hypothesis is consistent with both the increased overall levels of PHB mRNA in the dominant mutant and the increased activity of a dominant mutant phb mRNA on the abaxial, rather than the adaxial, domain of the leaf primordium (McConnell and Barton 1998; McConnell et al. 2001).
miR165 or miR166 is present in wheat germ extracts (Fig. 6A). miR165 and miR166 differ by a single C-to-U transition that decreases the complementarity of miR166 to PHV and PHB by changing a G:C base pair to a G:U wobble. Rice (Oryza) is the sequenced genome most closely related to wheat. Although the rice genome encodes no miR165 homolog, it encodes six copies of miR166 (Reinhart et al. 2002). Because our Northern hybridization conditions cannot distinguish between miR165 and miR166, we refer to the endogenous wheat miRNA as miR165/166.
Figure 6.
miR165/166 in wheat germ extract. (A) A wheat ortholog of miR165 or miR166 is present in wheat germ extract. Quantitative Northern hybridization analysis using synthetic miR165 RNA concentration standards, antisense miR165 RNA, and total RNA prepared from 30 μL of wheat germ extract or Drosophila embryo lysate. (B) Quantitation of the data in A. Closed circles, synthetic miR165 standards; open circle, RNA extracted from 30 μL of wheat germ extract. The line shows a linear fit of the four highest concentration standards. (C) Schematic of the RNA targets, indicating the sequences of the miR165/166-complementary regions of wild-type PHV and mutant phv mRNAs, miR165, miR166, and the siRNA antisense strands used in Figure 7C.
To begin to test the hypothesis that plant miRNAs function to regulate target gene expression by an RNAi-like mechanism, we prepared target RNAs encoding a portion of the wild-type sequence of Arabidopsis PHV or the dominant G → A point mutation, which falls within the PHV sequences proposed to pair with miR165/166. The target RNAs and relevant miRNAs are shown in Figure 6C. 5′-radiolabeled target RNAs were incubated with wheat germ extract, then analyzed on a denaturing sequencing gel. In the absence of any other exogenous RNA, the wild-type PHV target RNA, but not the dominant G → A mutant, was efficiently cleaved within the region complementary to miR165/166 (Fig. 7A,B). This 21-nt region is identical in PHV and PHB, and a target RNA that contained sequence from the Arabidopsis PHB mRNA was also cleaved within the sequences complementary to miR165/166 upon incubation in the wheat germ extract (data not shown). In the RNAi pathway, a key feature of small RNA-directed target destruction is that pretreatment with the single-stranded nucleic acid-specific enzyme, micrococcal nuclease, abolishes RISC activity (Hammond et al. 2000). Cleavage of the PHV target RNA was likewise abolished by pretreatment of the extract with micrococcal nuclease (data not shown), consistent with the view that miR165/166 acts as a guide to direct target cleavage. The difference in cleavage rate between wild-type and mutant target RNAs, which differ only at a single nucleotide, was >14-fold (Fig. 7B). Thus, the resistance of the mutant phv RNA to cleavage by an endogenous RNAi-like nuclease can explain why the mutation is dominant.
Figure 7.
An endogenous wheat nuclease efficiently cleaves wild-type but not mutant PHV target RNAs. (A) When incubated in wheat germ extract, 5′-radiolabeled target RNA containing wild-type PHV sequences was cleaved within the PHV sequences complementary to miR165 and miR166. In contrast, a dominant G → A mutant target RNA was cleaved inefficiently. (B) Quantification of the data in A. (Circles) Wild-type PHV sequences; (squares) mutant sequences; (filled symbols) full-length target RNA; (open symbols) 5′ cleavage product. The difference in cleavage rates is ∼14-fold. (C) Analysis of PHV cleavage in an in vitro RNAi reaction programmed with siRNA duplexes and Drosophila embryo lysate. The identity of the antisense strand of the siRNA duplex and the RNA target used is indicated above the gel and described in Figure 6C.
Next, we analyzed cleavage of the PHV target RNA by various siRNAs in Drosophila embryo lysate (Fig. 6C). An siRNA with perfect complementarity to the site predicted to pair with miR165/166 and an siRNA duplex in which one strand had the sequence of miR165 or miR166 directed cleavage of the PHV target RNA, yielding the predicted 514-nt 5′ cleavage product (Fig. 7C). None of these three siRNAs efficiently cleaved the PHV mutant target (Fig. 7C). The failure of the miR165–siRNA duplex to cleave mutant PHV was a direct consequence of its reduced complementarity to the target RNA at position 6 (with respect to the 5′ end of the antisense siRNA strand), because an siRNA with perfect complementarity to the mutant sequence (Fig. 6B) efficiently cleaved the mutant RNA (Fig. 7C). The 5′ cleavage product produced in the siRNA-programmed RNAi reactions comigrated with that produced when the PHV target RNA was incubated in wheat germ extract without exogenous siRNA (Fig. 7C).
