Abstract
With the discovery of RNA interference (RNAi) and related phenomena, new regulatory roles attributed to RNA continue to emerge. Here we show, in mammalian tissue culture, that a short interfering RNA (siRNA) can repress expression of a target mRNA with partially complementary binding sites in its 3′ UTR, much like the demonstrated function of endogenously encoded microRNAs (miRNAs). The mechanism for this repression is cooperative, distinct from the catalytic mechanism of mRNA cleavage by siRNAs. The use of siRNAs to study translational repression holds promise for dissecting the sequence and structural determinants and general mechanism of gene repression by miRNAs.
Keywords: siRNA, miRNA, RNAi, translational repression, 3′ UTR
The RNA interference (RNAi) pathway was first recognized in Caenorhabditis elegans as a response to exogenously introduced long double-stranded RNA (dsRNA; Fire et al. 1998). An RNase III enzyme, Dicer, cleaves the dsRNA into duplexes of 21–23 nucleotides (nt) termed short interfering RNAs (siRNAs), which then guide a multicomponent complex known as RISC (RNA induced silencing complex) to mRNAs sharing perfect complementarity and target their cleavage (Hamilton and Baulcombe 1999; Tuschl et al. 1999; Hammond et al. 2000; Zamore et al. 2000; Bernstein et al. 2001; Elbashir et al. 2001a). The RNAi pathway has been implicated in silencing transposons in the C. elegans germline (Ketting et al. 1999; Tabara et al. 1999), silencing Stellate repeats in the Drosophila germline (Aravin et al. 2001), and serving as an immune response against invading viruses in plants (for review, see Baulcombe 2001). Very little, however, is known about the intrinsic biological role of RNAi in mammalian systems; indeed, no endogenous siRNAs have been identified in mammals. Nevertheless, transfection of mammalian cells with exogenous siRNAs has rapidly been adopted as a technology for targeted gene silencing (Elbashir et al. 2001a).
A related short RNA species, microRNAs (miRNAs), has been identified in organisms ranging from plants to nematodes to mammals (Lagos-Quintana et al. 2001; Lau et al. 2001; Lee and Ambros 2001; Reinhart et al. 2002). These endogenous RNA species are first transcribed as a long RNA and then processed to a pre-miRNA of ∼70 nt (Lee et al. 2002). This pre-miRNA forms an imperfect hairpin structure, which is processed by Dicer to produce the mature, single-strand ∼22-nt miRNA (Grishok et al. 2001; Hutvagner et al. 2001; Ketting et al. 2001). Despite the large library of miRNAs now known in animals, only two have a known function; lin-4 and let-7 regulate endogenous genes involved in developmental timing in C. elegans by partially base-pairing to the 3′ UTR of target mRNAs such as lin-14 and lin-41, respectively (Lee et al. 1993; Wightman et al. 1993; Ha et al. 1996; Reinhart et al. 2000; Slack et al. 2000). This interaction does not affect the stability of the target mRNA but rather represses gene expression through an unknown mechanism known as translational repression (Olsen and Ambros 1999). The polysome profile of the target mRNA does not change upon gene silencing, suggesting that this repression occurs after initiation of translation, and potentially occurs posttranslationally (Olsen and Ambros 1999). This form of regulation is likely to be conserved in mammalian cells because overexpression of miR-30 can repress a reporter gene with partially complementary miR-30 binding sites in its 3′ UTR without affecting mRNA stability (Zeng et al. 2002).
The RNAi pathway of siRNA-directed mRNA cleavage and the miRNA-mediated translational repression pathway are genetically and biochemically distinct. In addition to different outcomes, the two pathways have differential requirements for Paz-Piwi domain (PPD) proteins in C. elegans. Translational repression by lin-4 and let-7 depends on alg-1 and alg-2 for miRNA processing and/or stability, yet these genes are not required for RNAi (Grishok et al. 2001), whereas rde-1 is needed in RNAi but is not necessary for translational repression (Tabara et al. 1999). In HeLa cells, Gemin 3 and Gemin 4 proteins immunoprecipitate with RISC activity (Hutvagner and Zamore 2002) and miRNAs (Mourelatos et al. 2002), but have not been detected as components of purified RISC activity from S100 extracts (Martinez et al. 2002).
