Abstract
The abundant and chromatin-associated protein HCF-1 is a critical player in mammalian cell proliferation as well as herpes simplex virus (HSV) transcription. We show here that separate regions of HCF-1 critical for its role in cell proliferation associate with the Sin3 histone deacetylase (HDAC) and a previously uncharacterized human trithorax-related Set1/Ash2 histone methyltransferase (HMT). The Set1/Ash2 HMT methylates histone H3 at Lys 4 (K4), but not if the neighboring K9 residue is already methylated. HCF-1 tethers the Sin3 and Set1/Ash2 transcriptional regulatory complexes together even though they are generally associated with opposite transcriptional outcomes: repression and activation of transcription, respectively. Nevertheless, this tethering is context-dependent because the transcriptional activator VP16 selectively binds HCF-1 associated with the Set1/Ash2 HMT complex in the absence of the Sin3 HDAC complex. These results suggest that HCF-1 can broadly regulate transcription, both positively and negatively, through selective modulation of chromatin structure.
Keywords: HCF-1, Sin3, Ash2, Set1, VP16, histone methyltransferase
Cells proliferate via a process of cell growth culminating in cell division. Both elements of this process are orchestrated by coordinated patterns of gene transcription. In eukaryotes, transcriptional regulation results from the ability of transcriptional activators and repressors to change chromatin structure and recruit RNA polymerase to specific promoters. Many of the changes in chromatin structure induced by transcriptional regulators involve complex patterns of histone modification by enzymes such as acetyltransferases (HATs), methyltransferases (HMTs), and kinases, and these modifications can often be removed by opposing activities such as deacetylases (HDACs) and phosphatases. These varying patterns of histone modification are often referred to as the histone code and lead to differing patterns of activated or repressed gene transcription (Jenuwein and Allis 2001).
Here, we describe an analysis of HCF-1, an important but poorly understood regulator of cell proliferation. HCF-1 (also referred to as C1, CFF, and VCAF) is a highly conserved protein in metazoans that was originally discovered as a host-cell factor required for herpes simplex virus (HSV) transcription (Herr 1998). It is an abundant chromatin-bound protein that plays a critical role in both cell growth and division. In humans, HCF-1 is unusual in that it is initially synthesized as a single large precursor molecule of 2035 amino acids that is subsequently cleaved at six centrally located HCF-1PRO repeats (Wilson et al. 1993a, 1995; Kristie et al. 1995). These cleavages result in stable N- (HCF-1N) and C- (HCF-1C) terminal subunits that remain noncovalently associated (Wilson et al. 1995).
In HSV infection, HCF-1 binds the virion protein VP16, a prototypical transcriptional activator, and stabilizes a complex with the cellular transcription factor Oct-1 on VP16-responsive elements found in HSV immediate-early (IE) promoters (for review, see O'Hare 1993; Herr 1998). To recruit HCF-1 to the VP16-induced complex, VP16 binds to an N-terminal region of HCF-1—called the Kelch domain owing to its similarity to the Drosophila protein Kelch—the same region that, in an uninfected cell, is responsible for tethering HCF-1 to chromatin (LaBoissière et al. 1997; Wilson et al. 1997; Wysocka et al. 2001). Although HCF-1 is known to stabilize the VP16-induced complex, what function it plays when bound to chromatin in an uninfected cell is poorly understood.
HCF-1 involvement in both cell growth and division is known because a single proline-to-serine substitution in the HCF-1 Kelch domain (P134S) causes a temperature-induced cell-growth arrest (Goto et al. 1997) and cytokinesis defect (Reilly and Herr 2002) in the hamster-cell line tsBN67, although the arrest only occurs 36 to 48 h after transfer to the nonpermissive temperature. This mutation also disrupts HCF-1 association with VP16 (Wilson et al. 1997) and with chromatin at the nonpermissive temperature (Wysocka et al. 2001). The disruption of chromatin association at nonpermissive temperature occurs prior to the tsBN67-cell proliferation arrest, suggesting that loss of HCF-1 chromatin association is a primary cause of the tsBN67-cell proliferation defect (Wysocka et al. 2001).
A transcriptional regulatory role of HCF-1 in cell proliferation has been emphasized by the finding that the SV40 large T antigen and adenoviral E1A oncoproteins can rescue tsBN67-cell proliferation at the nonpermissive temperature by inactivating the pRb family of proteins (Reilly et al. 2002). This regulatory role of HCF-1 involves at least two separate functional regions of the HCF-1N subunit: the Kelch domain, which is both necessary and sufficient to direct HCF-1 to chromatin (Wysocka et al. 2001), and a neighboring “Basic” region (Wilson et al. 1997), which is hypothesized to recruit important regulatory proteins to the sites targeted by the Kelch domain (Wysocka et al. 2001).
We show here that HCF-1 naturally associates with both HDAC and HMT chromatin-modifying activities: the Sin3 HDAC complex commonly involved in repression of gene transcription and a heretofore uncharacterized human histone H3 Lys 4 (K4) HMT complex that is related to the Saccharomyces cerevisiae Set1/Ash2 trithorax-group-related HMT complex—an important activator of protein-encoding gene transcription in yeast. These results suggest that, as an abundant chromatin-associated factor, HCF-1 accomplishes at least some of its key roles in cell proliferation by broadly activating and repressing transcription, at least in part, through the control of chromatin states.
Results
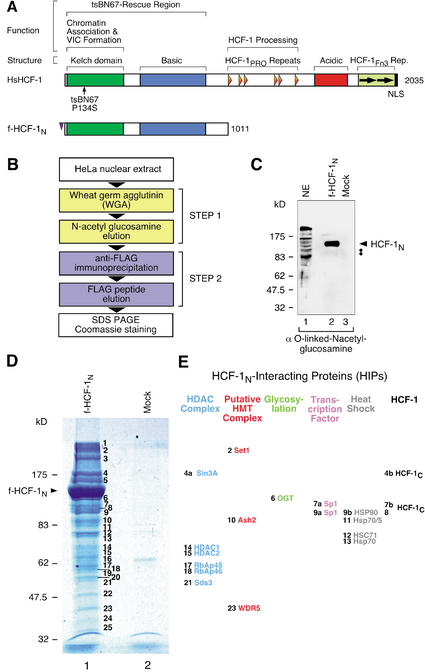
Figure 1A shows a schematic of human HCF-1 in which structural and functional features of this protein are highlighted. To identify effector molecules responsible for HCF-1's ability to promote cell proliferation, we used a previously described HeLa-cell line stably expressing a Flag-epitope-tagged HCF-1N subunit (f-HCF-1N; Fig. 1A) at levels similar to the endogenous HCF-1 protein (Wysocka et al. 2001). Purification of f-HCF-1N subunit-associated proteins presented several advantages: (1) it identifies proteins that associate with the HCF-1N subunit, which is sufficient to promote proper tsBN67-cell proliferation (Wilson et al. 1997; Reilly and Herr 2002); (2) the f-HCF-1N subunit obviates the complicated pattern of endogenous HCF-1N and HCF-1C subunits, which could obscure associated polypeptides; and (3) the Flag-epitope tag provides a very efficient purification step.
Figure 1.
Identification of the HCF-1-interacting proteins (HIPs). (A) Overall structures of HCF-1 and f-HCF-1N. (Upper diagram) Human (Hs) HCF-1. Above are shown structural or sequence elements and functional regions. Below, the position of the tsBN67-cell P134S missense mutation is indicated. (Lower diagram) The Flag-epitope-tagged HCF-1N subunit used in this study. VIC, VP16-induced complex; HCF-1PRO, HCF-1 proteolytic processing repeats; HCF-1Fn3 Rep., HCF-1 fibronectin type 3 repeats. (B) Two-step purification scheme for f-HCF-1N-associated polypeptides. (C) f-HCF-1N is modified by O-linked N-acetyl-glucosamine residues and is a major glycoprotein in the HIPs purification. Two-step purified f-HCF-1N or mock samples were analyzed by immunoblotting with an anti-O-linked N-acetyl-glucosamine antibody. The position of the f-HCF-1N subunit is indicated by the arrowhead. A weak doublet, displaying the correct migration for Sp1 polypeptides, is indicated by the two dots. NE, HeLa nuclear extract. (D) f-HCF-1N-associated proteins were purified from 2.4 × 1010 f-HCF-1N or mock HeLa cells by the two-step procedure outlined. Polypeptides were resolved by SDS-PAGE and stained with Coomassie blue. The positions of the molecular weight markers and f-HCF-1N species are indicated on the left. The 25 additional bands present in the f-HCF-1N-purified material are labeled 1–25 and were analyzed by mass spectrometry. Only two polypeptides could be visualized in the mock-purified sample, and they were subsequently identified as Hsp70 (minor species) and nuclear lamin (major species). (E) Identity of a subset of HIPs. Sixteen out of 33 unique polypeptides identified by HIP mass spectrometry analysis are grouped into six categories: members of the Sin3 HDAC complex (blue), human homologs of the Saccharomyces cerevisiae Set1/Ash2 HMT complex (red), O-linked N-acetyl-glucosamine transferase (OGT; green), transcription factor Sp1 (purple), heat-shock proteins (gray), and endogenous HCF-1C subunits (black). The number to the left of each species represents the stained band from which each protein originates.