The simplest explanation for the sequence-specificity of the nuclease is that it is guided by miR165/166: cleavage requires a nucleic acid component, occurs at the same site on the PHV target RNA as directed by an siRNA duplex with the sequence of miR165 or miR166 in Drosophila embryo lysate, and, like the siRNA, is inefficient with the G → A mutant phv RNA. In the RNAi pathway, an siRNA-programmed endonuclease complex is called an RISC (Hammond et al. 2000). Our data suggest that wheat miR165/166 is in an RISC, supporting the proposal that plant miRNAs regulate expression of their mRNA targets by endogenous RNAi.
miR165/166 directs multiple rounds of target cleavage
Does the miR165/166-programmed RISC act as an enzyme? Quantitative Northern hybridization demonstrates that the wheat germ extract reactions contained 0.083 nM miR165/166 (Fig. 6B). The target RNA concentration in these reactions was 5 nM, and more than half the target RNA was destroyed in 80 min (Fig. 7A). Thus, each miR165/166 RNA directed cleavage of ∼30 target RNA molecules. Therefore, the miR165/166-programmed RISC is a multiple-turnover enzyme.
Discussion
We have shown here that wheat germ extracts recapitulate in vitro many aspects of RNA silencing in plants. Wheat germ extracts convert exogenous dsRNA into two distinct classes of small RNAs. Detailed analysis of these small RNAs indicates that they are bona fide siRNAs. Thus, plant siRNAs are derived directly from longer dsRNA, just as in animals. A particularly appealing hypothesis is that distinct Dicer-like enzymes generate the two functionally distinct classes of siRNAs. Cloned endogenous small RNAs from Arabidopsis likewise form two distinct length classes, whose 5′ ends might suggest they are made by distinct enzymes. An alternative view, that one or more Dicer-like enzymes may generate both classes of small RNAs, with the different lengths a byproduct of local sequence context, is not consistent with our observation that production of 24–25-nt RNAs in wheat germ extract was inhibited by synthetic siRNA duplexes, whereas ∼21-nt siRNA production was not. If the production of siRNAs is tightly coupled to the assembly of downstream effector complexes, then their production by different Dicer orthologs may ensure that the two classes of siRNAs function in different cellular pathways, as proposed by Baulcombe and colleagues (Hamilton et al. 2002).
A hallmark of PTGS in plants and RNAi in nematodes is the spreading of silencing signals along the length of the mRNA target. In plants, spreading occurs in both the 5′ and 3′ directions and requires the putative RdRP gene, SGS2. Spreading is observed even when silencing is initiated by a single siRNA sequence (Klahre et al. 2002). One hypothesis is that 5′ spreading is initiated by the antisense siRNA strand priming copying of the target mRNA by an RdRP, thereby producing dsRNA. 3′ spreading cannot be explained by such a mechanism. Both 5′ and 3′ spreading might instead be catalyzed by the conversion of mRNA fragments into dsRNA by an RdRP that initiates synthesis at the 3′ end of the two fragments generated when an RISC cleaves the target RNA. This dsRNA would then be cleaved by a Dicer-like enzyme to produce secondary siRNAs (Lipardi et al. 2001; Sijen et al. 2001). Such RNA synthesis would occur without the involvement of a primer. We have demonstrated here that exogenous single-stranded RNA is copied into cRNA in the extract by a wheat RdRP that acts without the aid of an exogenous primer. The resulting dsRNA is cleaved preferentially into the longer class of siRNAs, suggesting the RdRP is physically linked to a specific Dicer ortholog. We do not yet know the biochemical function of the 24–25-nt siRNAs generated in this reaction.
miRNAs function as siRNAs in plants
Our data show that miRNAs in plants function in much the same way that siRNA duplexes function in Drosophila and humans: as guides for an endonuclease complex. Each endonuclease complex can catalyze multiple rounds of target cleavage, indicating that the miRNA is not consumed in the reaction. Entry of a miRNA into a multiple-turnover RNAi enzyme complex is not unprecedented; in human cells, the miRNA let-7 is a component of an RISC, although the human genome does not appear to contain any mRNA sequences with sufficient complementarity to be cleaved by this RISC (Hutvágner and Zamore 2002). Like the plant miR165/166-programmed RISC, the human let-7-programmed RISC can catalyze multiple rounds of target cleavage.
Additional support for the idea that plant miRNAs direct cleavage of complementary mRNA targets comes from the work of Carrington and colleagues, who recently showed that a family of Arabidopsis mRNAs encoding SCARECROW-LIKE (SCL) transcription factors is cleaved by an RNAi-like process directed by miR171, an miRNA that is fully complementary to its mRNA targets, unlike miR165/166 (Llave et al. 2002b). Like wheat miR165/166, Arabidopsis miR171 appears to direct the endonucleolytic cleavage of its target mRNAs. In this respect, miR171 functions as if it were a single-stranded siRNA. Single-stranded siRNAs can trigger RNAi in both Drosophila and mammalian cell extracts and in vivo in HeLa cells (Martinez et al. 2002a; Schwarz et al. 2002), although much higher concentrations of single-stranded siRNA is required than for duplex (Schwarz et al. 2002). Furthermore, an individual human RISC contains only one strand of the exogenous siRNA duplex used to trigger RNAi (Martinez et al. 2002a).
