Abstract
Plants possess several photoreceptors to sense the light environment. In Arabidopsis cryptochromes and phytochromes play roles in photomorphogenesis and in the light input pathways that synchronize the circadian clock with the external world. We have identified SRR1 (sensitivity to red light reduced), a gene that plays an important role in phytochrome B (phyB)-mediated light signaling. The recessive srr1 null allele and phyB mutants display a number of similar phenotypes indicating that SRR1 is required for normal phyB signaling. Genetic analysis suggests that SRR1 works both in the phyB pathway but also independently of phyB. srr1 mutants are affected in multiple outputs of the circadian clock in continuous light conditions, including leaf movement and expression of the clock components, CCA1 and TOC1. Clock-regulated gene expression is also impaired during day–night cycles and in constant darkness. The circadian phenotypes of srr1 mutants in all three conditions suggest that SRR1 activity is required for normal oscillator function. The SRR1 gene was identified and shown to code for a protein conserved in numerous eukaryotes including mammals and flies, implicating a conserved role for this protein in both the animal and plant kingdoms.
Keywords: Circadian rhythm, light-signaling, phytochrome B, Arabidopsis thaliana
Most organisms are sensitive to various aspects of their light environment. To anticipate the day–night cycles, several activities are under the control of the circadian clock. Clock-regulated processes are physiologically diverse, including the regulation of body temperature in mammals, conidiation in fungi, petal opening in plants, and eclosion in flies. For example, it is estimated that ∼5% of all Arabidopsis genes are clock controlled at the transcriptional level, which could account for diurnal regulation of a large number of metabolic pathways (Harmer et al. 2000; Schaffer et al. 2001). Global expression profiling in animals has also revealed a large number of circadian-regulated genes (e.g., see McDonald and Rosbash 2001; Storch et al. 2002). In a few cases it has been demonstrated that a functional clock enhances the fitness of the organism, suggesting an evolutionary advantage for anticipating dawn and dusk (Green et al. 2002). In the simplest model, the circadian clock is composed of three main constituents: an input pathway that conveys resetting information to the oscillator, a central oscillator that generates an ∼24-h period and clock-controlled output pathways that translate this into physiological rhythms (Young and Kay 2001). The oscillator is regularly reset by the photoperiod; in addition, light-regulated activities can be restricted by the circadian clock so that a pulse of light in the middle of the night does not have the same effect as the same light cue given during the day (Millar and Kay 1996). Both of these phenomena require that the plant responds to light cues in a time-of-day-specific manner. It is therefore apparent that light perception and circadian regulation are intimately linked (Devlin and Kay 2001).
Molecular genetic analysis of clock-controlled activities has identified several key components of the central oscillator in various organisms, including Drosophila melanogaster, Neurospora crassa, Synechococcus elongatus, Mus musculus, and Arabidopsis thaliana (Young and Kay 2001). In all those organisms transcriptional–translational negative feedback loops are important elements of the oscillator (Young and Kay 2001). Despite this mechanistic similarity, there is little conservation of the biochemical components of the central oscillator. In Arabidopsis, for example, three gene products have a proposed role in the circadian pacemaker: TOC1 and two related transcription factors, CCA1 and LHY (Alabadi et al. 2002; Green and Tobin 2002; Mizoguchi et al. 2002). These three proteins are unrelated to clock components of Neurospora, mice, or Drosophila (Young and Kay 2001). In contrast, the cryptochromes (CRYs) play important, but distinct, circadian functions in plants, flies, and mammals (Devlin and Kay 2001).
Analysis of the light input pathways to the clock reveals that several distinct photoreceptor systems are important for clock entrainment (Devlin and Kay 2001). Melanopsin was recently proposed as a circadian photoreceptor in mammals (Berson et al. 2002). The cryptochrome acts as an input photoreceptor resetting the clock in flies (Emery et al. 2000). In Arabidopsis, both cryptochromes and phytochromes mediate light input to the clock (Somers et al. 1998a). The phytochromes (PHY) are a class of red/far-red photoreceptors (Quail 2002b). Red light converts Pr, the red-light-absorbing form of phytochrome, into Pfr, the far-red-light-absorbing form, which is the active conformer for many phytochrome-mediated responses. Pfr can be converted back to Pr by irradiation with far-red light. The C-terminal portion of plant phytochromes is composed of two PAS repeats (PER, Arnt, SIM) and a histidine-kinase-related domain (Quail 2002b). A phytochrome-like protein is also an essential component of the light input pathway to the clock in cyanobacteria (Schmitz et al. 2000). In Neurospora, WC-1, a transcription factor with a PAS domain, is thought to act as light receptor to the clock (Froehlich et al. 2002; He et al. 2002). However, because wc-1 and wc-2 mutants can be entrained neither by light nor by temperature, WC-1 and the related protein WC-2 can also be classified as critical clock components (Crosthwaite et al. 1997). This illustrates the close association between light input pathways and the central oscillator. WC-1 and WC-2 share little sequence similarity with components of the light input pathway from other organisms. However, they contain PAS domains that are also found in the phytochromes and in the ZTL/LPK/FKF protein family. Arabidopsis ZTL/LPK/FKFs have been proposed to act in the light input pathway (Nelson et al. 2000; Somers et al. 2000; Schultz et al. 2001).
Signaling downstream of the CRY and PHY photoreceptors in plants has been extensively studied (Quail 2002a). Relatively few components working downstream of CRY during blue light perception are known at present (Lin 2002). In contrast, a large number of proteins have been proposed to act downstream from phytochromes (Quail 2002b). Arabidopsis has five phytochrome genes, PHYA–PHYE. phyA and phyB mutants have particularly obvious phenotypes. phyA mutants are blind to far-red light (a light quality found under a canopy of plants), and phyB appears to be the major photoreceptor for de-etiolation in red or white light (Quail 2002b). In addition to interconverting between its two spectral forms, light also affects the subcellular localization of phytochromes, their ability to interact with signaling partners, and the in vitro kinase activity of phyA (Fankhauser 2000; Quail 2002b). The best understood subpathway involves a direct interaction of phytochrome B with bHLH-domain transcription factors, which results in the activation of multiple genes, including CCA1 (Martinez-Garcia et al. 2000; Huq and Quail 2002). Light may therefore entrain the plant circadian clock both via transcriptional activation (as it does in Neurospora crassa and the mouse) and also via posttranslational mechanisms (as in Drosophila melanogaster; Somers et al. 2000).
In this report, we describe srr1, a new Arabidopsis mutant that is altered in multiple outputs of the circadian clock and in phytochrome-B-mediated light signaling. Cloning of the SRR1 gene reveals that SRR1 codes for a novel protein with homologs in numerous eukaryotes including mouse and humans. Our studies suggest that SRR1 might encode a regulator of the circadian clock that is conserved between plants and animals.