Purification of the HCF-1N subunit and associated polypeptides
To purify HCF-1N-associated proteins, we used a two-step purification procedure of salt-extracted nuclear proteins as shown in Figure 1B. For the first step, we established that, like endogenous HCF-1 (Wilson et al. 1993a), the f-HCF-1N subunit binds quantitatively to wheat-germ agglutinin (WGA) and can be efficiently eluted with N-acetyl glucosamine, providing a simple, efficient, and gentle first purification step. For the second step, we purified the f-HCF-1N subunit and its associated proteins by Flag-epitope-specific immunoprecipitation followed by Flag-peptide elution. To monitor the specificity of this two-step procedure, extracts from a matched cell line directing the synthesis of the Flag epitope alone were used in parallel mock purifications. Figure 1C indicates why the first step of the purification is probably so efficient: Purified material was probed by immunoblot with an O-linked N-acetyl-glucosamine antibody, and the major glycosylated species is the f-HCF-1N polypeptide. Our purification procedure was very reproducible, as evidenced by numerous independent purifications used in the studies described below.
Identity of the HCF-1N-interacting proteins (HIPs)
Following HCF-1N-complex purification, the resulting polypeptides were separated by SDS-PAGE and visualized by staining with Coomassie blue as shown in Figure 1D. In addition to the f-HCF-1N subunit, we identified 25 other bands (labeled 1–25; Fig. 1D, lane 1). These bands were not present in the parallel mock purification (Fig. 1D, cf. lanes 1 and 2) demonstrating that they represent specific protein association with the f-HCF-1N polypeptide. We refer to these HCF-1-interacting proteins as HIPs.
To identify the HIPs, we excised the stained bands from the polyacrylamide gel, subjected the entrapped polypeptides to in-gel digestion with trypsin, and analyzed the resulting peptides by MALDI-TOF and liquid chromatography tandem mass spectrometry (LC-MS/MS). We identified a total of 33 unique polypeptides, of which five are presently uncharacterized hypothetical proteins in the databases. Of the remaining 28 proteins, 16 are listed alongside the polyacrylamide gel in Figure 1E and are described here. The others will be described elsewhere. For each of the proteins listed in Figure 1E, we identified multiple peptides. These proteins fall into six categories: (1) endogenous HCF-1C subunits, (2) heat-shock proteins, (3) the transcription factor Sp1, (4) O-linked N-acetyl-glucosamine transferase (OGT), (5) human homologs of the Saccharomyces cerevisiae Set1/Ash2 HMT complex, and (6) members of the Sin3 HDAC complex.
Except for the heat-shock proteins, we confirmed the mass spectrometry results by immunoblot analysis for representatives of each category of associated protein using material purified separately. In all cases tested, proteins identified by mass spectrometry were present in the f-HCF-1N purification and absent in the mock-purified sample, demonstrating the robust HCF-1N-subunit specificity of each association. For example, Figure 2A shows that all of the different HCF-1C subunits, but not the HCF-1300 precursor, were present in the f-HCF-1N, but not the mock sample (Fig. 2A, panel a, cf. lanes 1 and 2), suggesting a low level of binding of processed HCF-1C subunits with the f-HCF-1N protein.
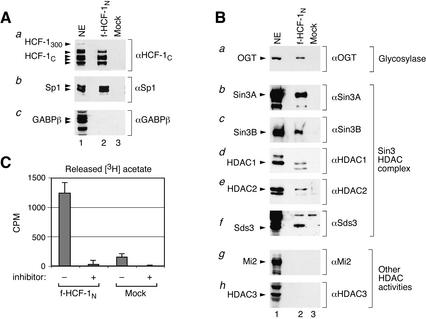
Figure 2.
Association of the f-HCF-1N subunit with endogenous HCF-1C subunits, Sp1, OGT, and the Sin3 HDAC complex, and histone deacetylase activity. (A) Two-step f-HCF-1N (lane 2) and mock (lane 3) purified samples were analyzed by immunoblotting with the antibodies against the HCF-1C subunit (panel a), Sp1 (panel b), and GABPβ (panel c). Labeled arrowheads identify polypeptides of interest. NE, HeLa nuclear extract (lane 1) used as a positive control. (B) The f-HCF-1N subunit specifically associates with the Sin3 HDAC complex. Samples as in A were analyzed by immunoblotting with antibodies against OGT (panel a), Sin3A, Sin3B, HDAC1, HDAC2, and Sds3 components of the Sin3 HDAC complex (panels b–f), a component of the NuRD complex Mi-2 (panel g), and the HDAC1/2-related class I histone deacetylase HDAC3 (panel h). Labeled arrowheads identify polypeptides of interest. NE, HeLa nuclear extract (lane 1) corresponding to 5% of the samples in lanes 2 and 3 was used as a positive control. (C) HCF-1 associates with HDAC activity. Two-step purified f-HCF-1N and mock-purified samples were added to reactions with [3H]-acetylated histone H4 peptide in the presence or absence of deacetylase inhibitor. Counts above the background CPM released from the acetylated peptide incubated alone are shown. The average of three experiments is shown.
Two well-characterized transcription factors have been shown to associate with the Basic region of HCF-1N in yeast two-hybrid assays: Sp1 (Gunther et al. 2000) and GABPβ (Vogel and Kristie 2000). We identified Sp1 in the HIP mass spectrometry analysis but not GABPβ. Consistent with these results, we detect Sp1 but not GABPβ by immunoblot analysis (Fig. 2A, lane 2, panels b,c). Thus, in contrast to Spl, the association of GABPβ with HCF-1, if it occurs in HeLa cells, apparently does not survive our high-salt purification conditions.
Association of O-linked N-acetyl-glucosamine transferase with the HCF-1N subunit
The regulatory role of nuclear protein glycosylation is poorly understood (Wells et al. 2001). Interestingly, although the f-HCF-1N protein had to be already glycosylated to have been purified during the first step of our two-step purification procedure (Fig. 1B), the OGT enzyme remains associated with the HCF-1N subunit (Fig. 2B, panel a, lane 2). Thus, OGT may play roles in the function of HCF-1 in addition to its glycosylation. Recently, OGT has been shown to associate with the Sin3 HDAC complex (Yang et al. 2002), which also associates with HCF-1. Perhaps HCF-1, OGT, and the Sin3 HDAC complex cooperate to effect specific regulatory outcomes.
HCF-1N-subunit association with the Sin3 HDAC complex and histone deacetylase activity
The HIP analysis identified six members of the Sin3 HDAC complex (Fig. 1E; for review, see Knoepfler and Eisenman 1999; Ng and Bird 2000): Sin3A, the HDAC catalytic subunits HDAC1 and HDAC2, the scaffolding proteins RbAp46 and RbAp48, and the recently described regulatory protein Sds3 (Alland et al. 2002). Figure 2B (panels b,d–f, lanes 2,3) confirms the specificity of the HCF-1N interaction with four of these members: Sin3A, HDAC1, HDAC2, and Sds3. Although not detected in the mass spectrometry analysis, immunoblot analyses showed that the Sin3-complex-associated protein SAP30 (data not shown) and the Sin3A-related protein Sin3B (Fig. 2B, panel c, lane 2) also associate with the HCF-1N subunit. Thus, it is likely that the entire core Sin3A and Sin3B HDAC complexes, referred to here generally as the Sin3 complex, are associated with the HCF-1N subunit. In contrast, other related HDAC class I complexes, in particular the NuRD complex represented here by the Mi2 subunit (Fig. 2B, panel g, lane 2) or the HDAC3 deacetylase (Fig. 2B, panel h, lane 2) are not associated with the HCF-1N subunit. Indeed, no other histone deacetylase-related or -associated protein was identified in the HIP analysis (data not shown), indicating that HCF-1 is specifically associated with the Sin3 HDAC complex.
To determine whether the HCF-1N-associated Sin3 HDAC complex possesses HDAC activity, we assayed deacetylation of an acetylated histone H4 peptide by the purified HCF-1N complex as shown in Figure 2C. Consistent with associated HDAC activity, the f-HCF-1N-purified material displayed significant sodium butyrate-sensitive deacetylase activity in comparison to the mock-purified material. Thus, when HCF-1 is associated with the Sin3 HDAC complex, it is probably able to direct histone deacetylation.
HCF-1N-Basic region interaction with the Sin3A PAH1 domain
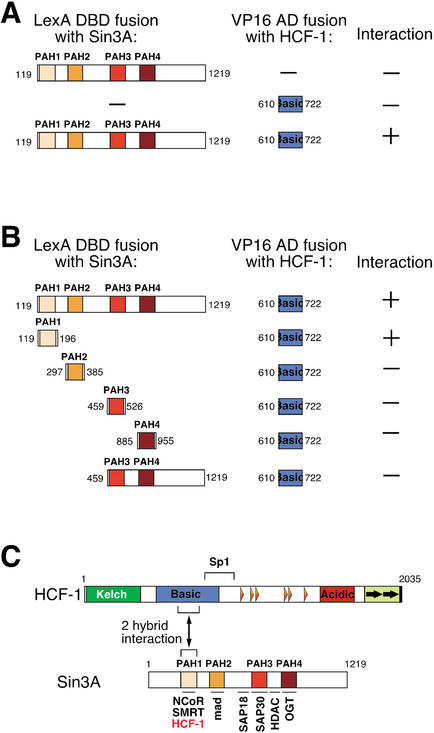
The two mammalian Sin3 proteins, Sin3A and Sin3B, are modular proteins containing four paired amphipathic α-helices (named PAH 1–4) that mediate specific sets of protein–protein interactions as shown in Figure 3C (for review, see Knoepfler and Eisenman 1999; Ng and Bird 2000). To identify Sin3A-interacting proteins, a yeast two-hybrid screen was performed. In this screen, an activation domain–mouse embryonic cDNA fusion library was probed using mouse Sin3A encompassing all four PAH domains fused to the lexA DNA-binding domain as bait (Laherty et al. 1998; see Materials and Methods). From this screen, four identical cDNA clones encoding mouse HCF-1 residues 610–722 were isolated (Fig. 3A). These residues, with one exception (Kristie 1997), are identical in human HCF-1 and lie in the center of the HCF-1 Basic region separate from the two-hybrid Sp1-interacting sequences (Gunther et al. 2000; Fig. 3C). As shown in Figure 3B, directed two-hybrid assays against the different Sin3A PAH1–4 domains demonstrated that this HCF-1 Basic region interacts selectively with the PAH1 domain. Thus, like the nuclear-hormone-receptor corepressors N-CoR and SMRT (Heinzel et al. 1997; Nagy et al. 1997), HCF-1 associates, probably directly, with the Sin3A PAH1 domain (Fig. 3C).