In addition to requiring Dicer processing to generate the short RNA, RNAi and translational repression share common components. The PPD protein eIF2C2 both immunoprecipitates with miRNAs from HeLa cells (Mourelatos et al. 2002) and copurifies with RISC activity (Martinez et al. 2002). Additionally, endogenous let-7 in HeLa extracts is capable of directing cleavage of a perfectly complementary target mRNA, suggesting that RNAi and translational repression share common entry points if not overlapping machinery (Hutvagner and Zamore 2002). Because of these similarities, we reasoned that siRNAs may be capable of repressing gene expression via the miRNA-mediated pathway.
Results and Discussion
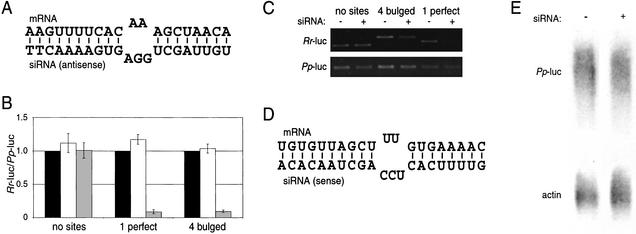
To test the ability of siRNAs to function like miRNAs in repressing gene expression, we designed a binding site that would base-pair to the antisense strand of a siRNA known to be active in vivo for cleavage of the cell-surface receptor CXCR4 mRNA (Fig. 1A). Notably, this binding site contains a central bulge, thereby precluding RISC-directed mRNA cleavage (Elbashir et al. 2001a; Holen et al. 2002). We introduced four of these binding sites as consecutive repeats separated by four nucleotides into the 3′ UTR of the Renilla reniformis luciferase reporter gene (Rr-luc); we also made a similar 3′ UTR construct with a single binding site with perfect complementarity, to serve as a positive control for RNAi activity. Transfection of HeLa cells and subsequent luciferase assays revealed that the CXCR4 siRNA induced at least 10-fold silencing of both of these constructs (Fig. 1B). Reverse transcriptase–PCR (RT–PCR) showed that the two constructs were suppressed by two different mechanisms, as the perfectly complementary antisense siRNA:mRNA interaction resulted in a significant decrease in the steady-state mRNA level, whereas the bulged interaction did not significantly reduce the mRNA level (Fig. 1C). Trace radio labeling of an independent RT–PCR experiment was also used to better quantitate RNA levels, normalizing first within a sample to the control Photinus pyralis luciferase (Pp-luc) and then across samples to the (−) siRNA transfection. Such quantitation revealed that the perfectly complementary construct, targeted for RNAi, showed a >10-fold decrease in RNA level, whereas the bulged construct showed only 1.2-fold reduction in RNA level (data not shown). Interestingly, the sense strand of the same CXCR4 siRNA was capable of repressing a mRNA with four bulged binding sites (Fig. 1D). However, in this case the level of repression was only 4-fold compared with the 10-fold repression observed above (data not shown). As an additional control, the four bulged CXCR4 binding sites (Fig. 1A) were introduced into the Pp-luc vector. Luciferase assays showed sixfold repression (data not shown). Northern analysis of cytoplasmic RNA confirmed that the bulged binding sites did not cause a decrease in mRNA levels, relative to the β-actin control (Fig. 1E). Thus, we conclude that a siRNA can function like a miRNA, repressing gene expression without a concordant decrease in mRNA stability.
Figure 1.
siRNAs translationally represses a target mRNA. (A) Schematic of the proposed interaction between a binding site engineered into the 3′ UTR of the target mRNA and the antisense strand of the CXCR4 siRNA. The thymidines at the 3′ end of the siRNA are deoxynucleotides. (B) Dual luciferase assay of transfected HeLa cells. Three Renilla reniformis luciferase (Rr-luc) constructs were used in this assay. One was unmodified (“no sites”), one contained a binding site perfectly complementary to the siRNA strand shown in A (“1 perfect”), and one contained four of the binding sites shown in A in tandem repeat (“4 bulged”). A Photinus pyralis luciferase (Pp-luc) served as an internal transfection control. The cells were transfected with no siRNA (black bars), a nonspecific (targeting GFP) siRNA (white bars), or the CXCR4 siRNA (gray bars). The ratios of Rr-luc to Pp-luc expression were normalized to the no siRNA transfections, ±S.E. from three independent experiments. (C) RT–PCR of harvested RNA. Total RNA was harvested from cells transfected with the constructs described in B, transfected with or without the CXCR4 siRNA. Control experiments demonstrate that DNA was successfully removed from the RNA preparation and that the PCR was in the linear range of amplification (data not shown). (D) Schematic of the proposed interaction between the sense strand of the CXCR4 siRNA and a designed binding site. (E) RNA analysis of Pp-luc with four bulged CXCR4 binding sites (shown in A), targeted for translational repression, transfected either with the CXCR4 siRNA (+) or no siRNA (−). RNA was detected by Northern analysis, probing for either Pp-luc or β-actin.