The observation that, in Drosophila embryo lysate, an siRNA with the sequence of miR165, which contains three mismatches with its target mRNA, is at least as potent as an siRNA with perfect complementarity to the same target sequence, demonstrates that mismatches per se do not block target cleavage. Rather, the specific position and sequence of siRNA:target RNA mismatches determine if they permit or disrupt RNAi. Our data also suggest that miRNAs in plants evolved to optimize cleavage efficiency rather than maximize complementarity to their targets. We predict that three or four mismatches between an miRNA (or the guide strand of an siRNA duplex) and its target RNA, properly placed so as to still permit mRNA cleavage, will facilitate the release of cleaved target RNA from the RISC complex, thereby increasing the rate of enzyme turnover.
miRNA function and the spread of silencing signals along a silenced sequence
Spreading of silencing signals along the length of a silenced mRNA sequence is a common feature of plant RNA silencing. Because miRNAs act as siRNAs, one might anticipate that they would also elicit spreading. However, miRNA-induced spreading is not consistent with the genetics of the PHV and PHB mutants; the very existence of a dominant PHV mutant excludes both 5′ and 3′ spreading. Spreading of the silencing signal—that is, the generation of new siRNAs 5′ or 3′ to the site of initial target cleavage—would produce siRNAs containing sequences common to both the wild-type and mutant PHV mRNAs. If such siRNAs were generated, they would direct destruction of the mutant PHV mRNA. In such a case, the PHV mutant could only have been recovered as a recessive, not a dominant allele. Genetic studies (McConnell et al. 2001) show that endonucleolytic cleavage of target RNAs by miRNA-directed RISC complexes does not trigger spreading in plants. This remains true even when the miRNA is the perfect complement of its mRNA target (Llave et al. 2002b).
How, then, can the well-documented spreading phenomenon observed for S-PTGS be reconciled with the absence of spreading in miRNA-directed target cleavage? Perhaps plants contain two separate mechanisms for target mRNA destruction. Endogenous mRNAs might be regulated by endonucleolytic cleavage directed by miRNA-programmed RISC complexes, whereas exogenous silencing triggers, such as transgenes or viruses, might initiate successive cycles of siRNA-primed, RdRP-catalyzed dsRNA synthesis, followed by cleavage of the dsRNA into siRNAs by Dicer-like enzymes, a mechanism termed random degradative PCR (Lipardi et al. 2001). RISC complexes would play no role in the execution of target RNAs in this cycle. We have not been able to test this idea biochemically, because we have as yet been unable to target an mRNA for degradation in wheat germ extract by adding corresponding exogenous dsRNA or siRNAs. However, the observation that a single siRNA sequence can trigger 3′ spreading (Klahre et al. 2002) is difficult to reconcile with a priming mechanism. Intriguingly, VIGS-mediated RNA silencing of endogenous genes is not associated with spreading of silencing into regions of the target sequence 5′ or 3′ to the initial silencing trigger (Vaistij et al. 2002), although such silencing clearly must involve siRNAs derived from viral dsRNA, not endogenous miRNAs.
An alternative hypothesis is that the absolute concentration of an RNA target might determine if the 5′ and 3′ cleavage fragments generated by target cleavage are converted into dsRNA by an RdRP. Only when the products of RISC-mediated target cleavage accumulate to a sufficiently high concentration would they serve as substrates for the RdRP and consequently trigger spreading. Experiments with polygalacturonase-silenced tomatoes support this view (Han and Grierson 2002). In these plants, siRNAs were produced from the silencing-inducing transgene but not the corresponding silenced endogene. The siRNAs were preferentially produced from the 3′ end of the transgene, consistent with the idea that plant RdRPs act without aid of a primer. Furthermore, these authors detected mRNA degradation products consistent with endonucleolytic cleavage of the targeted polygalacturonase endogene. Thus, RISC-mediated cleavage per se does not appear to trigger spreading along the target RNA sequence. More likely, the endonucleolytic cleavage of transgenic mRNA produces a sufficiently high concentration of mRNA fragments to recruit an RdRP, resulting in the production of siRNAs from the 3′ cleavage product. miRNA-directed cleavage of natural plant regulatory targets would not lead to spreading, because endogenous mRNA targets are not present at sufficiently high concentrations to recruit the RdRP. This model predicts that the putative RdRP SGS2 (SDE1) required for PTGS, will not be required for miRNA-directed destruction of endogenous mRNA targets. In fact, no developmental abnormalities have been reported for SGS2 mutants (Mourrain et al. 2000), including mutations likely to be strongly hypomorphic or functionally null (Dalmay et al. 2000), suggesting that plants lacking SGS2 protein have normal miRNA biogenesis and function.
Materials and methods
Lysate preparation
Fly embryo lysates were prepared as previously described (Tuschl et al. 1999). Wheat germ extracts were prepared from frozen or vacuum-packed raw wheat germ (e.g., Fearn Nature Fresh Raw Wheat Germ, Bread and Circus) as described (Erickson and Blobel 1983). The extract was centrifuged at 14,500g at 4°C for 25 min; the supernatant was then frozen in aliquots in liquid nitrogen and stored at −80°C. For cauliflower extract, the outer layer of fresh cauliflower (Shaws Supermarket) was harvested with a razor blade and ground to a powder under liquid nitrogen in a mortar and pestle, then homogenized with 3 mL of 1× lysis buffer (100 mM potassium acetate, 30 mM HEPES-KOH at pH 7.4, 2 mM magnesium acetate) containing 5 mM dithiothreitol (DTT) and 1 mg/mL Pefabloc SC (Boehringer Mannheim) per gram of plant tissue. The extract was centrifuged, and the supernatant was stored as described for the Drosophila embryo lysate.