Figure 3.
Association of Sin3A PAH1 with the HCF-1 Basic region in a yeast two-hybrid assay. (A) The LexA DNA-binding domain fused to mouse Sin3A amino acids 119–1219 (PAH1-E) was used as a bait to screen a library of mouse embryo cDNAs fused to the VP16 transactivation domain. Four identical positive clones encoding a portion of the HCF-1 Basic region were identified. (B) Directed two-hybrid assays were performed to map the Sin3A-interaction domain of HCF-1 Basic region using LexA fusions containing different regions of Sin3A: PAH1, PAH2, PAH3, PAH4, and PAH3-E (see Materials and Methods). (C) Schematic representations of HCF-1 and Sin3A are shown. The HCF-1 Sp1-interaction region is indicated above the diagram. Four paired amphipatic PAH1–4 helices of Sin3A are depicted. Regions of Sin3A involved in association with various protein partners are shown below the Sin3A diagram. Regions of HCF-1 and Sin3A that interact in the yeast two-hybrid assay are indicated.
The HCF-1N subunit binds human homologs of the yeast Set1/Ash2 HMT complex
In addition to the Sin3 HDAC complex, we discovered f-HCF-1N-associated proteins that share sequence similarity to a recently described S. cerevisiae trithorax-group-related Set1/Ash2 HMT complex (Miller et al. 2001; Roguev et al. 2001; Nagy et al. 2002). Components of the yeast Set1/Ash2 complex are shown in Figure 4A. Although related protein sequences can be found in the human protein databases or have been molecularly characterized to a limited extent on an individual basis (Ikegawa et al. 1999; Gori et al. 2001; Wang et al. 2001; Zhang and Reinberg 2001; Kouzarides 2002), no human Set1/Ash2 HMT activity has been reported.
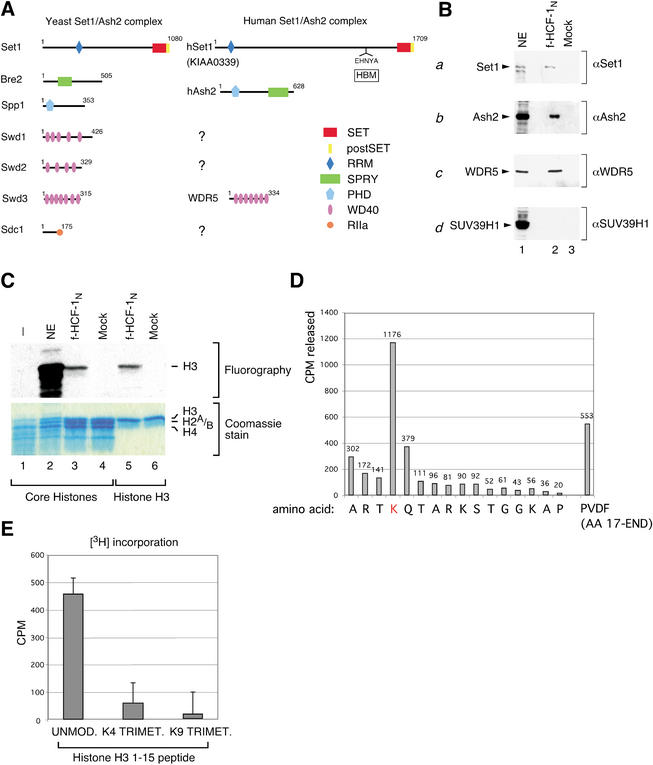
Figure 4.
Association of HCF-1 with the human homologs of components of the Saccharomyces cerevisiae Set1/Ash2 HMT complex and HMT activity. (A) Similarity between components of the S. cerevisiae Set1/Ash2 complex and human proteins identified among the HIPs. Schematic representations of yeast Set1/Ash2 complex proteins, and related HIPs (hSet1, accession no. gi 6683126; hAsh2, accession no. gi 4757790; and WDR5, accession no. gi 16554627) are shown on the left and right, respectively. Structural domains and motifs are shown in each protein and defined by the key on the bottom right. RRM, RNA recognition motif; SET, Set domain; postSET, Set domain-associated cysteine-rich motif; HBM, HCF-1-binding motif; PHD, PHD zinc-finger motif; SPRY, domain in SPla and the RYanodine receptor; WD40, WD40 repeats; RIIa, protein kinase A regulatory subunit dimerization domain motif. (B) Two-step purified f-HCF-1N (lane 2) and mock (lane 3) samples were analyzed by immunoblot with anti-human Set1 (panel a), Ash2 (panel b), WDR5 (panel c), and SUV39H1 (panel d) antibodies. Labeled arrowheads indicate polypeptides of interest; NE, HeLa nuclear extract (lane 1) corresponding to 5% of the samples in lanes 1 and 2 was used as a positive control. (C) HCF-1 associates with HMT activity. Two-step f-HCF-1N and mock HeLa-cell purified samples were incubated with core histones or recombinant histone H3 in the presence of [3H]AdoMet. Proteins were resolved by 15% SDS-PAGE and examined by Coomassie blue staining (lower panel) or fluorography (upper panel). The positions of histones are indicated on the right. NE, HeLa nuclear extract used as a positive control. (D) Histone H3-K4 specificity of human Set1/Ash2 HMT. Recombinant histone H3 was in vitro methylated using f-HCF-1N two-step purified material and subjected to 16 Edman degradation cycles. The radioactivity released in each cycle is indicated. (E) Interplay between K9 methylation and K4 methylation by Set1/Ash2 HMT. Two-step f-HCF-1N and mock-purified samples were incubated with a histone H3 peptide corresponding to amino acids 1–15, or the corresponding peptide trimethylated at K4 or K9. 3H incorporation for each substrate peptide was defined as the difference in 3H CPM obtained with the f-HCF-1N and mock samples. The average of five experiments is shown.
Significantly, sequence similarity analysis revealed that three of the identified HIPs are the closest human sequence homologs of the Set1, Bre2/Ash2, and Swd3 components of the yeast Set1/Ash2 HMT complex, referred to here as Set1, Ash2, and WDR5 (Fig. 4A). Like its yeast homolog, the human Set1 protein contains highly conserved SET and postSET domains at its C terminus that are related to those of the human and Drosophila trithorax protein. Additionally, both of the yeast and human proteins contain an N-terminal region with similarity to the RRM RNA-recognition motif found in many RNA-binding proteins (Perez-Canadillas and Varani 2001), indicating an important conserved role in Set1/Ash2 HMT function. Consistent with the presence of HCF-1 in animals but not yeast (Kristie and Sharp 1990; Wilson et al. 1993b), the human and Caenorhabditis elegans, but not the yeast, Set1 proteins also contain the sequence EHNY (EHCY in C. elegans; data not shown), which matches the HCF-1-binding motif (labeled HBM in Fig. 4A) found in HCF-1 Kelch-domain-binding proteins such as VP16 and the cellular leucine-zipper protein LZIP/Luman (Freiman and Herr 1997; Lu et al. 1998).
The human Ash2 homolog, which is related to the Drosophila trithorax-group protein Ash2, contains the SPRY domain present in the yeast Ash2 homolog Bre2 (Fig. 4A). In contrast to the Bre2 protein, however, human Ash2 additionally contains a PHD-related zinc-finger motif. Although not present in the Bre2 protein, a PHD finger-related motif is contained within the Spp1 component of the yeast Set1/Ash2 HMT. Perhaps the human Ash2 homolog functionally corresponds to both of the Bre2 and Spp1 yeast proteins.
Of the three WD40-repeat-containing proteins, Swd1–3, only Swd3 is evidently represented among the HIPs by the WDR5 protein (Fig. 4A), the likely human homolog of the mouse osteoblastic differentiation protein BIG-3 (Gori et al. 2001). A human homolog of the DPY30-related yeast protein Sdc1 was probably not identified as a HIP because the two closest human homologs of Sdc1 are too small to have been captured in our electrophoretic separation (Fig. 1D). The reason for the absence of homologs of the Swd1 and Swd2 WD40-repeat proteins is not known. Perhaps, in the human Set1/Ash2 HMT complex, these two yeast WD40-repeat proteins have been replaced by the RpAp46 and RpAp48 WD40-repeat proteins found associated with the HCF-1N subunit; consistent with this idea, the human Enhancer of Zeste HMT has been recently shown to contain RpAp46 and RpAp48 (Kuzmichev et al. 2002).