Cloning efforts in many labs have revealed a large library of miRNAs, yet C. elegans lin-4 and let-7 remain the only miRNAs with known mRNA targets for translational repression in animals, and no such interactions are known in mammals. Computational prediction of targets is difficult because the rules for miRNA:mRNA pairing which function in translational repression have not been determined. Systematic manipulation of genes encoding miRNAs to explore these rules is complicated because the mutant genes must be processed by Dicer, and the rules for this cleavage are not known. However, the ability of a siRNA to function by a miRNA-type pathway allows direct investigation of sequence and structure requirements for translational repression in the absence of Dicer processing.
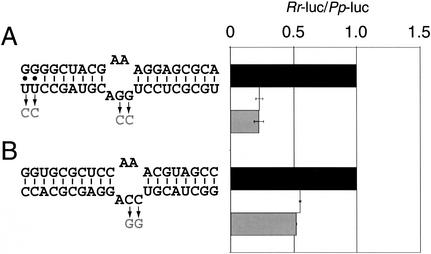
To begin to define these rules, different siRNA sequences were tested for their ability to repress reporters in the luciferase assay. Because both the more effective strand of the CXCR4 siRNA (Fig. 1A) and the only previously studied example of miRNA repression in mammalian cells (Zeng et al. 2002) had a 3′-AGG-5′ bulge in the siRNA strand when paired to the target mRNA, we tested the importance of this sequence. Two constructs were designed which would base-pair to the sense or antisense strand of a siRNA previously used to effectively target GFP mRNA for cleavage. The siRNA:mRNA interaction with the AGG bulge was twofold more effective than that with the ACC bulge (Fig. 2, cf. A and B). By using a different siRNA, the AGG bulge of the siRNA: mRNA interaction in Figure 2A was replaced with an ACC bulge, and the ACC bulge of the siRNA:mRNA interaction in Figure 2B was replaced with an AGG bulge. (We note that in Fig. 2A the two 3′ bases of the siRNA were changed from UU to CC.) Surprisingly, none of these changes had an effect on the degree of repression. Therefore, by this assay the sequence of the bulge is not the major determinant of translational repression activity.
Figure 2.
Analysis of sequence and structure rules for siRNA: mRNA interaction. HeLa cells were transfected with constructs containing four binding sites in tandem repeat with imperfect complementarity to either the antisense (A) or sense (B) strand of a GFP siRNA. The effect on luciferase expression is shown by the white bars, ±S.E. from two independent experiments, normalized to cells transfected with no siRNA (black bars). A different siRNA was then used to produce different bulges, shown in gray with arrows. These new interactions were assayed and are depicted with gray bars.
Since in Drosophila embryo extracts the antisense strand of the siRNA sets the ruler for cleavage of target mRNA, at the ninth nucleotide from its paired 5′ end (Elbashir et al. 2001b), the position of the bulge may be a critical determinant of translational repression activity. However, both the most effective and least effective bulges tested (Figs. 1A and 2B, respectively) position the bulge 8 bp from the 5′ end of the siRNA. Furthermore, another active construct positioned the bulge 9 bp from the 5′ end (Fig. 2A). We speculate that a combination of these sequence and structural parameters govern the ability of a siRNA/miRNA to induce translational repression, but that an expanded study will be necessary to define them.