Analysis of dsRNA processing
For analysis of dsRNA processing, 5 nM internally α-32P-UTP-labeled dsRNA was incubated in a 10-μL reaction containing 5 μL of Drosophila embryo lysate (Tuschl et al. 1999) or wheat germ extract, 100 μM GTP, 500 μM ATP, 10 mM creatine phosphate, 10 μg/mL creatine phosphokinase, 5 mM DTT, and 0.1 U/μL RNasin (Promega) at 25°C for 3 h. Reactions were stopped by the addition of 2× proteinase K buffer [200 mM Tris-HCl at pH 7.5, 25 mM EDTA, 300 mM NaCl, 2% (w/v) sodium dodecyl sulfate] and deproteinized with ∼2 mg/mL proteinase K at 65°C for 15 min. Products were precipitated with 3 volumes cold ethanol and analyzed by electrophoresis in a 15% polyacrylamide sequencing gel.
Gel filtration and RNAse protection
Internally α-32P-UTP-labeled dsRNAs were incubated in wheat germ extract, then deproteinized at room temperature with proteinase K (1 h) and RNA-precipitated with 3 volumes of cold ethanol. The RNA was resuspended in 1× lysis buffer and analyzed by gel filtration as described (Nykänen et al. 2001). For RNase protection, the RNA products of a 10-μL wheat germ extract reaction were deproteinized at room temperature and analyzed by RNAse protection essentially as described (Sambrook et al. 1989). Briefly, the siRNA pellets were dissolved in 10 μL of RNAse digestion buffer (300 mM NaCl, 10 mM Tris-HCl at pH 7.4, and 5 mM EDTA at pH 7.5) containing 10 mM β-glycerophosphate, 5 mM ATP, 0–6.6 U of RNAse A, and 0–1.1 U of RNAse T1. For control experiments, 5′-32P-radiolabeled synthetic, double-stranded siRNAs were mixed with the products of a wheat germ reaction performed with unlabeled dsRNA and coprecipitated with 3 volumes of cold ethanol. RNAse protection was at 25°C for 1 h, stopped by adding 0.6 μL of 10% SDS and 0.3 μL of 20 mg/mL proteinase K, then incubated at 25°C for 1 h. The reactions were then adjusted to 200 μL with 2× PK buffer containing 0.2 mg/mL Glycogen (Roche), extracted with an equal volume of phenol/chloroform/isoamylalcohol (25:24:1; v/v/v), precipitated with 3 volumes of cold ethanol, and analyzed in a 15% sequencing polyacrylamide gel.
Synthetic siRNAs used as inhibitors
The 21-nt siRNA inhibitor comprised CGUACGCGGAAUAC UUCGA(5-Iodo-U)U annealed with UCGAAGUAUUCCGC GUACGUG; the 25-mer comprised AUCACGUACGCGGAA UACUUCGA(5-Iodo-U)U annealed with UCGAAGUAUUCC GCGUACGUGAUUG. The 5-Iodo-U nucleotides were included to facilitate studies not presented here, and we have no evidence they enhance the effectiveness of the siRNAs as inhibitors.
Analysis of RdRP Activity
Assays were performed in a final volume of 10 μL containing 5 μL of lysate, 100 μM GTP, 100 μM CTP, 500 μM ATP, 20 μM UTP, 5 μCi of α-32P-UTP (25 Ci/mmole), 10 mM creatine phosphate, 10 μg/mL creatine phosphokinase, 5 mM DTT, 0.2 U/μL Super-RNasin (Ambion), and 7-methyl-G- or A-capped RNAs. After incubation at 25°C for 3 h, the reaction was deproteinized with proteinase K in 200 μL of 2× proteinase K buffer at 65°C for 15 min. After phenol/chloroform/isoamylalcohol extraction, the aqueous phase was precipitated with 3 volumes of cold ethanol, resuspended in 10 μL of 2× formamide loading buffer as described (Sambrook et al. 1989), and resolved on 10% or 15% polyacrylamide sequencing gels. For primed assays, capped RNAs were preincubated with single-stranded 21-nt RNA primers or siRNA duplexes at room temperature for 10 min before the remaining reaction components were added.
Arabidopsis PHV, PHB, and mutant PHV target RNAs
Arabidopsis PHV and PHB cDNA sequences containing the miR165/166 complementary sequences were amplified from an Arabidopsis flower cDNA library (CD4-6) by polymerase chain reaction (PCR) using the following primer pairs: 5′-PHV primer, GCGTAATACGACTCACTATAGGCGCCGGAACAAGTTG AAG, and 3′-PHV primer, GACAGTCACGGAACCAAGATG; or 5′-PHB primer, GCGTAATACGACTCACTATAGGTGA GTCTGTGGTCGTGAGTG, and 3′-PHB primer, GCTGCT GCTAAAGTCGTAGGA. The Arabidopsis G → A mutant phv template was initially amplified using the 5′-PHV primer and CCACTGCAGTTGCGTGAAACAGCTACGATACCAAT AGAATCCGGATCAGGCTTCATCCC. This PCR product was diluted 100-fold, then reamplified with the 5′-PHV primer and GACAGTCACGGAACCAAGATGGACGATCTTTGAG GATTTCAGCGACCTTCATGGGTTCTAAACTCACGAGG CCACAGGCACGTGCTGCTATTCCACTGCAGTTGCGTG AAACAGC. In vitro RNA transcription and cap labeling were as described (Tuschl et al. 1999; Zamore et al. 2000).