To verify the Set1/Ash2 HMT mass spectroscopy results, we raised antibodies against the human Set1, Ash2, and WDR5 proteins and performed immunoblot analyses of the two-step purified f-HCF-1N complexes as shown in Figure 4B. Consistent with the mass spectroscopy analysis, the human Set1, Ash2, and WDR5 proteins could be detected in the f-HCF-1N-containing material but not in the mock-purified material (Fig. 4B, panels a–c, cf. lanes 2 and 3), whereas the pRb-associated K9 HMT SUV39H1 protein was not detected (Fig. 4B, panel d). The absence of the SUV39H1 protein is consistent with the absence of any other SET-domain-containing proteins or other proteins with HMT similarity in our HIP analysis (data not shown).
HCF-1N subunit association with histone H3-K4 methyltransferase activity
Although in some rare instances it is associated with repression of transcription (e.g., the rDNA genes; Briggs et al. 2001; Bryk et al. 2002; Krogan et al. 2002), in yeast, the Set1/Ash2 HMT complex is generally associated with transcriptional activation of protein-encoding genes via methylation of histone H3 at K4 (Roguev et al. 2001; Bernstein et al. 2002; Nagy et al. 2002; Noma and Grewal 2002; Santos-Rosa et al. 2002), a mark associated with actively transcribed genes in a wide array of eukaryotic species (Strahl et al. 1999; Litt et al. 2001; Noma et al. 2001; Bernstein et al. 2002; Santos-Rosa et al. 2002). We therefore tested whether the two-step purified f-HCF-1N complexes containing human Set1 and Ash2 possess HMT activity as shown in Figure 4C with both purified core histones (lanes 1–4) and recombinant histone H3 (lanes 5,6; see Materials and Methods). With both substrates, the f-HCF-1N material displayed selective histone H3 HMT activity (Fig. 4C, lanes 3,5), whereas the control mock-purified material did not exhibit any activity (Fig. 4C, lanes 4,6). Because we found no other HMT-related proteins associated with the HCF-1N subunit, we conclude that the histone H3 methylation activity described here is due to the activity of the HCF-1N-associated human Set1/Ash2 HMT.
To map the site of histone H3 methylation, we in vitro methylated purified recombinant histone H3 by incubation with the f-HCF-1N two-step purified material in the presence of [H3]-S-adenosyl methionine and then subjected the radiolabeled histone H3 to Edman degradation. The analysis revealed that K4 is the only methylation target of human Set1/Ash2 HMT within the first 16 amino acids of histone H3 (Fig. 4D). Furthermore, the radioactivity remaining on the PVDF membrane after 16 cycles of Edman degradation indicates that K4 is the major, if not only, histone H3 methylation target for this HMT (Fig. 4D). Thus, similarly to its yeast counterpart, these results indicate that the human Set1/Ash2 is a histone H3-K4 HMT.
K9 methylation inhibits K4 methylation by the HCF-1N-associated Set1/Ash2 HMT
Histone modification can be regulated by prior modification of nearby residues as evidenced by the inhibition of histone H3-K9 methylation by Ser 10 phosphorylation (Rea et al. 2000), thus providing a mechanism for regulatory interplay by different histone “codes.” Histone H3 methylation of residues K4 and K9 is known to be associated with different transcriptional outcomes: transcriptional activation by K4 methylation and transcriptional repression by K9 methylation (Jenuwein and Allis 2001). To determine whether K4 methylation by the HCF-1N-associated Set1/Ash2 complex might be influenced by prior methylation of K9, we compared the ability of the f-HCF-1N- and mock-purified material to methylate either an unmodified peptide corresponding to the first 15 amino acids of histone H3 or a peptide premethylated (trimethylated) at K9; as a control, we included a corresponding peptide premethylated at K4, which should directly block Set1/Ash2 HMT activity. As shown in Figure 4E, consistent with Set1/Ash2 K4 methylation specificity, the unmodified peptide was an effective substrate for the f-HCF-1N-associated HMT activity, whereas the K4 trimethylated peptide was not. Interestingly, however, K9 premethylation blocked peptide methylation by the f-HCF-1N-associated HMT activity (Fig. 4E), indicating that there is a cross-talk between K9 methylation and K4 methylation by the Set1/Ash2 complex.
The Set1/Ash2 HMT and Sin3 HDAC complexes associate with separate functional regions of the HCF-1N subunit
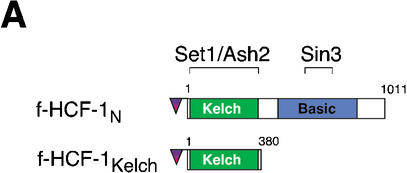
The HCF-1N subunit contains two functional regions required to promote cell proliferation (Wilson et al. 1997): the Kelch domain, which tethers HCF-1 to chromatin, and the Basic region, which we have hypothesized is involved in recruitment of coregulatory activities to chromatin (Wysocka et al. 2001). The yeast two-hybrid data presented in Figure 3 suggest that the Sin3 HDAC complex binds to the HCF-1 Basic region via the Sin3 PAH1 domain, but no other elements of the Sin3 complex or HCF-1N subunit were directly assayed for interaction in these experiments. To map sites of Sin3 HDAC and Set1/Ash2 HMT association with functional regions of HCF-1 in vivo, we immunopurified HCF-1-associated proteins from the f-HCF-1N HeLa-cell line and the previously described f-HCF-1Kelch HeLa-cell line, which expresses the HCF-1 Kelch domain alone (residues 2–380; Wysocka et al. 2001), as illustrated in Figure 5A. As a control, we performed parallel immunopurification from the cell line directing synthesis of the Flag epitope alone.
Figure 5.
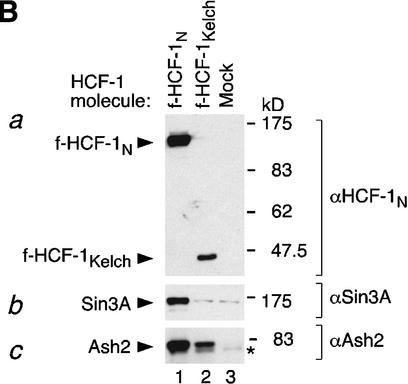
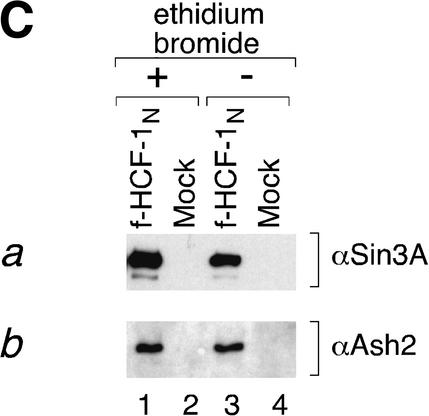
Independent Sin3 HDAC and Set1/Ash2 HMT association with separate functional regions of the HCF-1N subunit. (A) A schematic of f-HCF-1N and f-HCF-1Kelch is shown with Set1/Ash2- and Sin3-interaction regions depicted above the f-HCF-1N diagram. (B) Anti-Flag immunoprecipitations were performed from cells expressing f-HCF-1N, f-HCF-1Kelch, or transduced with the empty vector, and analyzed by immunoblotting with anti-HCF-1N (panel a), anti-Sin3 (panel b), and anti-Ash2 (panel c) antibodies. Samples were normalized to the cell equivalents. *, nonspecific anti-Ash2 cross-reacting species. (C) HCF-1N association with Sin3 and Ash2 is DNA-independent. Two-step purification was performed in parallel from f-HCF-1N and empty vector-transduced cells either in the presence (lanes 1,2) or in the absence (lanes 3,4) of 100 μg/mL ethidium bromide (see Materials and Methods). Purified material was analyzed by immunoblotting with anti-Sin3A (panel a) and anti-Ash2 (panel b) antibodies.
The anti-Flag-immunopurified complexes were first analyzed by immunoblot with an N-terminal anti-HCF-1N antibody (αN18) that recognizes both endogenous HCF-1N and the recombinant f-HCF-1N molecules as shown in Figure 5B (panel a). Only the Flag-tagged HCF-1 molecules were anti-Flag-immunopurified, as evidenced by the recovery of the f-HCF-1N (Fig. 5B, panel a, lane 1) and f-HCF-1Kelch (Fig. 5B, panel a, lane 2) proteins but not the endogenous HCF-1N proteins (Fig. 5B, panel a, lanes 1–3). In addition to demonstrating the efficacy of the purification procedure, we note that the lack of endogenous HCF-1N subunit copurification indicates that HCF-1N molecules do not associate with one another stably in the cell forming dimeric or multimeric HCF-1 complexes.
As monitored by anti-Sin3A immunoblotting, the Sin3 complex was specifically recovered with the f-HCF-1N but not the f-HCF-1Kelch protein (Fig. 5B, panel b, cf. lanes 1–3). Together with the yeast two-hybrid data, these results indicate that the Sin3 complex associates independently and exclusively with the HCF-1N Basic region. In contrast, as monitored by anti-Ash2 immunoblotting, the Set1/Ash2 complex was specifically recovered with both the f-HCF-1N and f-HCF-1Kelch proteins (Fig. 5B, panel c, cf. lanes 1–3), indicating that the Set1/Ash2 HMT complex associates independently with the HCF-1 Kelch domain. Thus, the two HCF-1-associated chromatin-modifying activities associate independently with two separate functional HCF-1N regions that are required for promoting cell proliferation: the Kelch domain and the Basic region.