Results
The srr1 mutant is impaired in phyB signaling
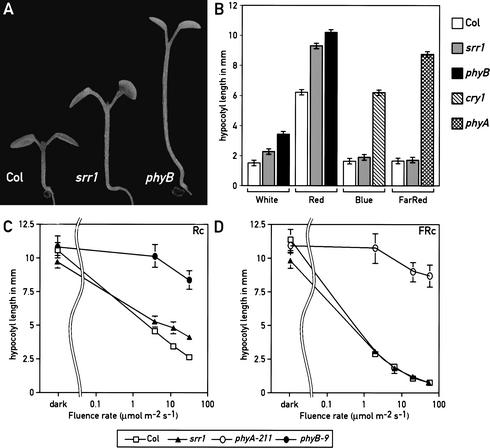
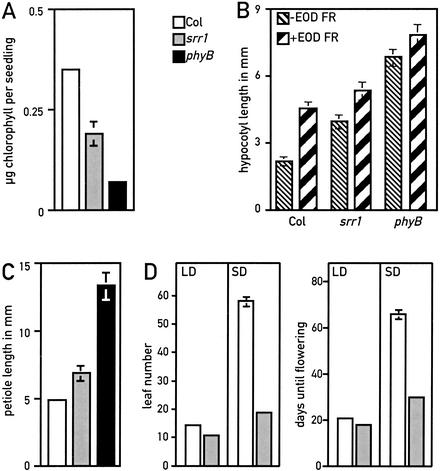
In an effort to identify novel components required for proper light-modulated development, we screened for Arabidopsis mutants that displayed reduced sensitivity to low intensities of white light. We selected seedlings that had a longer hypocotyl than the wild type in this light regime (Fig. 1A), and identified the srr1 (sensitivity to red light reduced) mutant. We determined that under monochromatic light conditions, srr1 mutants had a reduced sensitivity to red light specifically, but responded normally to both far-red and blue light (Fig. 1B–D). Because phytochrome B (phyB) is the major red light photoreceptor in Arabidopsis, we tested srr1 for other typical phyB-mediated responses. Arabidopsis phyB mutations lead to pleiotropic phenotypes. phyB mutants are pale because of reduced chlorophyll accumulation (Reed et al. 1993). We measured chlorophyll content in red-light-grown seedlings and determined that srr1 mutants accumulated intermediate levels of chlorophyll, between the levels of the wild type and a phyB null mutant (Fig. 2A). A saturating pulse of far-red light given at the end of the day induces several typical growth responses in Arabidopsis, including enhanced hypocotyl elongation (Robson et al. 1993; Aukerman et al. 1997). This end-of-day far-red (EOD FR) response is greatly diminished in phyB mutants (Fig. 2B; Robson et al. 1993; Aukerman et al. 1997), and srr1 seedlings also displayed a reduced EOD FR response (Fig. 2B). Later in development, phyB mutants had longer petioles, a phenotype that was also observed in the srr1 mutant (Fig. 2C). Moreover, srr1 mutants had a subtle early-flowering phenotype in long days but flowered much earlier than the wild type in short days (Fig. 2D). The difference in flowering time between long and short days was therefore much smaller in srr1 mutants than in the wild type (Fig. 2D). This suggests that srr1 mutants, like phyB mutants, have a reduced sensitivity to day length (Fig. 2D; Reed et al. 1993; Blazquez and Weigel 1999). Taken together, these data strongly suggest that srr1 mutants are defective in phyB-mediated signaling. To verify that srr1 mutants had normal levels of phytochrome, we determined by Western blots that the levels of phyA and phyB were similar in the srr1 mutant and the wild type (data not shown). These data suggest that srr1 mutants affect phyB-mediated signaling rather than stability or biosynthesis of the photoreceptor.
Figure 1.
srr1 shows reduced sensitivity to white and red light. (A) Wild-type (Col-7), srr1, and phyB-9 mutants grown in white light. (B) srr1 mutants have longer hypocotyls in white and red light. Data are means ± 2× S.E. of at least 12 seedlings for each light treatment. All seedlings were grown at 22°C in continuous light. Col (white bar), srr1 (gray bar), and the appropriate photoreceptor mutants were grown for 6 d in 50 μmoles m−2 sec−1 white (phyB-9, black bar), 30 μmoles m−2 sec−1 blue (cry1, hatched to the right bar), 15 μmoles m−2 sec−1 red (phyB-9, black bar), or 10 μmoles m−2 sec−1 far-red (phyA-211, hatched to the right and left bar). (C) Fluence rate response curve in continuous red light of Col, srr1, and phyB-9 seedlings. (D) Fluence rate response curve in continuous far-red light of Col, srr1, and phyA-211 seedlings. (Open square) Col; (closed triangles) srr1; (open circles) phyA-211; (closed circles) phyB-9.
Figure 2.
srr1 and phyB mutants have several similar phenotypes. (White bar) Col; (gray bar) srr1; (black bar) phyB-9. (A) Chlorophyll content (expressed as nanograms of chlorophyll per seedling) of 7-day-old seedlings grown in constant 35 μmoles m−2 sec−1 red light. Measurements were made in triplicate with 20 seedlings per sample. Values are means ± 2× S.E. (B) The srr1 mutant has a reduced end-of-day far-red (EOD FR) response. Seedlings were grown in control photoperiods (10 h light, 14 h dark, thin hatched bars to the right) or control photoperiods + 15 min FR light (thick hatched bar to the left). Values are means of 15 hypocotyls ± 2× S.E. (C) The srr1 mutant has longer petioles than the wild type. The petioles of 3-week-old plants grown in long days (16 h light, 8 h dark) were measured. Values are means of 16 petioles ± 2× S.E. (D) srr1 mutants flower early in short days. Long days (LD) are as in C; short days (SD) are 9 h light and 15 h dark. Values are the means ± 2× S.E, with at least 18 plants for each condition.
SRR1 also acts independently of phyB
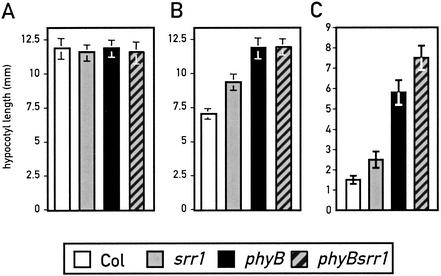
To verify that SRR1 operates in the phyB pathway, we made double mutants between srr1 and the null allele phyB-9. In red light (Fig. 3B), the phyB srr1 double mutants had hypocotyls of the same length as the phyB single mutant. However, when de-etiolation was studied under white light, the phyB srr1 double mutant had a stronger phenotype than the individual single mutants (Fig. 3C). In white light, we also observed an additive phenotype for cotyledon expansion in the phyB srr1 double mutant, which was not apparent in red light (data not shown). Collectively, these observations suggest that SRR1 works in the phyB pathway, but also acts independently of phyB.
Figure 3.
SRR1 works in the phyB signaling pathway. (A) Hypocotyl length in the dark. (B) Hypocotyl length at 40 μmoles m−2 sec−1 red light. (C) Hypocotyl length at 50 μmoles m−2 sec−1 white light. Col (white bar), srr1 mutants (gray bar), phyB-9 (black bar), and srr1 phyB-9 double mutants (hatched to the right bar with gray font). Data are means ± 2× S.E of at least 15 seedlings for each light treatment. All seedlings were grown at 22°C in continuous light.
SRR1 is required for multiple outputs of the circadian clock
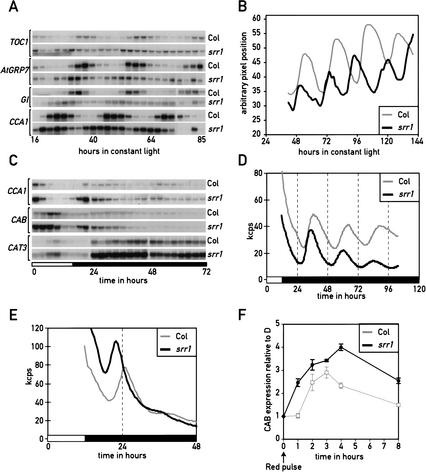
Arabidopsis is a facultative long-day plant, flowering earlier in long than short days. The srr1 flowering time phenotype suggests a defect in photoperiodic perception (Fig. 2D). A number of flowering-time mutants show a reduction or even an absence of sensitivity to day length (Mouradov et al. 2002). Such phenotypes are also observed in numerous circadian clock mutants; moreover, many of them also display an altered light sensitivity during seedling emergence (Mouradov et al. 2002). We therefore tested srr1 mutants for circadian phenotypes by performing Northern blots of RNA from wild type and srr1 mutants that were entrained in day–night cycles (LD) and released into constant light (LL). All circadian-regulated genes that were tested (CAB, AtGRP7/CCR2, CCA1, CAT3, TOC1, GI, and FKF1) showed shorter periods, more rapid dampening, and reduced amplitude of the oscillation in the srr1 mutant background (Fig. 4A; data not shown). Quantification of the Northern blots revealed that the period of AtGRP7/CCR2, CAB, and CCA1 oscillations was between 1.8 and 2.8 h shorter in the mutant than the wild type (data not shown). To test an unrelated clock-controlled phenotype, we recorded leaf movements in entrained srr1 mutants that were released into constant light conditions. Again the srr1 mutant had a period length of ∼2 h shorter than the wild type, 22.6 h in the srr1 mutant versus 24.5 h in the isogenic wild type (Fig. 4B). These results, together with the reduced sensitivity to day length during photoperiodic flower induction (Fig. 2C), indicate that SRR1 function is required for the timing of multiple outputs from the circadian clock.
Figure 4.