In vitro RNAi in fly embryo lysate and wheat germ extract
For RNAi in Drosophila embryo lysate, four siRNA duplexes were chemically synthesized (Dharmacon), annealed, and incubated in a standard RNAi reaction (Zamore et al. 2000). The sequences of siRNAs (sense and antisense strands) corresponding to miR165, miR166, PHV, and mutant phv target positions were miR165, UCGGACCAGGCUUCAUCCCCC and GG GAUGAAGCCUGGUCCGAGG; miR166, UCGGACCAGG CUUCAUUCCCC and GGAAUGAAGCCUGGUCCGAGA; PHV, CCGGACCAGGCUUCAUCCCAA and GGGAUGAAGC CUGGUCCGGAU; and mutant phv, CCGGAUCAGGCUUCA UCCCAA and GGGAUGAAGCCUGAUCCGGAU. Wheat germ extract target cleavage reactions were as standard Drosophila in vitro RNAi reactions, except that no exogenous siRNAs were added.
Total RNA isolation and Northern analysis
Total RNA was isolated from lysates, and Northern analysis was performed as described (Hutvágner and Zamore 2002). 5′-32P-radioalabeled synthetic miR165 antisense siRNA (above) was used as probe.
Acknowledgments
This work would not have been possible without the generosity of Elisabet C. Mandon and Reid Gilmore, who initially donated wheat germ extract and then taught us how to prepare our own. We also thank Hervé Vaucheret and the members of the Zamore lab for support, ideas, and comments on the manuscript; David Baulcombe for sharing data prior to publication; and Matthew Rhoades for help with sequence analysis. P.D.Z. is a Pew Scholar in the Biomedical Sciences and a W.M. Keck Foundation Young Scholar in Medical Research. This work was supported in part by a grant to P.D.Z. from the National Institutes of Health (GM62862-01).
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL phillip.zamore@umassmed.edu; FAX (508) 856-2003.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.1048103.
References
- Beclin C, Boutet S, Waterhouse P, Vaucheret H. A branched pathway for transgene-induced RNA silencing in plants. Curr Biol. 2002;12:684–688. doi: 10.1016/s0960-9822(02)00792-3. [DOI] [PubMed] [Google Scholar]
- Bernstein E, Caudy AA, Hammond SM, Hannon GJ. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature. 2001;409:363–366. doi: 10.1038/35053110. [DOI] [PubMed] [Google Scholar]
- Billy E, Brondani V, Zhang H, Muller U, Filipowicz W. Specific interference with gene expression induced by long, double-stranded RNA in mouse embryonal teratocarcinoma cell lines. Proc Natl Acad Sci. 2001;98:14428–14433. doi: 10.1073/pnas.261562698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Celotto AM, Graveley BR. Exon-specific RNAi: A tool for dissecting the functional relevance of alternative splicing. RNA. 2002;8:8–24. doi: 10.1017/s1355838202021064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu Y-L, Rana TM. RNAi in human cells: Basic structural and functional features of small interfering RNA. Mol Cell. 2002;10:549–561. doi: 10.1016/s1097-2765(02)00652-4. [DOI] [PubMed] [Google Scholar]
- Cogoni C, Macino G. Gene silencing in Neurospora crassa requires a protein homologous to RNA-dependent RNA polymerase. Nature. 1999;399:166–169. doi: 10.1038/20215. [DOI] [PubMed] [Google Scholar]
- Dalmay T, Hamilton A, Rudd S, Angell S, Baulcombe DC. An RNA-dependent RNA polymerase gene in Arabidopsis is required for posttranscriptional gene silencing mediated by a transgene but not by a virus. Cell. 2000;101:543–553. doi: 10.1016/s0092-8674(00)80864-8. [DOI] [PubMed] [Google Scholar]
- Djikeng A, Shi H, Tschudi C, Ullu E. RNA interference in Trypanosoma brucei: Cloning of small interfering RNAs provides evidence for retroposon-derived 24–26-nucleotide RNAs. RNA. 2001;7:1522–1530. [PMC free article] [PubMed] [Google Scholar]
- Elbashir SM, Harborth J, Lendeckel W, Yalcin A, Weber K, Tuschl T. Duplexes of 21-nucleotide RNAs mediate RNA interference in mammalian cell culture. Nature. 2001a;411:494–498. doi: 10.1038/35078107. [DOI] [PubMed] [Google Scholar]
- Elbashir SM, Lendeckel W, Tuschl T. RNA interference is mediated by 21- and 22-nucleotide RNAs. Genes & Dev. 2001b;15:188–200. doi: 10.1101/gad.862301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elbashir SM, Martinez J, Patkaniowska A, Lendeckel W, Tuschl T. Functional anatomy of siRNAs for mediating efficient RNAi in Drosophila melanogaster embryo lysate. EMBO J. 2001c;20:6877–6888. doi: 10.1093/emboj/20.23.6877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erickson AH, Blobel G. Cell-free translation of messenger RNA in a wheat germ system. Methods Enzymol. 1983;96:38–50. doi: 10.1016/s0076-6879(83)96007-x. [DOI] [PubMed] [Google Scholar]