As shown in Figure 5C, these Sin3 and Set1/Ash2 interactions are also independent of DNA or chromatin association because they are not sensitive to the DNA-binding inhibitor ethidium bromide (Fig. 5C, panels a,b, cf. lanes 1 and 3; Lai and Herr 1992). Consistent with these results, we note that these associations are not sensitive to high salt because the first WGA purification step of the HCF-1N complexes (Fig. 1B) is performed in buffer containing 420 mM KCl (see Materials and Methods). Thus, the association of HCF-1 with these two chromatin-modifying complexes can occur independently of the DNA-bound/chromatin-associated state of HCF-1 in the cell, but whether they naturally occur off the chromatin is not known.
Cosedimentation of HCF-1, Sin3A, and Ash2 in high-molecular-weight complexes
The ability of the generally activating Set1/Ash2 HMT and repressive Sin3 HDAC complexes to bind to different functional regions of HCF-1 independent of DNA suggests that they could bind HCF-1 simultaneously to create one or more “super” complexes with diverse regulatory activities. To test this hypothesis, we asked whether HCF-1 and the Sin3 HDAC and Set1/Ash2 HMT complexes cosediment during glycerol-gradient sedimentation. For this purpose, as shown in Figure 6A, we analyzed the glycerol-gradient sedimentation patterns of endogenous HCF-1 (as assayed with an HCF-1C subunit-specific antibody; Fig. 6A, panel a), f-HCF-1N (Fig. 6A, panel b), Sin3A (Fig. 6A, panel c), Ash2 (Fig. 6A, panel d), and Sp1 (Fig. 6A, panel e) proteins isolated from f-HCF-1N HeLa-nuclear extract enriched in HCF-1 molecules by the WGA purification step (Fig. 1B). To preserve high-molecular-weight complexes within the gradient, the samples were centrifuged in a dense gradient (25%–50% glycerol) for a relatively short period (see Materials and Methods). Under such conditions, protein complexes with an apparent molecular weight of 0.5 MD and less were present in the first five fractions (Fig. 6A, see markers at top).
Figure 6.
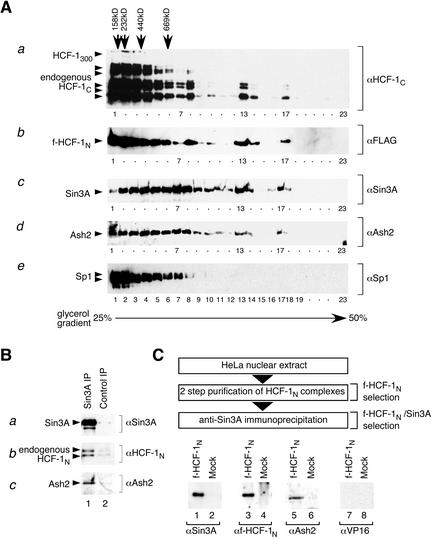
Sin3 HDAC and Set1/Ash2 HMT complex components coexist with HCF-1 in high-molecular-weight complexes. (A) Cosedimentation of HCF-1N, Sin3A, and Ash2 on glycerol gradients. HCF-1-enriched samples were prepared from cells synthesizing f-HCF-1N by the WGA purification step and applied to a 25%–50% glycerol gradient (see Materials and Methods). Fractions were analyzed by immunoblotting with antibodies recognizing endogenous HCF-1C (panel a), ectopic f-HCF-1N (panel b), Sin3A (panel c), Ash2 (panel d), and Sp1 (panel e). The position of molecular weight markers applied to a parallel gradient is indicated at the top. (B) Sin3A, endogenous HCF-1, and Ash2 copurify in nontranduced cells. Anti-Sin3A immunoprecipitation (lane 1) and control immunoprecipitation (lane 2) with a corresponding amount of the irrelevant antibody of the same isotype (12CA5) were performed from nontransduced HeLa cells. Immunoprecipitates were analyzed by immunoblotting with anti-Sin3A (panel a), anti-HCF-1 (panel b), and anti-Ash2 (panel c) antibodies. (C) Sin3A and Ash2 can associate with HCF-1 simultaneously. (Top) Purification scheme for f-HCF-1N/Sin3A complexes from f-HCF-1N/HA-VP16ΔC HeLa cells. Corresponding amounts of the two-step-purified samples from f-HCF-1N/HA-VP16ΔC and HA-VP16ΔC only HeLa cells fractionated by anti-Sin3A immunoprecipitation were analyzed by immunoblotting with Sin3A (lower panel, lanes 1,2), anti f-HCF-1N (lanes 3,4), Ash2 (lanes 5,6), and VP16 (lanes 7,8) antibodies.
In contrast to highly purified HCF-1300 and HCF-1N–HCF-1C heterodimeric complexes, which sediment at <100 kD (Wilson et al. 1993a), the majority of the WGA-purified endogenous HCF-1 protein and f-HCF-1N subunit sedimented with a much higher apparent molecular weight (Fig. 6A, panels a,b). Remarkably, the patterns of the endogenous HCF-1 and f-HCF-1N subunits are very similar (Fig. 6A, cf. panels a and b). This similarity cannot be explained by association of the recombinant f-HCF-1N subunit with the endogenous HCF-1C subunits because only very substoichiometric amounts of the HCF-1C subunit are purified with the f-HCF-1N subunit (Fig. 1D). Instead, these HCF-1 and HCF-1N-subunit cosedimentation results indicate that the HCF-1N subunit, and not the HCF-1C subunit, is primarily responsible for the mobility of endogenous HCF-1 during sedimentation, suggesting that the HCF-1N subunit has the major role in HCF-1 association with other cellular proteins. Importantly, these results additionally indicate that the recombinant f-HCF-1N subunit faithfully reproduces endogenous HCF-1 complexes in both nature and quantity, further validating the veracity of the HIP identification described in Figure 1.
To test whether the HCF-1 proteins cosediment with the Sin3 HDAC and Set1/Ash2 HMT complexes, we analyzed the same glycerol gradient by immunoblot analysis with the anti-Sin3A and anti-Ash2 antibodies. Although the Sin3A and Ash2 sedimentation patterns differ from the HCF-1 sedimentation pattern (e.g., Fig. 6A, cf. panels a–d, lane 7, which is relatively low for HCF-1 proteins but still high for Sin3A and Ash2 molecules), there is a remarkable cosedimentation of HCF-1, Sin3A, and Ash2 molecules among very-high-molecular-weight complexes (>1 MD; Fig. 6A, cf. panels a–d, fractions 13 and 17). This cosedimentation pattern is unlikely to be fortuitous because Sp1, revealed by immunoblot analysis (Fig. 6A, panel e), and other polypeptides, revealed by silver-stain analysis of the gradient fractions (data not shown), display very different sedimentation patterns. Together, these data indicate that both endogenous HCF-1 and the f-HCF-1N subunit cosediment with Sin3A and Ash2 in high-molecular-weight complexes, consistent with the presence of HCF-1-containing Sin3 HDAC and Set1/Ash2 HMT supercomplexes.
These cosedimentation analyses were performed with extracts from the stably expressing f-HCF-1N-subunit HeLa-cell line, which permitted the parallel analysis of endogenous and f-HCF-1N-subunit complexes. To demonstrate that HCF-1 naturally associates with the Sin3 HDAC complex in nontransduced HeLa cells, we prepared a WGA-purified fraction from a normal HeLa-cell nuclear extract and performed an anti-Sin3A immunoprecipitation from the HCF-1-enriched nuclear extract. As shown in Figure 6B, the Sin3A immunoprecipitation recovered endogenous HCF-1 polypeptides more effectively than a control immunoprecipitation with an unrelated antibody of the same isotype (Fig. 6B, panels a,b, cf. lanes 1 and 2). Thus, endogenous HCF-1 naturally associates with Sin3A. Ash2 was also specifically recovered in this Sin3A immunoprecipitation, suggesting that in native HeLa-cell extracts, HCF-1, Sin3 HDAC, and Set1/Ash2 HMT associate with one another.
The HCF-1N subunit tethers the Sin3 HDAC and Set1/Ash2 HMT components together
To extend the cosedimentation analyses described above, which do not directly address protein–protein association, we asked, by HCF-1N-subunit purification followed by Sin3A immunoprecipitation, whether the Sin3 HDAC and Set1/Ash2 HMT complexes associate with HCF-1 simultaneously. For this experiment, we took advantage of a HeLa-cell line coexpressing the f-HCF-1N subunit and VP16; as described further below, the VP16 protein provided a negative control for this experiment. We first purified the f-HCF-1N-containing complexes from these cells by the two-step protocol (Fig. 1B) and then isolated those f-HCF-1N-containing complexes containing Sin3A by anti-Sin3A immunoprecipitation as described in Figure 6C (top). A standard mock purification from matched cells lacking the f-HCF-1N subunit was performed in parallel and in all instances was negative for the proteins assayed (Fig. 6C, even-numbered lanes). The three-step f-HCF-1N/Sin3A-purified material contained Sin3A, f-HCF-1N, and Ash2 molecules [Fig. 6C, lanes 1,3,5; but not VP16 (Fig. 6C, lane 7), consistent with mutually exclusive Sin3 and VP16 association with HCF-1; see below]. Thus, at least Sin3A and Ash2—and probably their respective HDAC and HMT complexes—can associate with the HCF-1N subunit simultaneously. Given the independent Set1/Ash2 association with the HCF-1 Kelch domain and Sin3 association with the Basic region (Figs. 3, 5), these results strongly suggest that HCF-1 tethers the Set1/Ash2 HMT and Sin3 HDAC complexes together.