The srr1 mutation affects multiple outputs of the circadian clock. (A) srr1 mutants affect cycling of multiple circadian-regulated genes in LL. Plants grown in long days were transferred at zt0 (dawn) to constant light at 20°C. Plants were harvested at 3-h intervals over 3 d, starting at subjective dusk (LL16) on the first day in LL (LL16 to LL85). The cycling of TOC1, AtGRP7/CCR2, GI, and CCA1 was analyzed by Northern blotting. Actin was used as a loading control. (B) The srr1 mutation affects the period length of leaf movement in LL-grown plants. (C) Circadian phenotype of srr1 mutants in DD analyzed by Northern blotting. Plantlets were grown and analyzed as indicated in A, except that they were released into constant darkness. We also harvested samples during the last entrainment cycle. Northern blots were probed with CCA1, CAB3, and CAT3. (D) Circadian phenotype of srr1 mutants analyzed with CCR2∷LUC reporter lines. Col and srr1 seedlings were entrained in LD (12:12) for 7 d before being released into DD and analyzed for bioluminescence. (E) Circadian phenotype of srr1 mutants analyzed with CAB2∷LUC reporter lines. Col and srr1 seedlings were entrained in LD (12:12) for 7 d before being released into DD and analyzed for bioluminescence. (F) srr1 mutants have an enhanced acute response. Col and srr1 mutants were grown in the dark for 5 d on 1/2 MS 1.5% sucrose, then treated with a 3-min red light pulse at 50 μmoles m−2 sec−1 (LED light λmax = 670 nm) and returned into darkness. The Northern blot was probed with a CAB probe. Hybridization signals were quantified with a phosphorimager and normalized with an actin probe. The average of 3 experiments ± S.D. is shown.
SRR1 function is required for clock-regulated gene expression during day–night cycles and in constant darkness
Experiments conducted in constant light cannot discriminate between a defect in the light input pathway and a defect in the oscillator (Covington et al. 2001). To test whether srr1 is also required for clock function in the dark, we shifted entrained plants into constant dark (DD) conditions and tested the rhythmic expression of CAB, CCA1, and CAT3 during one LD cycle and 2 DD cycles by Northern blotting (Fig. 4C). This experiment showed that during LD conditions, the peak of expression of genes such as CAB or CCA1 is earlier in the srr1 mutant than in the wild type (Fig. 4C). This indicates that under entrained conditions, the srr1 mutants had a phenotype in circadian-regulated gene expression. phyB mutants also have an early phase of circadian gene expression (Hall et al. 2002; Salome et al. 2002).
In srr1 mutants, the early phase was also visible for CCA1 transcript oscillation in DD, but the amplitude of rhythmic gene expression was not high enough to determine the period length in DD by Northern blotting. However, this experiment showed very clearly that in DD conditions, the transcript level of CAT3 in srr1 mutants stabilizes to a high steady-state level. This stabilization at a high level is dependent on phyA and cry1 function but not phyB activity (Zhong et al. 1997). This observation is an additional piece of evidence linking SRR1 to phyB signaling rather than cry1 or phyA signaling.
To further test for a possible function of SRR1 in DD conditions, we generated srr1 CAB2∷LUC and srr1 CCR2∷LUC reporter lines. These plants were entrained in day–night cycles and released into constant darkness. These experiments showed that the phase of the circadian peak was 2 h earlier for both genes in the srr1 mutant (Fig. 4D,E). For the CCR2∷LUC line, the period length was shortened by 1.6 h in srr1 mutants relative to the wild-type control (Fig. 4D). The rapid dampening of CAB2∷LUC in DD prevented us from determining a period length of expression of this reporter gene. Taken together, these results suggest that SRR1 activity is required for normal oscillator function.
To test for a light-regulated gene expression phenotype when seedlings are transferred from the dark to the light without prior entrainment of the clock, we examined the induction of the CAB gene in etiolated seedlings in response to a pulse of red light. This early light induction of genes such as CAB is known as the acute response (Anderson and Kay 1995). Mutations in components of the Arabidopsis oscillator differentially affect this light response. toc1-1 mutants have a normal acute response, contrasting with cca1-1 and phyB seedlings, which show a reduced response (Somers et al. 1998b). The srr1 mutants had an enhanced acute CAB response (Fig. 4F).
SRR1 codes for a nuclear/cytoplasmic protein that is conserved in numerous eukaryotes
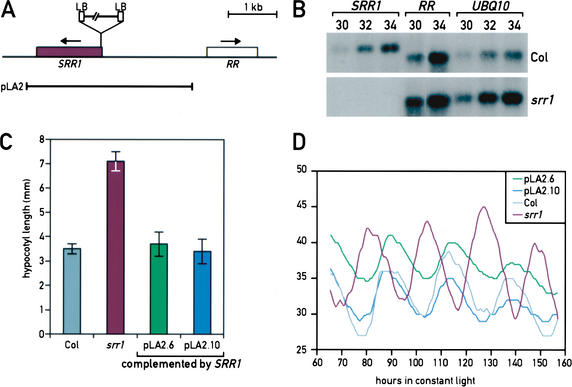
The srr1 mutant was identified in a collection of T-DNA-transformed Arabidopsis lines (Weigel et al. 2000). We crossed the mutant to an isogenic wild type and followed the segregation of the mutant phenotype and the T-DNA marker gene in >80 F2 plants. The mutation was recessive for all phenotypes tested. There was a single T-DNA insertion site that cosegregated with the srr1 mutant phenotype. We isolated genomic DNA flanking the left border (LB) of the T-DNA. The identified DNA matched the sequence of BAC F2O15 (nt 65296–65343), the insertion being in the 5′-untranslated region of a putative ORF coding for a protein of unknown function (F2O15.23, accession no. At5g59560). Using a combination of Southern blots and PCR, we determined that the T-DNA insertion is complex with two left borders (LB) flanking the insertion site (Fig. 5A). The insertion also resulted in the deletion of 82 nt. We identified and sequenced a cDNA coding for the gene that we call SRR1 from an EST collection (GenBank accession no. AY127047). We tested the expression of the SRR1 gene and of the gene most proximal to the T-DNA insertion (a gene coding for a response regulator RR, the ARR1-like protein AB025604) by RT–PCR. We found that SRR1 was not expressed in the srr1 mutant, whereas expression of the RR gene was unaffected (Fig. 5B). To demonstrate that loss of the SRR1 expression caused the srr1 mutant phenotype, we complemented the mutant by transforming it with a piece of genomic DNA including 1.8 kb of promoter sequence and 0.5 kb of sequence 3′ of the gene. Multiple independent lines were tested for de-etiolation and circadian phenotypes (Fig. 5C,D). These experiments confirmed that the lack of SRR1 expression caused the phenotypes of the srr1 mutant plants (Fig. 5C,D; data not shown).
Figure 5.
SRR1 gene structure and T-DNA insertion in the srr1 mutant. (A) The region corresponding to BAC F2O15 (nt 63000–69000) is schematically represented. Genes in this interval are boxed, SRR1 (in purple) and RR (a putative response regulator related to ARR1). The T-DNA insertion in the srr1 mutant is represented; it has one left border (LB) on each side. The piece of genomic DNA used for complementation of the srr1 mutant is indicated (pLA2). (B) The SRR1 gene is not expressed in the srr1 mutant. RT–PCR analysis of the two genes closest to the insertion site of the T-DNA in the srr1 mutant (30, 32, or 34 PCR cycles were performed). UBQ10 was used as a loading control. The number of PCR cycles performed before Southern blot analysis is indicated above each lane. (C) The SRR1 gene complements the srr1 mutation. More than 20 independent transformants of pLA2 in srr1 rescued the visible phenotypes of srr1. Complementation of the srr1 hypocotyl phenotype of two independent lines transformed with pLA2. (D) Two independent srr1 plants transformed with pLA2 were analyzed for the period length of leaf movement in LL. Col is shown in light gray, the srr1 mutant in purple, srr1 pLA2.6 in green, and srr1 pLA2.10 in blue.