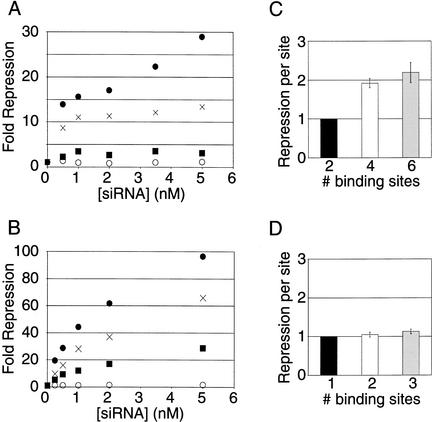
The number of miRNA binding sites in a target mRNA is a likely determinant of the effectiveness of translational repression. Indeed, the lin-14 3′ UTR contains seven potential lin-4 miRNA binding sites, and the lin-41 3′ UTR contains one lin-4 miRNA and two let-7 miRNA binding sites (for review, see Banerjee and Slack 2002). To investigate this possibility, a series of Rr-luc reporters with an increasing number of binding sites—zero, two, four, and six—were transfected into HeLa cells with increasing concentrations of CXCR4 siRNA. The level of repression increased with increasing number of binding sites and with increasing concentrations of siRNA (Fig. 3A). To compare the effectiveness of translational repression to mRNA cleavage by siRNAs, a series of Pp-luc reporters with an increasing number of binding sites—zero, one, two, and three—perfectly complementary to the CXCR4 siRNA were transfected with increasing concentrations of siRNA. Like the translational repression effect observed above, the level of gene silencing by RNAi increases with increasing number of perfectly complementary binding sites and with increasing concentration of siRNA (Fig. 3B). As might be expected from a mechanism that results in cleavage of the mRNA, RNAi silences gene expression to a greater extent than translational repression.
Figure 3.
Comparison of RNAi and translational repression. (A) Titration of Renilla reniformis luciferase (Rr-luc) constructs containing zero (○), two (▪), four (×), or six (●) of the bulged binding sites, for pairing with the antisense strand of the CXCR4 siRNA, as depicted in Figure 1A. The level of repression achieved is plotted, normalized to cells transfected with no siRNA. (B) Titration of Pp-luc constructs containing zero (○), one (▪), two (×), or three (●) binding sites perfectly complementary to the antisense strand of the CXCR4 siRNA (Fig. 1A). (C) Analysis of the relative repression each site contributes for the data presented in A, normalized to the construct with two binding sites, ±S.E. (D) Analysis of the relative repression each site contributes for the data presented in B, normalized to the construct with one binding site, ±S.E.
The mechanism of mRNA cleavage in RNAi implies that each siRNA:binding site interaction will function independently of another interaction; once a mRNA is cleaved, it is expected to be rapidly degraded. Thus, a second cleavage event would have little if any effect on gene expression. To assess this, we divided the repression observed for each construct in Figure 3B by the number of binding sites on that mRNA, at each concentration of siRNA. These values were then normalized to the repression observed for a single binding site to assess the relative contribution of each site (Fig. 3D). As expected, the relative effectiveness of each site remained the same as the number of binding sites increased. Addition of more binding sites likely only increases the probability of the single necessary cleavage event, and thus multiple binding sites function independently of one another. This same analysis was applied to the translational repression constructs in Figure 3A, normalizing to the construct with two binding sites (Fig. 3C). Strikingly, the degree of repression achieved by increasing the number of sites is not simply additive, as each site in the construct with four binding sites conferred twice as much repression as each site in the construct with two binding sites. The effectiveness of each binding site in the construct with six sites was similar to that of the construct with four sites. These results suggest that the effects of binding multiple miRNA complexes to the 3′ UTR are likely to be cooperative. Ribonucleoprotein complexes could either mutually stabilize one another or cooperatively interact to more effectively inhibit translation or both. As with other cooperative interactions in gene regulation, this would allow a cell to fine-tune the expression of a mRNA by regulating the degree of binding of different miRNAs to the 3′ UTR of the mRNA.
The discovery that siRNAs can function in translational repression as miRNAs, and that the sequence requirements for this interaction are less stringent than those for RNAi, may help to explain nonspecific effects sometimes observed in experiments using siRNAs for targeted gene silencing. Using an arbitrary 21-nt sequence, BLAST searches against the mRNAs predicted from the human genome identify multiple inexact matches with 16–18 nt complementarity. Combined with the potential of G-U wobble base pairs, and depending on the overall sequence rules for translational repression, there may be translational repression of a number of off-target genes by the introduction of a siRNA intended to knock down the expression of only the target gene. However, the mechanistic finding that several binding sites are needed to produce a significant effect on protein expression may make nonspecific siRNA effects the exception rather than the rule, and to date, siRNAs have certainly been used with ostensible specificity.