Preferential VP16 association with the HCF-1-Set1/Ash2 HMT complex
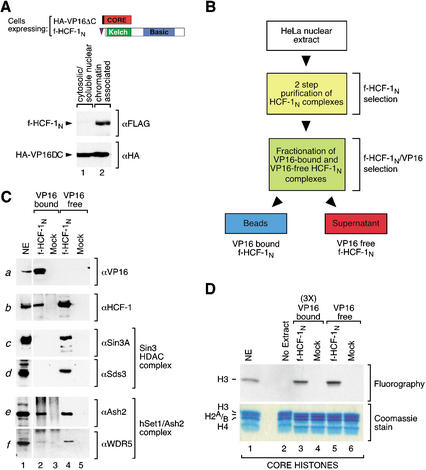
The simultaneous association of HCF-1 with chromatin-modifying complexes apparently involved in both activation (the Set1/Ash2 HMT) and repression (the Sin3 HDAC) of transcription is curious because, in the context of the VP16-induced complex, HCF-1 is only known to be involved in activation of transcription. We therefore asked which, if any, of these histone-modifying complexes is associated with HCF-1 when it is bound to VP16. To answer this question, we first prepared the cell lines briefly mentioned above: these are derivatives of wild-type and f-HCF-1N-containing HeLa cells that also contain a hemagglutinin-epitope (HA)-tagged VP16-core molecule called HA-VP16ΔC lacking the C-terminal transcriptional activation domain but containing all the necessary elements for VP16-induced complex formation as shown in Figure 7A (top; Greaves and O'Hare 1990). Chromatin fractionation of the f-HCF-1N/HA-VP16ΔC cells showed that nearly all of the f-HCF-1N molecules were present in the chromatin fraction (Fig. 7A, upper panel, cf. lanes 1 and 2). Even though these HSV uninfected cells do not contain HSV VP16-response elements, about half of the VP16 protein is recovered in the chromatin fraction, whereas the other half is present in the cytosolic fraction (Fig. 7A, lower panel, lanes 1,2). Thus, although the specific sites of chromatin binding are unknown, many of the VP16 molecules are in a proper location to associate with chromatin-bound f-HCF-1N and its associated factors.
Figure 7.
Preferential VP16 association with f-HCF-1N–Set1/Ash2 HMT over f-HCF-1N–Sin3 HDAC complexes. (A) A significant portion of HA-VP16ΔC is bound to chromatin in the f-HCF-1N/VP16ΔC cells. A schematic of HA-VP16ΔC and f-HCF-1N is shown. f-HCF-1N/HA-VP16ΔC HeLa cells were fractionated into cytosolic/soluble nuclear and chromatin-associated protein fractions and probed with anti-Flag (upper panel) and anti-HA (lower panel) epitope antibodies. (B) Schematic of the VP16-bound and VP16-free f-HCF-1N complexes fractionation. (C) Two-step f-HCF-1N and mock-purified samples from f-HCF-1N/HA-VP16ΔC and HA-VP16ΔC only cells, were fractionated by anti-HA coimmunoprecipitation into a VP16-bound f-HCF-1N fraction (pellet) and VP16-free f-HCF-1N fraction (supernatant). Samples were analyzed by immunoblotting with VP16 (panel a), HCF-1 (αN18; panel b), Sin3A (panel c), Sds3 (panel d), Ash2 (panel e), and WDR5 (panel f) antibodies. (D) VP16-bound HCF-1 complexes contain HMT activity. VP16-bound and VP16-free f-HCF-1N complexes were isolated as for C, except that VP16-bound complexes were eluted from the beads with excess of HA peptide. Fractions were then normalized to the amount of Ash2 present (threefold more of VP16-bound than VP16-free fraction was used, indicated by 3×) and assayed for the HMT activity using core histones as a substrate. Proteins were resolved by 15% SDS-PAGE and examined by Coomassie blue staining (lower panel) or fluorography (upper panel). The positions of the histones are indicated on the left. NE, HeLa nuclear extract (10-fold less than used in Fig. 4C) was used as a positive control.
To identify VP16 association with the f-HCF-1N subunit and its associated factors, we first isolated f-HCF-1N-containing complexes from the f-HCF-1N/HA-VP16ΔC cell line by the two-step protocol (Fig. 1B), performing a mock purification with HA-VP16ΔC-only HeLa cells in parallel, and then separated VP16-bound from VP16-free f-HCF-1N complexes by HA-epitope-specific HA-VP16ΔC immunoprecipitation as illustrated in Figure 7B. We then analyzed corresponding amounts of the VP16-bound and VP16-free f-HCF-1N fractions by immunoblot as shown in Figure 7C.
Consistent with VP16 association with the f-HCF-1N subunit and its effective recovery in the VP16 immunoprecipitation, all of the detectable VP16 was present in the VP16-bound and not in the VP16-free fraction (Fig. 7C, panel a, cf. lanes 2 and 4). This VP16 immunoprecipitation step resulted in recovery of ∼25% of the f-HCF-1N molecules (Fig. 7C, panel b, cf. lanes 2 and 4), indicating that a significant portion of the f-HCF-1N molecules in the coexpressing cells is associated with VP16. Of this portion, however, there is little, if any, coassociated Sin3A molecules or Sds3 molecules (Fig. 7C, panels c,d, cf. lanes 2 and 4). These results suggest that VP16 binds preferentially to f-HCF-1N molecules that are not bound to the repressive Sin3 HDAC complex.
In contrast, a significant portion of the Set1/Ash2 HMT complex members Ash2 and WDR5 cofractionate with the VP16-bound fraction (Fig. 7C, panels e,f, cf. lanes 2 and 4). Additionally, as shown in Figure 7D, the VP16-bound f-HCF-1N fraction contained histone H3 HMT activity as does the VP16-free fraction (Fig. 7D, cf. lanes 3 and 5), indicating that the Set1/Ash2 complex is not only present in the VP16-bound fraction but also active. Thus, the prototypical transcriptional activator VP16 distinguishes between coregulatory complexes bound to HCF-1, favoring association with the histone-modifying complex implicated in activation of transcription.
Discussion
We have defined a series of molecular activities associated with the N-terminal subunit of HCF-1 in HSV uninfected cells. Two of these activities are associated with opposing roles in the regulation of transcription through the modulation of chromatin structure: Sin3 HDAC and a novel human Set1/Ash2 HMT. HCF-1 tethers these two histone-modifying complexes together in a supercomplex by their independent association with separate regions of HCF-1 that cooperate in promoting cell proliferation. Although HCF-1 can bind the Sin3 HDAC and Set1/Ash2 HMT complexes simultaneously, VP16 does not stably associate with this supercomplex; instead, it is found selectively bound to HCF-1 with the activating Set1/Ash2 HMT complex. Thus, as a transcriptional activator, VP16 distinguishes between chromatin-modifying activities bound to a coregulator.
A human Set1/Ash2 HMT complex
Our analysis of HCF-1-associated proteins suggests that a K4 histone H3 HMT complex has been conserved from yeast to humans in both structure and activity: the Set1/Ash2 HMT. Although we have not analyzed the Set1/Ash2 complex independently of HCF-1, the structural and functional similarity of the HCF-1-associated Set1/Ash2 HMT activities with the yeast counterparts strongly suggest that a bona fide Set1/Ash2 HMT complex exists in human cells. The results presented here show that this Set1/Ash2 HMT complex, in mutually exclusive interactions, can associate with HCF-1 bound to the repressive Sin3 HDAC or the transcriptional activator VP16, indicating a diversity of transcriptional regulatory roles.
Simultaneous association of VP16 with the HCF-1-Set1/Ash2 complex was unexpected because the only known mechanism for binding the HCF-1 Kelch domain is via the (D/E)HXY tetrapeptide motif, for which there is only one binding site in the HCF-1 Kelch domain (Lu et al. 1997). Both VP16 and Set1 possess the (D/E)HXY tetrapeptide motif-binding sequence, readily explaining how these two proteins bind the HCF-1 Kelch domain independently. But how do they associate together? Perhaps, when the Set1/Ash2 HMT complex binds the HCF-1 Kelch domain it can contact two different surfaces—the (D/E)HXY-binding site and a second independent site—and adopts different configurations on the HCF-1 Kelch domain when bound alone or with an activator such as VP16.
The conserved nature of the Set1/Ash2 complex between yeast and humans suggests that it plays important roles in different species. The Ash2 component of the Set1/Ash2 HMT was originally identified as a trithorax-group protein in Drosophila (LaJeunesse and Shearn 1995), where this class of proteins plays an important role in maintaining expression of homeotic regulators of developmental differentiation and pattern formation (Kennison 1995). Furthermore, Set1 is related to another trithorax-group protein, Trx (HRX, MLL, or ALL in humans). The presence of the two trithorax-group-related proteins in human Set1/Ash2 HMT suggests that it plays a role in the regulation of human homeotic gene expression.
Here, we show that Set1/Ash2 histone H3-K4 HMT activity is blocked by prior methylation of the histone H3-K9 residue (Fig. 4). In S. cerevisiae, histone H3-K9 methylation has not been detected, and therefore it is unlikely that the mode of methyl K9 to K4 interplay described here plays a role in Set1/Ash2 function in S. cerevisiae. Thus, the Set1/Ash2 HMT may play more intricate regulatory activities in animals (and Schizosaccharomyces pombe, where K9 methylation is also present) than in budding yeasts.