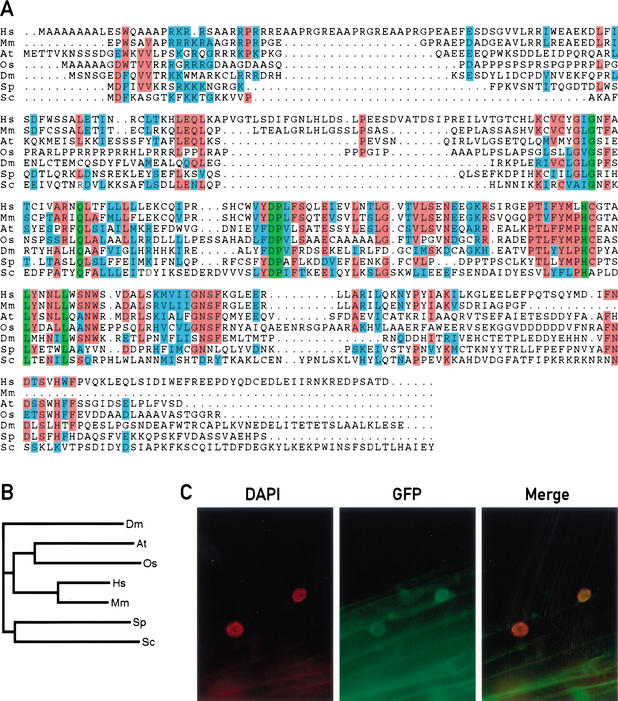
Using BLAST and PSI-BLAST (Altschul et al. 1997), we found sequences homologous to the predicted SRR1 protein in numerous eukaryotes including rice, mouse, human, Drosophila, and yeast (Fig. 6A). The function of these genes is presently unknown. A biochemical activity for these putative proteins is not immediately apparent because they contain no recognizable domains. A putative nuclear localization sequence is located between amino acids 26 and 30 of the Arabidopsis protein. This part of the putative protein is relatively well conserved between the animal and plant sequences (Fig. 6A,B). To test the subcellular localization of SRR1 in Arabidopsis, we generated plants that expressed an SRR1–GFP fusion protein under the control of the Cauliflower Mosaic Virus 35S promoter. Fluorescence microscopy indicated that the SRR1–GFP fusion was found both in the nucleus and the cytoplasm of these plants (Fig. 6C). This fusion protein when expressed from its own promoter was capable of complementing the phenotype of the srr1 mutants, indicating that the fusion to GFP did not alter SRR1 activity (data not shown).
Figure 6.
SRR1 is localized in the nucleus and the cytoplasm, and homologous proteins are found in numerous eukaryotes. (A) The alignment was generated using the ClustalW program (http://www2.ebi.ac.uk/clustalw). The sequences used are Arabidopsis SRR1 cDNA (accession no. AY127047); a mouse cDNA clone (accession no. AK012640.1); a human putative protein predicted from the genomic sequencing program (accession no. Z99714.2); a Drosophila melanogaster putative protein predicted from the genomic sequencing program (accession no. AE003721.1); a Schizosaccharomyces pombe putative protein (accession no. AL022305.1); the Saccharomyces cerevisiae hypothetical protein YLR412w (accession no. S59378); and a sequence from the rice genome (https://portal.tmri.org/rice/p/UserHome) that was found to have the highest homology to Arabidopsis SRR1 (Goff et al. 2002). Identical residues are boxed in green; red shading indicates residues identical in at least 4/7 sequences, similar residues in at least 4/7 sequences are shaded in blue. (B) Phylogenetic relationship between various SRR1 homologs. (C) DAPI fluorescence, GFP fluorescence, and merge picture of Col transformed with a SRR1–GFP fusion construct (CF325).
The SRR1 transcript is induced by light
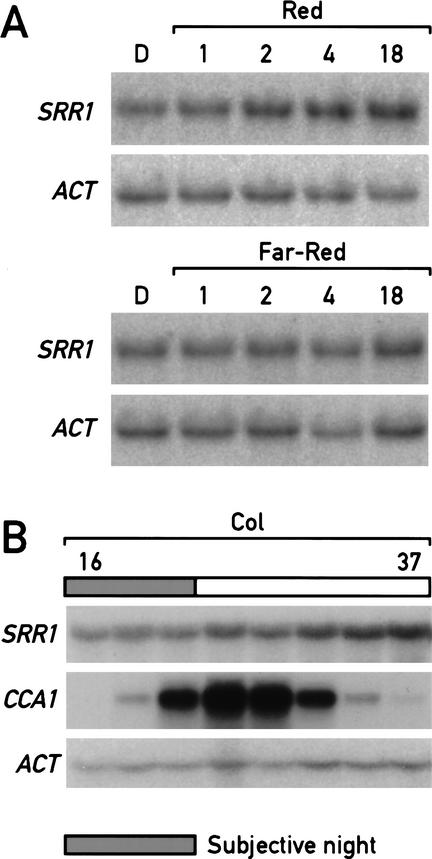
Numerous clock components are themselves under circadian regulation at the transcriptional level (e.g., see Wang and Tobin 1998). We tested this for SRR1, but found that steady-state SRR1 mRNA levels were constant across the circadian cycle in LL (Fig. 7B). Because SRR1 also plays a role in phytochrome-mediated light signaling, we tested whether the SRR1 message was up-regulated in response to light. The steady-state level of SRR1 mRNA was induced when dark-grown seedlings were transferred into red light. The induction was already visible 2 h after the shift into the light (Fig. 7A). Interestingly, the SRR1 transcript was not modulated by far-red light (Fig. 7A). This pattern of light induction is consistent with a role for SRR1 in phyB signaling.
Figure 7.
The mRNA level of SRR1 is light-regulated, but is not under clock control. (A) SRR1 mRNA is up-regulated in response to red light but not far-red light. Five-day-old etiolated seedlings grown on 1/2 MS 1.5% sucrose were shifted into red light or far-red light for the indicated amount of time; the Northern blot was probed with SRR1- and actin-specific probes. (B) SRR1 mRNA levels don't cycle in LL. The samples were as in Figure 4A corresponding to LL16 to LL37. The blot was probed with SRR1, followed by actin as a loading control and CCA1 as a cycling control.
Discussion
In Drosophila, CRY plays a role both in the light input pathway to the clock and in the core oscillator (Krishnan et al. 2001). Similarly, the detailed characterization of numerous Arabidopsis mutants makes it increasingly apparent that it is not simple to distinguish between mutants in the circadian system and photomorphogenic mutants (Devlin and Kay 2001). A common feature among these mutants is that they are affected in the photoperiodic induction of flowering, light sensitivity during seedling emergence, and clock-controlled gene expression. It should be noted that not all light-signaling mutants have major clock phenotypes and certain clock mutants de-etiolate quite normally. Photoreceptor mutants such as phyB show dramatic de-etiolation and flowering time phenotypes, but they have mild period length phenotypes of clock-controlled genes (Somers et al. 1998a). This has been demonstrated by using a phyAphyBcry1cry2 quadruple mutant that is practically blind to white light, but still shows robust cycling of leaf movements in constant light (Yanovsky et al. 2000). In contrast, the toc1-1 mutant shows normal de-etiolation phenotypes but has dramatic circadian defects (Somers et al. 1998b).
Mutants affecting the circadian system are often impaired in de-etiolation in red light
The characterization of the srr1 mutant demonstrates a role for SRR1 both in phyB signaling and as an important regulator of the circadian clock. srr1 mutants are affected in hypocotyl elongation, greening, the end-of-day far-red response, petiole growth, and flowering in a manner very similar to phyB mutants (Figs. 1, 2). In addition, dampening of CAT3 transcript oscillations to a high level in DD is not affected in the srr1 mutant (Fig. 4C). This result is interesting in view of the fact that this stabilization to a high level requires both phyA and cry1 but not phyB, reinforcing the notion that SRR1 works specifically in a phyB signaling pathway (Zhong et al. 1997). Moreover, srr1 and phyB mutants have an early phase of circadian gene expression (Fig. 4; Hall et al. 2002; Salome et al. 2002).
It is quite intriguing to note that ztl, elf3, and gi mutants all have a specific deficiency for de-etiolation in red, but not far-red or blue light (Huq et al. 2000; Reed et al. 2000; Somers et al. 2000). The same is true for srr1 mutants (Fig. 1). Our analysis indicates that similarly to GI and ELF3, this is caused by decreased phyB-mediated signaling. The analysis of double mutants reveals that SRR1, like ELF3, affects both phyB-mediated signaling and a pathway working in parallel with phyB (Liu et al. 2001). Hypocotyl elongation in the dominant lhy mutant and plants overexpressing CCA1 also show decreased sensitivity to white light, but it is not known whether this is restricted to one monochromatic light condition (Schaffer et al. 1998; Wang and Tobin 1998).
A role for SRR1 in phyB signaling
A number of mutants that define components in phyB-mediated signaling have been identified in recent years. The signaling events occurring after photoperception appear rather complex. A number of those components, including ELF3 and GI, have been shown to be nuclear-localized. The best understood branch of phyB-mediated signaling involves the bHLH transcription factors PIF3 and PIF4. PIF3 and PIF4 interact with phyB in a conformation-dependent manner (Martinez-Garcia et al. 2000; Huq and Quail 2002). In the case of PIF3, the interaction with phytochrome occurs when the transcription factor is bound to light-regulated promoters (Martinez-Garcia et al. 2000). Because phyB is translocated into the nucleus in response to light, this would potentially give a very short light-signaling cascade (Smith 2000). This picture contrasts with the fairly large number of mutants implicated in phyB-mediated signaling, and is likely to represent one facet of this complex network (Hudson 2000). The exact function of SRR1 in phyB-mediated light signaling is presently not known. We have no indication for a direct interaction between SRR1 and the phytochromes (data not shown). Double-mutant analysis with other phyB signaling mutants could shed some light on this. In addition, it will be interesting to see if the subcellular localization of phyB is affected in an srr1 background, as SRR1, like the phytochromes, can be found both in the nucleus and the cytoplasm (Fig. 6C).