- Fagard M, Boutet S, Morel J-B, Bellini C, Vaucheret H. AGO1, QDE-2, and RDE-1 are related proteins required for post-transcriptional gene silencing in plants, quelling in fungi, and RNA interference in animals. Proc Natl Acad Sci. 2000;97:11650–11654. doi: 10.1073/pnas.200217597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grishok A, Pasquinelli AE, Conte D, Li N, Parrish S, Ha I, Baillie DL, Fire A, Ruvkun G, Mello CC. Genes and mechanisms related to RNA interference regulate expression of the small temporal RNAs that control C. elegans developmental timing. Cell. 2001;106:23–34. doi: 10.1016/s0092-8674(01)00431-7. [DOI] [PubMed] [Google Scholar]
- Hamilton AJ, Baulcombe DC. A species of small antisense RNA in posttranscriptional gene silencing in plants. Science. 1999;286:950–952. doi: 10.1126/science.286.5441.950. [DOI] [PubMed] [Google Scholar]
- Hamilton A, Voinnet O, Chappell L, Baulcombe D. Two classes of short interfering RNA in RNA silencing. EMBO J. 2002;21:4671–4679. doi: 10.1093/emboj/cdf464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammond SM, Bernstein E, Beach D, Hannon GJ. An RNA-directed nuclease mediates post-transcriptional gene silencing in Drosophila cells. Nature. 2000;404:293–296. doi: 10.1038/35005107. [DOI] [PubMed] [Google Scholar]
- Hammond SM, Boettcher S, Caudy AA, Kobayashi R, Hannon GJ. Argonaute2, a link between genetic and biochemical analyses of RNAi. Science. 2001;293:1146–1150. doi: 10.1126/science.1064023. [DOI] [PubMed] [Google Scholar]
- Han Y, Grierson D. Relationship between small antisense RNAs and aberrant RNAs associated with sense transgene mediated gene silencing in tomato. Plant J. 2002;29:509–519. doi: 10.1046/j.1365-313x.2002.01236.x. [DOI] [PubMed] [Google Scholar]
- Hannon GJ. RNA interference. Nature. 2002;418:244–251. doi: 10.1038/418244a. [DOI] [PubMed] [Google Scholar]
- Holen T, Amarzguioui M, Wiiger MT, Babaie E, Prydz H. Positional effects of short interfering RNAs targeting the human coagulation trigger Tissue Factor. Nucleic Acids Res. 2002;30:1757–1766. doi: 10.1093/nar/30.8.1757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutvágner G, Zamore PD. A microRNA in a multiple-turnover RNAi enzyme complex. Science. 2002;297:2056–2060. doi: 10.1126/science.1073827. [DOI] [PubMed] [Google Scholar]
- Hutvágner G, McLachlan J, Pasquinelli AE, Balint É, Tuschl T, Zamore PD. A cellular function for the RNA-interference enzyme Dicer in the maturation of the let-7 small temporal RNA. Science. 2001;293:834–838. doi: 10.1126/science.1062961. [DOI] [PubMed] [Google Scholar]
- Jacobsen SE, Running MP, Meyerowitz EM. Disruption of an RNA helicase/RNase III gene in Arabidopsis causes unregulated cell division in floral meristems. Development. 1999;126:5231–5243. doi: 10.1242/dev.126.23.5231. [DOI] [PubMed] [Google Scholar]
- Kennerdell JR, Yamaguchi S, Carthew RW. RNAi is activated during Drosophila oocyte maturation in a manner dependent on aubergine and spindle-E. Genes & Dev. 2002;16:1884–1889. doi: 10.1101/gad.990802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ketting RF, Fischer SE, Bernstein E, Sijen T, Hannon GJ, Plasterk RH. Dicer functions in RNA interference and in synthesis of small RNA involved in developmental timing in C. elegans. Genes & Dev. 2001;15:2654–2659. doi: 10.1101/gad.927801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klahre U, Crete P, Leuenberger SA, Iglesias VA, Meins F., Jr High molecular weight RNAs and small interfering RNAs induce systemic posttranscriptional gene silencing in plants. Proc Natl Acad Sci. 2002;99:11981–11986. doi: 10.1073/pnas.182204199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kooter JM, Matzke MA, Meyer P. Listening to the silent genes: Transgene silencing, gene regulation and pathogen control. Trends Plant Sci. 1999;4:340–347. doi: 10.1016/s1360-1385(99)01467-3. [DOI] [PubMed] [Google Scholar]
- Lagos-Quintana M, Rauhut R, Lendeckel W, Tuschl T. Identification of novel genes coding for small expressed RNAs. Science. 2001;294:853–858. doi: 10.1126/science.1064921. [DOI] [PubMed] [Google Scholar]
- Lagos-Quintana M, Rauhut R, Yalcin A, Meyer J, Lendeckel W, Tuschl T. Identification of tissue-specific microRNAs from mouse. Curr Biol. 2002;12:735–739. doi: 10.1016/s0960-9822(02)00809-6. [DOI] [PubMed] [Google Scholar]
- Lau NC, Lim LP, Weinstein EG, Bartel DP. An abundant class of tiny RNAs with probable regulatory roles in Caenorhabditis elegans. Science. 2001;294:858–862. doi: 10.1126/science.1065062. [DOI] [PubMed] [Google Scholar]
- Lee RC, Ambros V. An extensive class of small RNAs in Caenorhabditis elegans. Science. 2001;294:862–864. doi: 10.1126/science.1065329. [DOI] [PubMed] [Google Scholar]