Materials and methods
DNA constructs and siRNAs
3′ UTR binding sites for the siRNAs were constructed by a multimerization of DNA oligonucleotides (IDT), gel purification, PCR amplification, and restriction digestion. The products were inserted into the XbaI site immediately downstream of the stop codon in either the pRL-TK vector coding for Rr-luc or the pGL3 control vector coding for Pp-luc (Promega). siRNAs were purchased as single strands, deprotected, and annealed according to the manufacturer (Dharmacon). All sequences for siRNAs and 3′ UTR constructs used in this study are available on the Phil Sharp Lab Web site at http://web.mit.edu/sharplab/RNAi/sequences.html.
Cell culture and transfections
Adherent HeLa cells were grown in 10% IFS in DMEM, supplemented with glutamine in the presence of antibiotics. For all transfections, except those noted below, cells were transfected with lipofectin and the PLUS reagent (Invitrogen). On the day before transfection, exponentially growing cells were trypsinized and plated into 24-well plates at a density of 3 × 104 cells/well in antibiotic-free media. The next day, the cells were transfected with 0.2 μg DNA and 25 nM siRNA in a final volume of 250 μL. For Figures 1E and 3, cells were transfected with Lipofectamine 2000 because during the course of this study we found that this reagent delivers effective doses of siRNAs at lower concentrations. On the day before transfection, cells were trypsinized and plated into 24-well plates at a density of 8 × 104 cells/well in antibiotic-free media. The next day cells were transfected with 0.8 μg DNA and 5 nM siRNA, unless noted, in a final volume of 500 μL.
Luciferase assays
Dual-luciferase assays (Promega) were performed 24 h after transfection according to the manufacturer's protocol and detected with an Optocomp I luminometer (MGM Instruments). Rr-luc target vectors were cotransfected with control pGL3, and Pp-luc target vectors were cotransfected with a pRL-CMV control (Promega). Transfections were harvested 24 h after transfection, and the two luciferase activities consecutively assayed.
RT–PCR
Total RNA was harvested from transfected HeLa cells by using the RNAeasy kit (Qiagen). Total RNA was DNase-treated twice with DNase-Free (Ambion) and was reverse-transcribed by using Omniscript reverse transcriptase (Qiagen) with a DNA primer complementary to a region near the SV40 polyadenylation sequence found in both the Pp-luc and Rr-luc reporter vectors (5′-GCATTCTAGTTGTGGTTTGTCC-3′). Trace radio-labeled PCR products were detected via autoradiography, and quantitated with ImageQuant software version 1.2 (Molecular Dynamics).
Northern analysis
Cytoplasmic RNA was harvested by hypotonic lysis without detergent and subsequent needle homogenization of HeLa cells 24 h after transfected using Lipofectamine 2000. Nuclei were pelleted at 1,500g for 15 min, and the supernatant was treated with proteinase K, extracted in phenol:chloroform and again in chloroform, precipitated with isopropanol, and washed with 70% ethanol. Samples were then treated with DNase-Free (Ambion). Northern analysis was performed by using the NorthernMax kit (Ambion). Ten micrograms of RNA from the (+) siRNA or (−) siRNA samples were separated by electrophoresis on a 1% formaldehyde agarose gel and transferred onto Hybond N+ nitrocellulose by downward transfer (Amersham Pharmacia). The 1.5-kb ORF of the Pp-luc cDNA was generated by restriction digest of the pGL3 control vector with XbaI and HindIII (New England Biolabs), and used with DECA-Prime II (Ambion) in the presence of 32P-dATP to generate a random-primed DNA used to probe the membrane. The membrane was stripped and reprobed with β-actin probe, generated from DECAtemplate-β-actin-mouse (Ambion).
Acknowledgments
We thank A. Grishok for helpful discussion and C. Novina, D. M. Dykxhoorn, H. Houbaviy, D. Tantin, and R. Bodner for comments on the manuscript. J.G.D. is a Howard Hughes Medical Institute Predoctoral Fellow. C.P.P. is a National Science Foundation Predoctoral Fellow. This work was supported by U.S. Public Health Service MERIT Award R37-GM34277 from the National Institutes of Health and PO1-CA42063 from the National Cancer Institute to P.A.S., and partially by Cancer Center Support (core) grant P30-CA14051 from the National Cancer Institute.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL sharppa@mit.edu; FAX (617) 253-3867.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.1064703.