SRR1 and ELF3 are important for distinct aspects of the circadian clock
The comparison of srr1 and elf3 mutants shows that they have both similar and distinct phenotypes. The circadian phenotype of srr1 mutants in LL is less dramatic than the one observed for elf3-1 null mutants (Reed et al. 2000). This is not caused by allele strength, because srr1 is a null allele (Fig. 5B). srr1 mutants clearly have a short period phenotype in LL, and dampening of clock-controlled genes is more rapid than in the wild type (Fig. 4). srr1 and elf3-1 mutants also show an early phase of circadian gene expression in LD and an enhanced acute response for CAB expression in response to light (Fig. 4; Hicks et al. 1996; Covington et al. 2001). Moreover, ELF3 works in part in the phyB pathway as indicated by the phenotypic analysis of the mutant and the direct interaction of phyB and ELF3 in vitro (Liu et al. 2001). ELF3 has been shown to control the light input pathway to the clock. elf3 mutants show arrhythmia in the light but remain rhythmic in the dark (McWatters et al. 2000). This contrasts with srr1 mutants, which have a circadian phenotype both in the dark and in the light (Fig. 4). Therefore, despite certain similarities between srr1 and elf3 mutants, SRR1 activity is required for normal oscillator function in contrast to ELF3, which is required for light input to the clock specifically.
SRR1 is required for normal circadian oscillator function
Single null mutations can unconditionally abolish overt rhythms in some species, indicating that the cognate, wild-type genes are required for clock function (Young and Kay 2001). No such recessive, arhythmic single mutant has been reported in Arabidopsis (Green and Tobin 2002). In this regard, the elf4 mutant has the most dramatic phenotype, but elf4 still retains low-amplitude cycling (Doyle et al. 2002). srr1 null mutants display rhythmicity in DD or LL but with an early phase and a shorter period (Fig. 4). These data suggest that there is redundancy at the level of SRR1 or that SRR1 activity is required for proper clock function rather than being a key element of the clock itself. The circadian phenotype of srr1 mutants, like the one reported for toc1, cca1, or lhy loss-of-function alleles, results in a shortening of the period length in constant conditions (Green and Tobin 1999). toc1-2, a putative null allele, causes dampened rhythms with a short period in constant light (Strayer et al. 2000). The dampening of the oscillation does not seem to occur in cca1 loss-of-function mutants (Green and Tobin 1999). This relatively mild phenotype of cca1-1 is caused by the redundancy of CCA1 and LHY (Alabadi et al. 2002). Interestingly both cca1-1 and lhy loss of function mutants have flowering-time phenotypes specifically in short days. The double mutant is almost day neutral and has an early phase of circadian gene expression during day–night cycles (Mizoguchi et al. 2002). It has been proposed that this circadian phenotype affects the phase of CO expression during short days, thereby leading to early flowering (Mizoguchi et al. 2002; Roden et al. 2002; Yanovsky and Kay 2002). Interestingly, the circadian and flowering-time phenotypes of srr1 mutants are very similar to those of cca1, lhy, and toc1 loss-of-function alleles, clearly suggesting a role for SRR1 in the clock (Figs. 2, 4; Strayer et al. 2000).
Mutants affecting clock components have distinct phenotypes for the acute light response of CAB gene expression. cca1 mutants affect both the phytochrome-mediated acute response and the circadian peak of CAB expression (Green and Tobin 1999). Similarly, srr1 mutants show a phenotype during light induction of CAB expression (Fig. 4E). The enhanced acute response phenotype observed in the srr1 mutant was also reported for elf3, a mutant in the light input pathway to the clock (Covington et al. 2001). This contrasts with the situation in toc1-1, which affects the period of light-regulated genes without effects on the acute light response of CAB expression (Somers et al. 1998b).
A circadian clock function for SRR1 homologs in animals?
Many extremely diverse organisms have a circadian clock. In the systems in which it has been studied, the mechanism generating an ∼24-h period appears to be conserved mechanistically; however, the components of the machinery are mostly unrelated (Young and Kay 2001). The cryptochromes are an exception in this context because they are important for circadian regulation both in plants and animals (Cashmore et al. 1999). Their function, however, is not identical in different species, and it has been proposed that they have arisen twice during evolution (Cashmore et al. 1999). Cloning of SRR1 in Arabidopsis identifies a putative protein of unknown biochemical function but with clearly related proteins found in numerous eukaryotes. Despite the involvement of phytochromes in the light input pathway to the clock in cyanobacteria, we have not found any clear SRR1 homolog in prokaryotes (Schmitz et al. 2000).
In the context of circadian biology, the putative homologs in human, mouse, and Drosophila are of particular interest. It must be pointed out that in most organisms these genes are only described based on annotation of sequenced genomes. Because there are ESTs (expressed sequence tags) in mouse and given the good conservation between mammals, it is likely that the human equivalent is a functional gene. The mouse SRR1 homolog maps in proximity to the FRP2 short-period circadian rhythm QTL (quantitative trait locus; Shimomura et al. 2001). This is noteworthy in light of the fact that among the numerous mouse QTLs affecting the circadian clock, only two have a short period, and one of them, FRP2, maps close to the mouse SRR1 homolog. Cloning this gene is one way to address the possible conservation of SRR1 function across species. The mouse SRR1 homolog is expressed at very low levels in all tested tissues with no circadian oscillation of its mRNA levels (S.A. Brown and U. Schibler, pers. comm.). The same is true for SRR1 in Arabidopsis; however, this is not exceptional for factors required for proper clock function (Fig. 7; Sugano et al. 1999; Somers et al. 2000). We have not found an SRR1-related protein in Neurospora, but given that the entire genome has not been sequenced yet, this does not exclude the existence of a related protein in this species. More distant relatives of SRR1 have also been found in the yeasts Schizosaccharomyces pombe and Saccharomyces cerevisiae with significantly better homologies between SRR1 and the S. pombe sequence than between SRR1 and YLR412w from S. cerevisiae. There are a few reports on circadian rhythms in yeast (Kippert and Hunt 2000). The yeast genes are of unknown function, and systematic yeast two-hybrid analysis performed with YLR412w has not been informative concerning a putative function of this protein in S. cerevisiae. Despite a lack of conservation of the elements that constitute a central oscillator in animals, fungi, and plants, there are some common features such as regulated nuclear translocation, phosphorylation, and transcriptional–translational negative feedback loops (Young and Kay 2001). SRR1 might modulate one of those processes.
Material and methods
Plant material and light sources
The progeny of 2600 independent Arabidopsis ecotype Col-7 transformants (Weigel et al. 2000) were screened in 50 μmoles m−2 sec−1 white light. We screened ∼10 T2 seeds per original transformant. Seeds were surface-sterilized and plated on Petri dishes on 1/2 MS, 0.7% phytagar. Plates were stored in the dark at 4°C for 3 d and moved into constant white light at 22°C. For monochromatic light conditions, germination was induced by a white light treatment; the seedlings were then grown in the appropriate light conditions for 4–6 d depending on the experiment. Experiments in monochromatic light were performed in a Percival E-30LED using either the blue (λmax = 469 nm), red (λmax = 667 nm), or the FR (λmax = 739 nm) diodes, at 22°C in continuous light. Light intensities were determined with a LiCor LI-1800 spectroradiometer or with an International light IL1400A photometer equipped with an SEL033 probe with appropriate light filters.
Petiole-length and flowering-time determination
Petiole length was determined as described (Aukerman et al. 1997). Flowering time was measured according to Blazquez and Weigel (1999). Briefly, seeds were stratified at 4°C for 3 d, and plants were grown at 23°C in long (16 h light, 8 h dark) or short (9 h light, 15 h dark) days under a mixture of 3:1 cool-white and Gro-Lux fluorescent lights.
Hypocotyl length and chlorophyll accumulation
Measurements of hypocotyl length were performed as described (Neff and Chory 1998). For chlorophyll determination, triplicates of 20 seedlings were each incubated in 1 mL of 80% acetone overnight in the dark at room temperature under constant shaking. The OD of the solution was determined at 647 nm and 663 nm, and the chlorophyll content was calculated with the following formula: micrograms per microliter of total chlorophyll = 18.71 × (OD647) + 7.15 × (OD663). The effect of the end-of-day far-red treatment was determined as in Aukerman et al. (1997).