- Lee RC, Feinbaum RC, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–854. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- Lee Y, Jeon K, Lee JT, Kim S, Kim VN. MicroRNA maturation: Stepwise processing and subcellular localization. EMBO J. 2002;21:4663–4670. doi: 10.1093/emboj/cdf476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li WX, Ding SW. Viral suppressors of RNA silencing. Curr Opin Biotechnol. 2001;12:150–154. doi: 10.1016/s0958-1669(00)00190-7. [DOI] [PubMed] [Google Scholar]
- Lipardi C, Wei Q, Paterson BM. RNAi as random degradative PCR. siRNA primers convert mRNA into dsRNAs that are degraded to generate new siRNAs. Cell. 2001;107:297–307. doi: 10.1016/s0092-8674(01)00537-2. [DOI] [PubMed] [Google Scholar]
- Llave C, Kasschau KD, Rector MA, Carrington JC. Endogenous and silencing-associated small RNAs in plants. Plant Cell. 2002a;14:1605–1619. doi: 10.1105/tpc.003210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llave C, Xie Z, Kasschau KD, Carrington JC. Cleavage of Scarecrow-Like mRNA targets directed by a class of Arabidopsis miRNA. Science. 2002b;297:2053–2056. doi: 10.1126/science.1076311. [DOI] [PubMed] [Google Scholar]
- Mallory AC, Reinhart BJ, Bartel D, Vance VB, Bowman LH. A viral suppressor of RNA silencing differentially regulates the accumulation of short interfering RNAs and micro-RNAs in tobacco. Proc Natl Acad Sci. 2002;99:15228–15233. doi: 10.1073/pnas.232434999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martens H, Novotny J, Oberstrass J, Steck TL, Postlethwait P, Nellen W. RNAi in Dictyostelium: The role of RNA-directed RNA polymerases and double-stranded RNase. Mol Biol Cell. 2002;13:445–453. doi: 10.1091/mbc.01-04-0211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez J, Patkaniowska A, Urlaub H, Lührmann R, Tuschl T. Single-stranded antisense siRNAs guide target RNA cleavage in RNAi. Cell. 2002a;110:563. doi: 10.1016/s0092-8674(02)00908-x. [DOI] [PubMed] [Google Scholar]
- Martinez LA, Naguibneva I, Lehrmann H, Vervisch A, Tchénio T, Lozano G, Harel-Bellan A. Synthetic small inhibiting RNAs: Efficient tools to inactivate oncogenic mutations and restore p53 pathways. Proc Natl Acad Sci. 2002b;99:14849–14854. doi: 10.1073/pnas.222406899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matzke MA, Matzke AJ, Pruss GJ, Vance VB. RNA-based silencing strategies in plants. Curr Opin Genet Dev. 2001;11:221–227. doi: 10.1016/s0959-437x(00)00183-0. [DOI] [PubMed] [Google Scholar]
- McConnell JR, Barton MK. Leaf polarity and meristem formation in Arabidopsis. Development. 1998;125:2935–2942. doi: 10.1242/dev.125.15.2935. [DOI] [PubMed] [Google Scholar]
- McConnell JR, Emery J, Eshed Y, Bao N, Bowman J, Barton MK. Role of PHABULOSA and PHAVOLUTA in determining radial patterning in shoots. Nature. 2001;411:709–713. doi: 10.1038/35079635. [DOI] [PubMed] [Google Scholar]
- Mourelatos Z, Dostie J, Paushkin S, Sharma AK, Charroux B, Abel L, Rappsilber J, Mann M, Dreyfuss G. miRNPs: A novel class of ribonucleoproteins containing numerous microRNAs. Genes & Dev. 2002;16:720–728. doi: 10.1101/gad.974702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mourrain P, Beclin C, Elmayan T, Feuerbach F, Godon C, Morel JB, Jouette D, Lacombe AM, Nikic S, Picault N, et al. Arabidopsis SGS2 and SGS3 genes are required for posttranscriptional gene silencing and natural virus resistance. Cell. 2000;101:533–542. doi: 10.1016/s0092-8674(00)80863-6. [DOI] [PubMed] [Google Scholar]
- Nykänen A, Haley B, Zamore PD. ATP requirements and small interfering RNA structure in the RNA interference pathway. Cell. 2001;107:309–321. doi: 10.1016/s0092-8674(01)00547-5. [DOI] [PubMed] [Google Scholar]
- Pal-Bhadra M, Bhadra U, Birchler JA. RNAi related mechanisms affect both transcriptional and posttranscriptional transgene silencing in Drosophila. Mol Cell. 2002;9:315–327. doi: 10.1016/s1097-2765(02)00440-9. [DOI] [PubMed] [Google Scholar]
- Park W, Li J, Song R, Messing J, Chen X. CARPEL FACTORY, a Dicer homolog, and HEN1, a novel protein, act in microRNA metabolism in Arabidopsis thaliana. Curr Biol. 2002;12:1484–1495. doi: 10.1016/s0960-9822(02)01017-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parrish S, Fire A. Distinct roles for RDE-1 and RDE-4 during RNA interference in Caenorhabditis elegans. RNA. 2001;7:1397–1402. [PMC free article] [PubMed] [Google Scholar]
- Parrish S, Fleenor J, Xu S, Mello C, Fire A. Functional anatomy of a dsRNA trigger. Differential requirement for the two trigger strands in RNA interference. Mol Cell. 2000;6:1077–1087. doi: 10.1016/s1097-2765(00)00106-4. [DOI] [PubMed] [Google Scholar]