Role of HCF-1 in VP16 transcriptional activation
HCF-1 may play a more active role in transcriptional activation of the HSV IE genes by VP16 than previously appreciated: Luciano and Wilson (2002) have shown that the HCF-1C subunit contains a transcriptional activation domain that cooperates with VP16 to activate transcription, and the studies described here show that HCF-1 can provide a chromatin-modifying activity associated with transcriptional activation. The preferential association of the Set1/Ash2-HCF-1 complex with VP16 suggests that chromatin structure plays a role in transcriptional regulation of the HSV IE genes during lytic infection. This suggestion is surprising, because it is generally thought that chromatin structure does not play an important role in HSV gene transcription, largely because, unlike other viruses (e.g., SV40), newly replicated HSV genomes are not packaged into chromatin (Leinbach and Summers 1980). But little, if anything, is known about the nucleosome-bound status of the newly deposited genome(s) of the infecting HSV prior to activation of IE-gene transcription by the VP16-induced complex during lytic infection. Perhaps newly deposited genomes are, indeed, assembled into chromatin by the host cell. Alternatively, nucleosomes may not be the sole target of the Set1/Ash2 HMT—for example, the VP16-induced complex itself or other site-specific activators might be methylation targets.
Involvement of varied chromatin-modifying activities in HCF-1's role in cell proliferation
By isolating proteins that associate with the HCF-1N subunit in nuclear extracts, that is, HIPs, our analysis has been biased toward major HCF-1-interaction partners. Thus, chromatin-modifying activities appear to be major HCF-1 effectors. Interestingly, the two complexes identified bind to separate regions of the HCF-1N subunit that cooperate to support cell proliferation. It is likely that HCF-1 is able to promote cell proliferation by regulating chromatin structure. Previous estimates have indicated that a typical HeLa cell contains about a half million molecules of chromatin-bound HCF-1 or about one molecule of HCF-1 for every 10 kb of genomic DNA (Wysocka et al. 2001). Additionally, we estimate that ∼5%–10% of endogenous HCF-1 molecules are bound to the Sin3 HDAC complex in HeLa cells (J. Wysocka and W. Herr, unpubl.). Thus, we suggest that HCF-1 is a widespread regulator of chromatin structure.
A regulatory oscillator?
One of the unexpected findings described here is that HCF-1 tethers two chromatin-modifying activities associated with opposing transcriptional regulatory activities together. Although generally associated with opposite transcriptional activities, they may cooperate in transcriptional regulation. Indeed, in S. cerevisiae, Rpd3, the homolog of human Sin3-complex HDACs 1 and 2, and the Set1 HMT are both found associated with common transcriptional targets, and the levels of their chromatin-modifying activities inversely correlate (Bernstein et al. 2002; Kurdistani et al. 2002). How these different activities are corecruited to genes in yeast is not known. Perhaps in animals, HCF-1 can provide such a function by tethering the HDAC and HMT activities simultaneously to genes that it regulates, thus providing a platform that can lead to oscillating on and off states of transcription. A similar view is emerging from studies of inducible transcription in S. cerevisiae (Papamichos-Chronakis et al. 2002; Proft and Struhl 2002), suggesting that such coordinated dual modes of regulation may be common in eukaryotes.
Materials and methods
Cell lines
HeLa cells stably synthesizing N-terminally Flag-epitope-tagged HCF-1 residues 2–1011 (f-HCF-1N) or 2–380 (f-HCF-1Kelch) or transduced with the empty pBabeFlag-Puro vector were described previously (Wysocka et al. 2001). Sequences encoding HA-VP16ΔC (residues 5–411) were cloned into the pBabe-Hygro retroviral vector after PCR amplification. Populations of HeLa cells stably synthesizing both HA-VP16ΔC and f-HCF-1N or HA-VP16ΔC alone were obtained by retroviral gene transfer with the Phoenix amphotropic virus packaging line (Serrano et al. 1997) and the HA-VP16ΔC vector into HeLa cells already directing synthesis of f-HCF-1N or the Flag epitope alone. Appropriate cells were selected by culturing in 10% fetal bovine serum (FBS) in Dulbecco's modified Eagle's medium (DMEM) containing 1 mg/mL hygromycin and 2 μg/mL puromycin for 10 d.
Purification of the f-HCF-1N complexes
For the two-step f-HCF-1N purification protocol, nuclear extracts (Dignam et al. 1983) were prepared from ∼2.4 × 1010 HeLa cells directing synthesis of the f-HCF-1N subunit or the Flag epitope alone and immediately incubated at 4°C for 1 h with WGA agarose beads (Vector Laboratories). The WGA agarose beads were equilibrated with buffer D (20 mM HEPES at pH 7.9, 20% glycerol, 0.2 mM EDTA, 0.1% NP-40, and freshly added protease inhibitors, Pefabloc and Complete; Roche) containing 420 mM KCl (buffer D/420 mM KCl); 1 mL of beads was used per 4.3 × 109 cell equivalents (Wilson et al. 1993a). After incubation, the WGA beads were washed three times in buffer D/420 mM KCl and 4 times in buffer D/100 mM KCl. WGA-bound proteins were eluted at room temperature three times for 20 min each time with buffer D/100 mM KCl saturated with N-acetylglucosamine (∼400 mM). Eluates were combined and immediately incubated at 4°C with Flag M2 agarose beads (Sigma) for 3 to 12 h. About 1 mL of anti-Flag epitope beads was used per 7 × 109 cells. After incubation, the beads were washed five times with buffer D/100 mM KCl and three times with buffer D/100 mM NaCl and 10% glycerol. Immunoprecipitates were eluted at room temperature three times for 20 min each time with buffer D/100 mM NaCl, 10% glycerol, and 0.5 mg/mL Flag peptide. Eluates were combined, concentrated with Microcon columns, and fractionated by SDS-PAGE. Proteins were visualized by colloidal Coomassie blue staining (Invitrogen). For the ethidium bromide control, WGA eluates were preincubated for 30 min with 100 μg/mL ethidium bromide, and anti-Flag immunoprecipitation followed by Flag peptide elution was performed as described above, except that all the buffers contained in addition ethidium bromide at a final concentration of 100 μg/mL.
Purification of f-HCF-1N/VP16ΔC, f-HCF-1N/Sin3A, and endogenous HCF-1/Sin3A complexes
Chromatin fractionation was performed as previously described (Wysocka et al. 2001). f-HCF-1N complexes were purified by the two-step purification protocol, from each of the HeLa-cell lines stably expressing HA-VP16ΔC and f-HCF-1N or HA-VP16ΔC alone. Combined Flag peptide eluates were immediately incubated at 4°C with 12CA5 anti-HA monoclonal antibody covalently cross-linked to the Protein G agarose (Roche). After incubation, the supernatant was collected for further analysis, and the 12CA5 anti-HA beads were washed six times with buffer D/100 mM NaCl; immunoprecipitates were recovered from the beads by boiling in SDS-Laemmli buffer. Corresponding amounts of supernatant and immunoprecipitates were subsequently analyzed by immunoblotting. Alternatively, immunoprecipitates were eluted from the 12CA5 anti-HA beads with buffer D/100 mM NaCl and 0.5 mg/mL HA peptide and eluates analyzed in the HMT assay.
To purify the f-HCF-1N/Sin3A complexes, f-HCF-1N complexes purified by the two-step purification protocol were incubated at 4°C with anti-Sin3A mouse monoclonal antibody. Immunoprecipitates were collected after addition of protein-G agarose beads, washed six times with buffer D/100 mM NaCl, recovered from the beads by boiling in SDS-Laemmli buffer, and analyzed by immunoblotting. To purify the endogenous HCF-1/Sin3A complexes, nuclear extract was prepared from nontransduced HeLa cells, enriched for HCF-1 by the WGA purification step, and incubated with the anti-Sin3A monoclonal antibody. Immunoprecipitates were collected and washed as described above.
Protein identification by mass spectrometry
Stained f-HCF-1N complex components after SDS-PAGE fractionation were excised and processed for in-gel trypsin digestion following standard protocols. The resulting peptides were extracted and purified on C18-Ziptips (Millipore) according to the manufacturer's protocol and resuspended in 10 μL of 30% methanol, 0.5% acetic acid. A fraction of the purified peptides was analyzed by MALDI-TOF mass spectrometry, and the data were analyzed using the software programs m/z (Proteometrics) and the PROFOUND search engine (Zhang and Chait 2000). The remaining sample was analyzed by LC-MS/MS using an LCQDeca mass spectrometer (Thermo-Finnigan). Samples judged by MALDI-TOF spectrometry to contain relatively large amounts of peptide were separated using a 0.3 mm × 50 mm C18 column (VYDAC) developed at 8 μL/min. Samples judged to be of low abundance were analyzed using a 0.075 mm × 50 mm Aquasil-C18 picofrit column (Newobjective) developed at 0.5 μL/min. The resulting spectra were analyzed with the SONARS software package (Proteometrics).