Plant DNA and RNA preparation
Plant DNA was prepared either according to Neff et al. (1998) or with Plant DNAzol Reagent (GIBCO-BRL) according to the manufacturer's instructions.
RNA was extracted as described (Heintzen et al. 1997). We ran formaldehyde MOPS gels loaded with 10–15 μg of total RNA per lane and transferred the RNA with 10× SSC. Probes generated by random priming were described in Leutwiler et al. (1986). Northern and Southern blots were hybridized with Church buffer at 62°C and washed according to the manufacturer's instructions. For quantification, we used a PhosphorImager, Imagequant (Molecular Dynamics). RNA levels for clock-regulated genes were normalized to the levels of actin RNA. Normalized data were analyzed using FFT-NLLS, to provide objective period estimates. This software returns a valid best-fit period from such short and sparse time series, although the associated estimates of uncertainty of fit must be treated with caution.
Cloning of the SRR1 gene
Because the srr1 mutant came from a T-DNA insertion collection, we determined that the phenotype cosegregated with the T-DNA insertion marker by analyzing 80 F2 progeny of a cross between the homozygous srr1 mutant and wild-type Col-7. The srr1 phenotype is recessive, and the original mutant contained a single T-DNA insertion site. For subsequent experiments the mutant was back-crossed twice.
To obtain DNA sequence flanking the T-DNA insertion site, we used a PCR technique related to the Genome Walker Kit (Clonetech). Sequencing identified the T-DNA left border sequence flanked by 70 bp of sequence of unknown origin and 48 bp of BAC F2O15 (nt 65296–65343), finishing with an HindIII site ligated to the adaptor sequence. The insertion site was then confirmed by PCR with various primers. Using Southern blots, we determined that the insertion is complex, with the other end of the T-DNA also containing a Left Border as determined by PCR followed by sequencing. The other end flanks nt 65426 from BAC F2O15, and the T-DNA insertion therefore deletes 82 nt corresponding to nt 65344–65425 of BAC F2O15. This T-DNA insertion is just upstream of a putative ORF for which we found the following EST clones: SQ058e07F (AV557017) and SQ091c08F (AV558172; Kasuza DNA Research Institute). The longer of the two, SQ091c08F, encompasses nt 64054–65328 of BAC F2O15 including a putative ORF identified by the Arabidopsis genome project.
RT–PCR
For the analysis of transcript levels, a reverse transcription reaction was performed with total RNA extracted from wild-type and srr1 mutant plants. An oligo(dT)(12–18) primer was hybridized to 1 μg of RNA. The reverse transcription was performed with Superscript II (GIBCO-BRL) according to the manufacturer's instructions. After the RT reaction, the cDNA was diluted threefold with dH2O, and 1 μL of diluted cDNA was used for PCR reactions with gene-specific primers for UBQ10, SRR1, and RR. PCR reactions were stopped after 30, 32, and 34 cycles, and the reactions were analyzed by Southern blotting. To test for DNA contamination, a mock RT reaction was performed in the absence of oligo(dT)(12–18). The product was then PCR-amplified and Southern blotted as described above.
Complementation of the srr1 mutant
For complementation of the srr1 mutant, we gel-purified the 3.4-kb SpeI fragment from BAC F2O15 encompassing the SRR1 gene (nt 63753–67173). The fragment was ligated into an XbaI-digested and phosphatase-treated pPZP211 vector (Hajdukiewicz et al. 1994). The resulting construct (pLA2) was introduced into Agrobacterium tumefaciens, strain GV3101. srr1 plants were transformed with the spray method (Weigel et al. 2000). T2 seeds from multiple independent transformants were used to confirm the complementation.
Microscopy
Using standard cloning techniques, we fused a modified GFP (Friedrichsen et al. 2000) in frame after the last codon of the SRR1 gene. This fusion gene was either expressed under the 35S promoter by cloning it into pCHF3 (Jarvis et al. 1998) to create pCF325, or expressed from its own promoter to create pCF327. Col plants transformed with CF325 and srr1 mutants transformed with pCF327 were generated as described above. We performed microscopy with 5-day-old dark-grown homozygous single-insertion Col pCF325 plants. The seedlings were mounted in 20% glycerol, 1/2 MS, and 0.5 μg/mL DAPI (Sigma) and were observed with a DAPI filter or a narrow band GFP filter (Chroma 41020) on a Zeiss Axioplan 2 fluorescence microscope. Images were recorded with a Quantix CCD camera and pseudocolored using Photoshop.
Analysis of circadian phenotypes
Leaf movement rhythms were monitored in a Kujata imaging system and analyzed using FFT-NLLS as described (Swarup et al. 1999). Experiments comparing srr1 with Col-7 were repeated three times, with very similar results. Figure 4 presents data from a representative experiment. Leaf movement rhythms were analyzed in a single experiment for two srr1 mutant lines that were independently transformed with the complementing construct, pLA2. Seedlings were arranged randomly with respect to genotypes within each experiment, to avoid positional bias in the imaging arrays. Transcriptional rhythms of CAB2 and CCR2 expression were assayed using luciferase reporter gene fusions as described (Doyle et al. 2002). The CCR2∷LUC and CAB2∷LUC constructions were transformed into Col-7 and the srr1 mutant (Weigel et al. 2000), and progeny from three independent transformants were analyzed for each genotype/transformation combination. Rhythmic data on the T2 and T3 generations were collected on a TopCount (Packard). Period estimates were generated using FFT-NLLS, as described above.
Acknowledgments
We thank Nicolas Roggli for artwork, Consuelo Salomon and Victoria Ravenscroft for technical support, Detlef Weigel (The Salk Institute) for providing the T-DNA lines, Kasuza DNA Research Institute for EST clones, and the Arabidopsis stock center for BAC F2O15. We thank Klaus Apel (ETH), in whose lab some of the experiments were performed. We thank Syngenta for access to the rice genome data. This work was supported by grants from the Swiss National Science Foundation (631-58 151.99 to C.F. and FN 3100-052475.97/1 to D.S.), the state of Geneva (to C.F.), the EMBO young investigator program (to C.F.), ETH (grant ETH- 34./00-3 to D.S.), a BBSRC studentship to N.S., the Life Sciences Research Foundation (Department of Energy Bioscience fellowship to S.J.D.), the BBSRC (grant G15231 to A.J.M.), and the U.S. National Institutes of Health (2RO1 GM52413 to J.C.). J.C. is an Investigator of the Howard Hughes Medical Institute.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL christian.fankhauser@molbio.unige.ch; FAX 41-22-702-6868.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.244103.