- Pasquinelli AE, Reinhart BJ, Slack F, Martindale MQ, Kuroda MI, Maller B, Hayward DC, Ball EE, Degnan B, Muller P, et al. Conservation of the sequence and temporal expression of let-7 heterochronic regulatory RNA. Nature. 2000;408:86–89. doi: 10.1038/35040556. [DOI] [PubMed] [Google Scholar]
- Plasterk RH. RNA silencing: The genome’s immune system. Science. 2002;296:1263–1265. doi: 10.1126/science.1072148. [DOI] [PubMed] [Google Scholar]
- Reinhart BJ, Slack FJ, Basson M, Pasquinelli AE, Bettinger JC, Rougvie AE, Horvitz HR, Ruvkun G. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature. 2000;403:901–906. doi: 10.1038/35002607. [DOI] [PubMed] [Google Scholar]
- Reinhart BJ, Weinstein EG, Rhoades MW, Bartel B, Bartel DP. MicroRNAs in plants. Genes & Dev. 2002;16:1616–1626. doi: 10.1101/gad.1004402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhoades MW, Reinhart BJ, Lim LP, Burge CB, Bartel B, Bartel DP. Prediction of plant microRNA targets. Cell. 2002;110:513–520. doi: 10.1016/s0092-8674(02)00863-2. [DOI] [PubMed] [Google Scholar]
- Roberts BE, Paterson BM. Efficient translation of tobacco mosaic virus RNA and rabbit globin 9S RNA in a cell-free system from commercial wheat germ. Proc Natl Acad Sci. 1973;70:2330–2334. doi: 10.1073/pnas.70.8.2330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roignant, J.-Y., Carré, C., Mugat, B., Szymczak, D., Lepesant, J.-A., and Antoniewski, C. 2003. Absence of transitive and systemic pathways allows cell-specific and isoform-specific RNAi in Drosophila. RNA (In press). [DOI] [PMC free article] [PubMed]
- Ruvkun G. Molecular biology. Glimpses of a tiny RNA world. Science. 2001;294:797–799. doi: 10.1126/science.1066315. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch E, Maniatis T. Molecular cloning: A laboratory manual. 2nd ed. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Schiebel W, Pelissier T, Riedel L, Thalmeir S, Schiebel R, Kempe D, Lottspeich F, Sanger HL, Wassenegger M. Isolation of an RNA-directed RNA polymerase-specific cDNA clone from tomato. Plant Cell. 1998;10:2087–2101. doi: 10.1105/tpc.10.12.2087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz DS, Hutvágner G, Haley B, Zamore PD. Evidence that siRNAs function as guides, not primers, in the Drosophila and human RNAi pathways. Mol Cell. 2002;10:537–548. doi: 10.1016/s1097-2765(02)00651-2. [DOI] [PubMed] [Google Scholar]
- Sijen T, Fleenor J, Simmer F, Thijssen KL, Parrish S, Timmons L, Plasterk RH, Fire A. On the role of RNA amplification in dsRNA-triggered gene silencing. Cell. 2001;107:465–476. doi: 10.1016/s0092-8674(01)00576-1. [DOI] [PubMed] [Google Scholar]
- Smardon A, Spoerke J, Stacey S, Klein M, Mackin N, Maine E. EGO-1 is related to RNA-directed RNA polymerase and functions in germ-line development and RNA interference in C. elegans. Curr Biol. 2000;10:169–178. doi: 10.1016/s0960-9822(00)00323-7. [DOI] [PubMed] [Google Scholar]
- Tabara H, Sarkissian M, Kelly WG, Fleenor J, Grishok A, Timmons L, Fire A, Mello CC. The rde-1 gene, RNA interference, and transposon silencing in C. elegans. Cell. 1999;99:123–132. doi: 10.1016/s0092-8674(00)81644-x. [DOI] [PubMed] [Google Scholar]
- Tijsterman M, Ketting RF, Okihara KL, Sijen T, Plasterk RH. RNA helicase MUT-14-dependent gene silencing triggered in C. elegans by short antisense RNAs. Science. 2002;295:694–697. doi: 10.1126/science.1067534. [DOI] [PubMed] [Google Scholar]
- Tuschl T, Zamore PD, Lehmann R, Bartel DP, Sharp PA. Targeted mRNA degradation by double-stranded RNA in vitro. Genes & Dev. 1999;13:3191–3197. doi: 10.1101/gad.13.24.3191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaistij FE, Jones L, Baulcombe DC. Spreading of RNA targeting and DNA methylation in RNA silencing requires transcription of the target gene and a putative RNA-dependent RNA polymerase. Plant Cell. 2002;14:857–867. doi: 10.1105/tpc.010480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaucheret H, Beclin C, Fagard M. Post-transcriptional gene silencing in plants. J Cell Sci. 2001;114:3083–3091. doi: 10.1242/jcs.114.17.3083. [DOI] [PubMed] [Google Scholar]
- Waterhouse PM, Wang MB, Lough T. Gene silencing as an adaptive defence against viruses. Nature. 2001;411:834–842. doi: 10.1038/35081168. [DOI] [PubMed] [Google Scholar]
- Williams RW, Rubin GM. ARGONAUTE1 is required for efficient RNA interference in Drosophila embryos. Proc Natl Acad Sci. 2002;99:6889–6894. doi: 10.1073/pnas.072190799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zamore PD, Tuschl T, Sharp PA, Bartel DP. RNAi: Double-stranded RNA directs the ATP-dependent cleavage of mRNA at 21 to 23 nucleotide intervals. Cell. 2000;101:25–33. doi: 10.1016/S0092-8674(00)80620-0. [DOI] [PubMed] [Google Scholar]