References
- Aravin AA, Naumova NM, Tulin AV, Vagin VV, Rozovsky YM, Gvozdev VA. Double-stranded RNA-mediated silencing of genomic tandem repeats and transposable elements in the D. melanogaster germline. Curr Biol. 2001;11:1017–1027. doi: 10.1016/s0960-9822(01)00299-8. [DOI] [PubMed] [Google Scholar]
- Banerjee D, Slack F. Control of developmental timing by small temporal RNAs: A paradigm for RNA-mediated regulation of gene expression. Bioessays. 2002;24:119–129. doi: 10.1002/bies.10046. [DOI] [PubMed] [Google Scholar]
- Baulcombe D. RNA silencing: Diced defence. Nature. 2001;409:295–296. doi: 10.1038/35053256. [DOI] [PubMed] [Google Scholar]
- Bernstein E, Caudy AA, Hammond SM, Hannon GJ. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature. 2001;409:363–366. doi: 10.1038/35053110. [DOI] [PubMed] [Google Scholar]
- Elbashir SM, Harborth J, Lendeckel W, Yalcin A, Weber K, Tuschl T. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature. 2001a;411:494–498. doi: 10.1038/35078107. [DOI] [PubMed] [Google Scholar]
- Elbashir SM, Martinez J, Patkaniowska A, Lendeckel W, Tuschl T. Functional anatomy of siRNAs for mediating efficient RNAi in Drosophila melanogaster embryo lysate. EMBO J. 2001b;20:6877–6888. doi: 10.1093/emboj/20.23.6877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391:806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- Grishok A, Pasquinelli AE, Conte D, Li N, Parrish S, Ha I, Baillie DL, Fire A, Ruvkun G, Mello CC. Genes and mechanisms related to RNA interference regulate expression of the small temporal RNAs that control C. elegans developmental timing. Cell. 2001;106:23–34. doi: 10.1016/s0092-8674(01)00431-7. [DOI] [PubMed] [Google Scholar]
- Ha I, Wightman B, Ruvkun G. A bulged lin-4/lin-14 RNA duplex is sufficient for Caenorhabditis elegans lin-14 temporal gradient formation. Genes & Dev. 1996;10:3041–3050. doi: 10.1101/gad.10.23.3041. [DOI] [PubMed] [Google Scholar]
- Hamilton AJ, Baulcombe DC. A species of small antisense RNA in posttranscriptional gene silencing in plants. Science. 1999;286:950–952. doi: 10.1126/science.286.5441.950. [DOI] [PubMed] [Google Scholar]
- Hammond SM, Bernstein E, Beach D, Hannon GJ. An RNA-directed nuclease mediates post-transcriptional gene silencing in Drosophila cells. Nature. 2000;404:293–296. doi: 10.1038/35005107. [DOI] [PubMed] [Google Scholar]
- Holen T, Amarzguioui M, Wiiger MT, Babaie E, Prydz H. Positional effects of short interfering RNAs targeting the human coagulation trigger tissue factor. Nucleic Acids Res. 2002;30:1757–1766. doi: 10.1093/nar/30.8.1757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutvagner G, Zamore PD. A microRNA in a multiple-turnover RNAi enzyme complex. Science. 2002;297:2056–2060. doi: 10.1126/science.1073827. [DOI] [PubMed] [Google Scholar]
- Hutvagner G, McLachlan J, Pasquinelli AE, Balint E, Tuschl T, Zamore PD. A cellular function for the RNA-interference enzyme Dicer in the maturation of the let-7 small temporal RNA. Science. 2001;293:834–838. doi: 10.1126/science.1062961. [DOI] [PubMed] [Google Scholar]
- Ketting RF, Haverkamp TH, van Luenen HG, Plasterk RH. Mut-7 of C. elegans, required for transposon silencing and RNA interference, is a homolog of Werner syndrome helicase and RNaseD. Cell. 1999;99:133–141. doi: 10.1016/s0092-8674(00)81645-1. [DOI] [PubMed] [Google Scholar]