References
- Alabadi D, Yanovsky MJ, Mas P, Harmer SL, Kay SA. Critical role for CCA1 and LHY in maintaining circadian rhythmicity in Arabidopsis. Curr Biol. 2002;12:757–761. doi: 10.1016/s0960-9822(02)00815-1. [DOI] [PubMed] [Google Scholar]
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson S, Kay S. Functional dissection of circadian clock and phytochrome-regulated transcription of the Arabidopsis CAB2 gene. Proc Natl Acad Sci. 1995;92:1500–1504. doi: 10.1073/pnas.92.5.1500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aukerman MJ, Hirschfeld M, Wester L, Weaver M, Clack T, Amasino RM, Sharrock RA. A deletion in the PHYD gene of the Arabidopsis Wassilewskija ecotype defines a role for phytochrome D in red/far-red light sensing. Plant Cell. 1997;9:1317–1326. doi: 10.1105/tpc.9.8.1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berson DM, Dunn FA, Takao M. Phototransduction by retinal ganglion cells that set the circadian clock. Science. 2002;295:1070–1073. doi: 10.1126/science.1067262. [DOI] [PubMed] [Google Scholar]
- Blazquez MA, Weigel D. Independent regulation of flowering by phytochrome B and gibberellins in Arabidopsis. Plant Physiol. 1999;120:1025–1032. doi: 10.1104/pp.120.4.1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cashmore AR, Jarillo JA, Wu YJ, Liu D. Cryptochromes: Blue light receptors for plants and animals. Science. 1999;284:760–765. doi: 10.1126/science.284.5415.760. [DOI] [PubMed] [Google Scholar]
- Covington MF, Panda S, Liu XL, Strayer CA, Wagner DR, Kay SA. Elf3 modulates resetting of the circadian clock in Arabidopsis. Plant Cell. 2001;13:1305–1316. doi: 10.1105/tpc.13.6.1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crosthwaite SK, Dunlap JC, Loros JJ. Neurospora wc-1 and wc-2: Transcription, photoresponses, and the origins of circadian rhythmicity. Science. 1997;276:763–769. doi: 10.1126/science.276.5313.763. [DOI] [PubMed] [Google Scholar]
- Devlin PF, Kay SA. Circadian photoperception. Annu Rev Physiol. 2001;63:677–694. doi: 10.1146/annurev.physiol.63.1.677. [DOI] [PubMed] [Google Scholar]
- Doyle MR, Davis SJ, Bastow RM, McWatters HG, Kozma-Bognar L, Nagy F, Millar AJ, Amasino RM. The ELF4 gene controls circadian rhythms and flowering time in Arabidopsis thaliana. Nature. 2002;419:74–77. doi: 10.1038/nature00954. [DOI] [PubMed] [Google Scholar]
- Emery P, Stanewsky R, Hall JC, Rosbash M. A unique circadian-rhythm photoreceptor. Nature. 2000;404:456–457. doi: 10.1038/35006558. [DOI] [PubMed] [Google Scholar]
- Fankhauser C. Phytochromes as light-modulated protein kinases. Semin Cell Dev Biol. 2000;11:467–473. doi: 10.1006/scdb.2000.0201. [DOI] [PubMed] [Google Scholar]
- Friedrichsen DM, Joazeiro CA, Li J, Hunter T, Chory J. Brassinosteroid-insensitive-1 is a ubiquitously expressed leucine-rich repeat receptor serine/threonine kinase. Plant Physiol. 2000;123:1247–1256. doi: 10.1104/pp.123.4.1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Froehlich AC, Liu Y, Loros JL, Dunlap JC. White Collar-1, a circadian blue light photoreceptor, binding to the frequency promoter. Science. 2002;297:815–819. doi: 10.1126/science.1073681. [DOI] [PubMed] [Google Scholar]
- Goff SA, Ricke D, Lan TH, Presting G, Wang R, Dunn M, Glazebrook J, Sessions A, Oeller P, Varma H, et al. A draft sequence of the rice genome (Oryza sativa L. ssp. japonica) Science. 2002;296:92–100. doi: 10.1126/science.1068275. [DOI] [PubMed] [Google Scholar]
- Green RM, Tobin EM. Loss of the circadian clock-associated protein 1 in Arabidopsis results in altered clock-regulated gene expression. Proc Natl Acad Sci. 1999;96:4176–4179. doi: 10.1073/pnas.96.7.4176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ————— The role of CCA1 and LHY in the plant circadian clock. Dev Cell. 2002;2:516–518. doi: 10.1016/s1534-5807(02)00184-3. [DOI] [PubMed] [Google Scholar]
- Green RM, Tingay S, Wang ZY, Tobin EM. Circadian rhythms confer a higher level of fitness to Arabidopsis plants. Plant Physiol. 2002;129:576–584. doi: 10.1104/pp.004374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajdukiewicz P, Svab Z, Maliga P. The small, versatile pPZP family of Agrobacterium binary vectors for plant transformation. Plant Mol Biol. 1994;25:989–994. doi: 10.1007/BF00014672. [DOI] [PubMed] [Google Scholar]
- Hall A, Kozma-Bognár L, Bastow RM, Nagy F, Millar AJ. Distinct regulation of CAB and PHYB gene expression by similar circadian clocks. Plant J. 2002;32:529–537. doi: 10.1046/j.1365-313x.2002.01441.x. [DOI] [PubMed] [Google Scholar]
- Harmer SL, Hogenesch JB, Straume M, Chang HS, Han B, Zhu T, Wang X, Kreps JA, Kay SA. Orchestrated transcription of key pathways in Arabidopsis by the circadian clock. Science. 2000;290:2110–2113. doi: 10.1126/science.290.5499.2110. [DOI] [PubMed] [Google Scholar]
- He Q, Cheng P, Yang Y, Wang L, Gardner KH, Liu Y. White Collar-1, a DNA binding transcription factor and a light sensor. Science. 2002;297:840–843. doi: 10.1126/science.1072795. [DOI] [PubMed] [Google Scholar]
- Heintzen C, Nater M, Apel K, Staiger D. AtGRP7, a nuclear RNA-binding protein as a component of a circadian-regulated negative feedback loop in Arabidopsis thaliana. Proc Natl Acad Sci. 1997;94:8515–8520. doi: 10.1073/pnas.94.16.8515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hicks KA, Millar AJ, Carre IA, Somers DE, Straume M, Meeks-Wagner DR, Kay SA. Conditional circadian dysfunction of the Arabidopsis early-flowering 3 mutant. Science. 1996;274:790–792. doi: 10.1126/science.274.5288.790. [DOI] [PubMed] [Google Scholar]
- Hudson ME. The genetics of phytochrome signalling in Arabidopsis. Semin Cell Dev Biol. 2000;11:475–483. doi: 10.1006/scdb.2000.0204. [DOI] [PubMed] [Google Scholar]
- Huq E, Quail PH. PIF4, a phytochrome-interacting bHLH factor, functions as a negative regulator of phytochrome B signaling in Arabidopsis. EMBO J. 2002;21:2441–2450. doi: 10.1093/emboj/21.10.2441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huq E, Tepperman JM, Quail PH. GIGANTEA is a nuclear protein involved in phytochrome signaling in Arabidopsis. Proc Natl Acad Sci. 2000;97:9789–9794. doi: 10.1073/pnas.170283997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarvis P, Chen LJ, Li H, Peto CA, Fankhauser C, Chory J. An Arabidopsis mutant defective in the plastid general protein import apparatus. Science. 1998;282:100–103. doi: 10.1126/science.282.5386.100. [DOI] [PubMed] [Google Scholar]
- Kippert F, Hunt P. Ultradian clocks in eukaryotic microbes: From behavioural observation to functional genomics. Bioessays. 2000;22:16–22. doi: 10.1002/(SICI)1521-1878(200001)22:1<16::AID-BIES5>3.0.CO;2-1. [DOI] [PubMed] [Google Scholar]
- Krishnan B, Levine JD, Lynch MK, Dowse HB, Funes P, Hall JC, Hardin PE, Dryer SE. A new role for cryptochrome in a Drosophila circadian oscillator. Nature. 2001;411:313–317. doi: 10.1038/35077094. [DOI] [PubMed] [Google Scholar]
- Leutwiler L, Meyerowitz E, Tobin E. Structure and expression of three light-harvesting chlorophyll a/b-binding protein genes in Arabidopsis thaliana. Nucleic Acids Res. 1986;14:4051–4064. doi: 10.1093/nar/14.10.4051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin C. Blue light receptors and signal transduction. Plant Cell. 2002;14:S207–S225. doi: 10.1105/tpc.000646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu XL, Covington MF, Fankhauser C, Chory J, Wagner DR. ELF3 encodes a circadian clock-regulated nuclear protein that functions in an Arabidopsis PHYB signal transduction pathway. Plant Cell. 2001;13:1293–1304. doi: 10.1105/tpc.13.6.1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Garcia JF, Huq E, Quail PH. Direct targeting of light signals to a promoter element-bound transcription factor. Science. 2000;288:859–863. doi: 10.1126/science.288.5467.859. [DOI] [PubMed] [Google Scholar]
- McDonald MJ, Rosbash M. Microarray analysis and organization of circadian gene expression in Drosophila. Cell. 2001;107:567–578. doi: 10.1016/s0092-8674(01)00545-1. [DOI] [PubMed] [Google Scholar]
- McWatters HG, Bastow RM, Hall A, Millar AJ. The ELF3 zeitnehmer regulates light signalling to the circadian clock. Nature. 2000;408:716–720. doi: 10.1038/35047079. [DOI] [PubMed] [Google Scholar]
- Millar AJ, Kay SA. Integration of circadian and phototransduction pathways in the network controlling CAB gene transcription in Arabidopsis. Proc Natl Acad Sci. 1996;93:15491–15496. doi: 10.1073/pnas.93.26.15491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizoguchi T, Wheatley K, Hanzawa Y, Wright L, Mizoguchi M, Song HR, Carre IA, Coupland G. LHY and CCA1 are partially redundant genes required to maintain circadian rhythms in Arabidopsis. Dev Cell. 2002;2:629–641. doi: 10.1016/s1534-5807(02)00170-3. [DOI] [PubMed] [Google Scholar]
- Mouradov A, Cremer F, Coupland G. Control of flowering time: Interacting pathways as a basis for diversity. Plant Cell. 2002;14 Suppl.:S111–S130. doi: 10.1105/tpc.001362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neff MM, Chory J. Genetic interactions between phytochrome A, phytochrome B, and cryptochrome 1 during Arabidopsis development. Plant Physiol. 1998;118:27–35. doi: 10.1104/pp.118.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neff MM, Neff JD, Chory J, Pepper AE. dCAPS, a simple technique for the genetic analysis of single nucleotide polymorphisms: Experimental applications in Arabidopsis thaliana genetics. Plant J. 1998;14:387–392. doi: 10.1046/j.1365-313x.1998.00124.x. [DOI] [PubMed] [Google Scholar]
- Nelson DC, Lasswell J, Rogg LE, Cohen MA, Bartel B. FKF1, a clock-controlled gene that regulates the transition to flowering in Arabidopsis. Cell. 2000;101:331–340. doi: 10.1016/s0092-8674(00)80842-9. [DOI] [PubMed] [Google Scholar]
- Quail PH. Photosensory perception and signalling in plant cells: New paradigms? Curr Opin Cell Biol. 2002a;14:180–188. doi: 10.1016/s0955-0674(02)00309-5. [DOI] [PubMed] [Google Scholar]
- ————— Phytochrome photosensory signalling networks. Nat Rev Mol Cell Biol. 2002b;3:85–93. doi: 10.1038/nrm728. [DOI] [PubMed] [Google Scholar]
- Reed JW, Nagpal P, Poole DS, Furuya M, Chory J. Mutations in the gene for the red/far-red light receptor phytochrome B alter cell elongation and physiological responses throughout Arabidopsis development. Plant Cell. 1993;5:147–157. doi: 10.1105/tpc.5.2.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed JW, Nagpal P, Bastow RM, Solomon KS, Dowson-Day MJ, Elumalai RP, Millar AJ. Independent action of ELF3 and phyB to control hypocotyl elongation and flowering time. Plant Physiol. 2000;122:1149–1160. doi: 10.1104/pp.122.4.1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robson PRH, Whitelam GC, Smith H. Selected components of the shade-avoidance syndrome are displayed in a normal manner in mutants of Arabidopsis thaliana and Brassica rapa deficient in phytochrome B. Plant Physiol. 1993;102:1179–1184. doi: 10.1104/pp.102.4.1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roden LC, Song HR, Jackson S, Morris K, Carre IA. Floral responses to photoperiod are correlated with the timing of rhythmic expression relative to dawn and dusk in Arabidopsis. Proc Natl Acad Sci. 2002;99:13313–13318. doi: 10.1073/pnas.192365599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salome PA, Michael TP, Kearns EV, Fett-Neto AG, Sharrock RA, McClung CR. The out of phase 1 mutant defines a role for PHYB in circadian phase control in Arabidopsis. Plant Physiol. 2002;129:1674–1685. doi: 10.1104/pp.003418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaffer R, Ramsay N, Samach A, Corden S, Putterill J, Carre IA, Coupland G. The late elongated hypocotyl mutation of Arabidopsis disrupts circadian rhythms and the photoperiodic control of flowering. Cell. 1998;93:1219–1229. doi: 10.1016/s0092-8674(00)81465-8. [DOI] [PubMed] [Google Scholar]
- Schaffer R, Landgraf J, Accerbi M, Simon VV, Larson M, Wisman E. Microarray analysis of diurnal and circadian-regulated genes in Arabidopsis. Plant Cell. 2001;13:113–123. doi: 10.1105/tpc.13.1.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitz O, Katayama M, Williams SB, Kondo T, Golden SS. CikA, a bacteriophytochrome that resets the cyanobacterial circadian clock. Science. 2000;289:765–768. doi: 10.1126/science.289.5480.765. [DOI] [PubMed] [Google Scholar]
- Schultz TF, Kiyosue T, Yanovsky M, Wada M, Kay SA. A role for LKP2 in the circadian clock of Arabidopsis. Plant Cell. 2001;13:2659–2670. doi: 10.1105/tpc.010332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimomura K, Low-Zeddies SS, King DP, Steeves TD, Whiteley A, Kushla J, Zemenides PD, Lin A, Vitaterna MH, Churchill GA, et al. Genome-wide epistatic interaction analysis reveals complex genetic determinants of circadian behavior in mice. Genome Res. 2001;11:959–980. doi: 10.1101/gr.171601. [DOI] [PubMed] [Google Scholar]
- Smith H. Phytochromes and light signal perception by plants—An emerging synthesis. Nature. 2000;407:585–591. doi: 10.1038/35036500. [DOI] [PubMed] [Google Scholar]
- Somers DE, Devlin PF, Kay SA. Phytochromes and cryptochromes in the entrainment of the Arabidopsis circadian clock. Science. 1998a;282:1488–1490. doi: 10.1126/science.282.5393.1488. [DOI] [PubMed] [Google Scholar]
- Somers DE, Webb AA, Pearson M, Kay SA. The short-period mutant, toc1-1, alters circadian clock regulation of multiple outputs throughout development in Arabidopsis thaliana. Development. 1998b;125:485–494. doi: 10.1242/dev.125.3.485. [DOI] [PubMed] [Google Scholar]
- Somers DE, Schultz TF, Milnamow M, Kay SA. ZEITLUPE encodes a novel clock-associated PAS protein from Arabidopsis. Cell. 2000;101:319–329. doi: 10.1016/s0092-8674(00)80841-7. [DOI] [PubMed] [Google Scholar]
- Storch KF, Lipan O, Leykin I, Viswanathan N, Davis FC, Wong WH, Weitz CJ. Extensive and divergent circadian gene expression in liver and heart. Nature. 2002;417:78–83. doi: 10.1038/nature744. [DOI] [PubMed] [Google Scholar]
- Strayer C, Oyama T, Schultz TF, Raman R, Somers DE, Mas P, Panda S, Kreps JA, Kay SA. Cloning of the Arabidopsis clock gene TOC1, an autoregulatory response regulator homolog. Science. 2000;289:768–771. doi: 10.1126/science.289.5480.768. [DOI] [PubMed] [Google Scholar]
- Sugano S, Andronis C, Ong MS, Green RM, Tobin EM. The protein kinase CK2 is involved in regulation of circadian rhythms in Arabidopsis. Proc Natl Acad Sci. 1999;96:12362–12366. doi: 10.1073/pnas.96.22.12362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swarup K, Alonso-Blanco C, Lynn JR, Michaels SD, Amasino RM, Koornneef M, Millar AJ. Natural allelic variation identifies new genes in the Arabidopsis circadian system. Plant J. 1999;20:67–77. doi: 10.1046/j.1365-313x.1999.00577.x. [DOI] [PubMed] [Google Scholar]
- Wang ZY, Tobin EM. Constitutive expression of the CIRCADIAN CLOCK ASSOCIATED 1 (CCA1) gene disrupts circadian rhythms and suppresses its own expression. Cell. 1998;93:1207–1217. doi: 10.1016/s0092-8674(00)81464-6. [DOI] [PubMed] [Google Scholar]
- Weigel D, Ahn JH, Blazquez MA, Borevitz JO, Christensen SK, Fankhauser C, Ferrandiz C, Kardailsky I, Malancharuvil EJ, Neff MM, et al. Activation tagging in Arabidopsis. Plant Physiol. 2000;122:1003–1014. doi: 10.1104/pp.122.4.1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanovsky MJ, Kay SA. Molecular basis of seasonal time measurement in Arabidopsis. Nature. 2002;419:308–312. doi: 10.1038/nature00996. [DOI] [PubMed] [Google Scholar]
- Yanovsky MJ, Mazzella MA, Casal JJ. A quadruple photoreceptor mutant still keeps track of time. Curr Biol. 2000;10:1013–1015. doi: 10.1016/s0960-9822(00)00651-5. [DOI] [PubMed] [Google Scholar]
- Young MW, Kay SA. Time zones: A comparative genetics of circadian clocks. Nat Rev Genet. 2001;2:702–715. doi: 10.1038/35088576. [DOI] [PubMed] [Google Scholar]
- Zhong HH, Resnick AS, Straume M, Robertson McClung C. Effects of synergistic signaling by phytochrome A and cryptochrome 1 on circadian clock-regulated catalase expression. Plant Cell. 1997;9:947–955. doi: 10.1105/tpc.9.6.947. [DOI] [PMC free article] [PubMed] [Google Scholar]