- Ketting RF, Fischer SE, Bernstein E, Sijen T, Hannon GJ, Plasterk RH. Dicer functions in RNA interference and in synthesis of small RNA involved in developmental timing in C. elegans. Genes & Dev. 2001;15:2654–2659. doi: 10.1101/gad.927801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagos-Quintana M, Rauhut R, Lendeckel W, Tuschl T. Identification of novel genes coding for small expressed RNAs. Science. 2001;294:853–858. doi: 10.1126/science.1064921. [DOI] [PubMed] [Google Scholar]
- Lau NC, Lim LP, Weinstein EG, Bartel DP. An abundant class of tiny RNAs with probable regulatory roles in Caenorhabditis elegans. Science. 2001;294:858–862. doi: 10.1126/science.1065062. [DOI] [PubMed] [Google Scholar]
- Lee RC, Ambros V. An extensive class of small RNAs in Caenorhabditis elegans. Science. 2001;294:862–864. doi: 10.1126/science.1065329. [DOI] [PubMed] [Google Scholar]
- Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–854. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- Lee Y, Jeon K, Lee JT, Kim S, Kim VN. MicroRNA maturation: Stepwise processing and subcellular localization. EMBO J. 2002;21:4663–4670. doi: 10.1093/emboj/cdf476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez J, Patkaniowska A, Urlaub H, Luhrmann R, Tuschl T. Single-stranded antisense siRNAs guide target RNA cleavage in RNAi. Cell. 2002;110:563–574. doi: 10.1016/s0092-8674(02)00908-x. [DOI] [PubMed] [Google Scholar]
- Mourelatos Z, Dostie J, Paushkin S, Sharma A, Charroux B, Abel L, Rappsilber J, Mann M, Dreyfuss G. miRNPs: A novel class of ribonucleoproteins containing numerous microRNAs. Genes & Dev. 2002;16:720–728. doi: 10.1101/gad.974702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen PH, Ambros V. The lin-4 regulatory RNA controls developmental timing in Caenorhabditis elegans by blocking LIN-14 protein synthesis after the initiation of translation. Dev Biol. 1999;216:671–680. doi: 10.1006/dbio.1999.9523. [DOI] [PubMed] [Google Scholar]
- Reinhart BJ, Slack FJ, Basson M, Pasquinelli AE, Bettinger JC, Rougvie AE, Horvitz HR, Ruvkun G. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature. 2000;403:901–906. doi: 10.1038/35002607. [DOI] [PubMed] [Google Scholar]
- Reinhart BJ, Weinstein EG, Rhoades MW, Bartel B, Bartel DP. MicroRNAs in plants. Genes & Dev. 2002;16:1616–1626. doi: 10.1101/gad.1004402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slack FJ, Basson M, Liu Z, Ambros V, Horvitz HR, Ruvkun G. The lin-41 RBCC gene acts in the C. elegans heterochronic pathway between the let-7 regulatory RNA and the LIN-29 transcription factor. Mol Cell. 2000;5:659–669. doi: 10.1016/s1097-2765(00)80245-2. [DOI] [PubMed] [Google Scholar]
- Tabara H, Sarkissian M, Kelly WG, Fleenor J, Grishok A, Timmons L, Fire A, Mello CC. The rde-1 gene, RNA interference, and transposon silencing in C. elegans. Cell. 1999;99:123–132. doi: 10.1016/s0092-8674(00)81644-x. [DOI] [PubMed] [Google Scholar]
- Tuschl T, Zamore PD, Lehmann R, Bartel DP, Sharp PA. Targeted mRNA degradation by double-stranded RNA in vitro. Genes & Dev. 1999;13:3191–3197. doi: 10.1101/gad.13.24.3191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wightman B, Ha I, Ruvkun G. Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans. Cell. 1993;75:855–862. doi: 10.1016/0092-8674(93)90530-4. [DOI] [PubMed] [Google Scholar]
- Zamore PD, Tuschl T, Sharp PA, Bartel DP. RNAi: double-stranded RNA directs the ATP-dependent cleavage of mRNA at 21 to 23 nucleotide intervals. Cell. 2000;101:25–33. doi: 10.1016/S0092-8674(00)80620-0. [DOI] [PubMed] [Google Scholar]
- Zeng Y, Wagner EJ, Cullen BR. Both natural and designed micro RNAs can inhibit the expression of cognate mRNAs when expressed in human cells. Mol Cell. 2002;9:1327–1333. doi: 10.1016/s1097-2765(02)00541-5. [DOI] [PubMed] [Google Scholar]