HDAC and HMT assays
The HDAC assay was performed with the HDAC Assay Kit (Upstate Biotechnology) according to the manufacturer's instructions using chemically [3H]-acetylated histone H4 peptide (amino acids 1–20) as a substrate. The HMT assay was performed as described previously (Strahl et al. 1999). One of the following substrates: 10 μg of purified core histones (Upstate Biotechnology), 2 μg of recombinant histone H3 purified from Escherichia coli (Upstate Biotechnology), or 1 μg of a histone H3 peptide corresponding to amino acids 1–15, either unmodified or trimethylated on residue K4 or K9 (kind gifts of C.D. Allis, University of Virginia, Charlottesville, VA), was used for methyltransferase reactions. Reactions were analyzed by SDS-PAGE followed by Coomassie blue staining and fluorography or by spotting on P-81 paper and scintillation counting.
Edman degradation
f-HCF-1N purified material containing ∼1 ng of Set1 was used to in vitro methylate recombinant histone H3 as described above. Proteins were separated by SDS-PAGE, transferred to the PVDF membrane, and stained with Coomassie blue. The band corresponding to histone H3 was excised from the membrane and subjected to 16 cycles of Edman degradation reaction. Radioactivity released in each cycle and remaining on the membrane after 16 cycles was measured by scintillation counting.
Glycerol gradients
HCF-1-enriched nuclear extract was prepared from cells synthesizing endogenous HCF-1 and f-HCF-1N by the WGA purification step as described above and applied to a 5-mL 25%–50% buffer D/100 mM KCl glycerol gradient and centrifuged in a Vti65 rotor at 32,500 rpm for 10 h. Fractions (∼200 μL) were collected and analyzed by immunoblotting. Molecular markers were applied to a parallel gradient, and their migration was analyzed by Coomassie blue staining.
Two-hybrid assay
A yeast two-hybrid screen was performed using a bait containing the lexA DNA-binding domain fused to mouse Sin3A cDNA encoding amino acids 119–1219 (PAH1-E) to screen a library of mouse embryo cDNAs fused to the VP16 transactivation domain (Vojtek et al. 1993; Hollenberg et al. 1995). Transformants expressing both HIS3 and lacZ were cured of bait plasmids and tested for binding specificity by mating assays with AMR70 strains containing the original bait or a control plasmid. Of the 150 HIS3- and lacZ-positive transformants tested for binding specificity, 33 clones were subsequently sequenced. Of these sequenced clones, 4 were identified as containing identical cDNA inserts corresponding to mouse HCF-1. Directed two-hybrid assays were performed to map the Sin3A-interaction domains of positive clones using lexA fusions containing different regions of Sin3A: PAH1 (residues 119–196), PAH2 (residues 297–385), PAH3 (residues 459–526), PAH4 (residues 885–955), and PAH3-E (residues 459–1219).
Immunoblot analysis
Polyclonal anti-HCF-1N subunit (αN18; Goto et al. 1997) and anti-HCF-1C subunit (αH12; Wilson et al. 1993a) antisera have been described previously. The anti-Flag (mouse monoclonal M2), anti-Flag (rabbit polyclonal), and anti-HA (rabbit polyclonal) antibodies were obtained from Sigma; anti-Sin3A (rabbit polyclonal K-20), anti-HDAC1 (H-11, mouse monoclonal), anti-HDAC2 (C-8, mouse monoclonal), and anti-Sp1 (1C6, mouse monoclonal) antibodies were obtained from Santa Cruz Biotechnology; anti-Mi2 (rabbit polyclonal), and anti-HDAC3 (rabbit polyclonal) antibodies were obtained from Upstate Biotechnology; and anti-O-linked N-acetylglucosamine (mouse monoclonal) antibody from Affinity Bioreagents. The rabbit polyclonal anti-GABPβ antibody was a kind gift of H. Handa (Tokyo Institute of Technology, Yokohama, Japan); rabbit polyclonal anti-OGT antibody was a kind gift of R.S. Haltiwanger (State University of New York at Stony Brook); rabbit polyclonal anti-mSds3 antibody was a kind gift of G. David (Dana-Farber Cancer Institute, Harvard Medical School, Boston, MA) and R.A. DePinho (Dana-Farber Cancer Institute, Harvard Medical School, Boston, MA); and anti-SUV39H1 mouse monoclonal antibody was a kind gift of M.L. Cleary (Stanford University School of Medicine, CA). Rabbit polyclonal antisera were raised against peptides corresponding to the C-terminal sequences of the respective proteins: TESCRGSLN (αSet1C), Ash2 CHVET EVDGRRSPPWEP (αAsh2C), and WDR5 LENDKTIKLWKSDC (αWDR5C), using the ResGen antibody production facility. Immunoblots were prepared by liquid transfer and developed by the chemiluminescence reaction (SuperSignal; Pierce).
Coimmunoprecipitation
Nuclear extracts were prepared from cells expressing f-HCF-1N, f-HCF-1Kelch, or as a control, from cells transduced with the empty vector. Extracts were diluted to the concentration of 100 mM KCl and directly incubated with Flag M2 agarose beads (Sigma). Immunoprecipitates were washed and eluted from the beads with the specific peptide as described above and analyzed by immunoblotting.
Acknowledgments
We thank C.D. Allis for methylated H3 peptides; M.L. Cleary, G. David, R.A. DePinho, R.S. Haltiwanger, and H. Handa for antibodies; S.I. Grewal, N. Hernandez, E. Julien, T. Swigut, and R.M. Xu for critical readings of the manuscript; R. Kobayashi for his involvement in the initial phase of the project; members of the Herr laboratory for helpful discussions and suggestions; J. Duffy for artwork; and J. Reader for help with the preparation of the manuscript. M.P.M. thanks B.T. Chait and J.C. Padovan for sharing their expertise; and J.W. thanks T. Swigut, P.T. Reilly, and A. Bubulya for their support and encouragement. This work was supported by PHS grants R01GM 54598 to W.H., P01CA13106 to W.H. and M.P.M., and R01CA57138 to R.N.E.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL herr@cshl.edu; FAX (516) 367-6919.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.252103.
References
- Alland L, David G, Shen-Li H, Potes J, Muhle R, Lee HC, Hou H, Jr, Chen K, DePinho RA. Identification of mammalian Sds3 as an integral component of the Sin3/histone deacetylase corepressor complex. Mol Cell Biol. 2002;22:2743–2750. doi: 10.1128/MCB.22.8.2743-2750.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein BE, Humphrey EL, Erlich RL, Schneider R, Bouman P, Liu JS, Kouzarides T, Schreiber SL. Methylation of histone H3 Lys 4 in coding regions of active genes. Proc Natl Acad Sci. 2002;99:8695–8700. doi: 10.1073/pnas.082249499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briggs SD, Bryk M, Strahl BD, Cheung WL, Davie JK, Dent SY, Winston F, Allis CD. Histone H3 lysine 4 methylation is mediated by Set1 and required for cell growth and rDNA silencing in Saccharomyces cerevisiae. Genes & Dev. 2001;15:3286–3295. doi: 10.1101/gad.940201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryk M, Briggs SD, Strahl BD, Curcio MJ, Allis CD, Winston F. Evidence that Set1, a factor required for methylation of histone H3, regulates rDNA silencing in S. cerevisiae by a Sir2-independent mechanism. Curr Biol. 2002;12:165–170. doi: 10.1016/s0960-9822(01)00652-2. [DOI] [PubMed] [Google Scholar]
- Dignam JD, Lebovitz RM, Roeder RG. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 1983;11:1475–1489. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freiman RN, Herr W. Viral mimicry: Common mode of association with HCF by VP16 and the cellular protein LZIP. Genes & Dev. 1997;11:3122–3127. doi: 10.1101/gad.11.23.3122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gori F, Divieti P, Demay MB. Cloning and characterization of a novel WD-40 repeat protein that dramatically accelerates osteoblastic differentiation. J Biol Chem. 2001;276:46515–46522. doi: 10.1074/jbc.M105757200. [DOI] [PubMed] [Google Scholar]
- Goto H, Motomura S, Wilson AC, Freiman RN, Nakabeppu Y, Fukushima K, Fujishima M, Herr W, Nishimoto T. A single-point mutation in HCF causes temperature sensitive cell-cycle arrest and disrupts VP16 function. Genes & Dev. 1997;11:726–737. doi: 10.1101/gad.11.6.726. [DOI] [PubMed] [Google Scholar]
- Greaves RF, O'Hare P. Structural requirements in the herpes simplex virus type 1 transactivator Vmw65 for interaction with the cellular octamer-binding proteins and target TAATGARAT sequences. J Virol. 1990;64:2716–2724. doi: 10.1128/jvi.64.6.2716-2724.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunther M, Laithier M, Brison O. A set of proteins interacting with transcription factor Sp1 identified in a two-hybrid screening. Mol Cell Biochem. 2000;210:131–142. doi: 10.1023/a:1007177623283. [DOI] [PubMed] [Google Scholar]
- Heinzel T, Lavinsky RM, Mullen TM, Soderstrom M, Laherty CD, Torchia J, Yang WM, Brard G, Ngo SD, Davie JR, et al. A complex containing N-CoR, mSin3 and histone deacetylase mediates transcriptional repression. Nature. 1997;387:43–48. doi: 10.1038/387043a0. [DOI] [PubMed] [Google Scholar]
- Herr W. The herpes simplex virus VP16-induced complex: mechanisms of combinatorial transcriptional regulation. Cold Spring Harb Symp Quant Biol. 1998;63:599–607. doi: 10.1101/sqb.1998.63.599. [DOI] [PubMed] [Google Scholar]
- Hollenberg SM, Sternglanz R, Cheng PF, Weintraub H. Identification of a new family of tissue-specific basic helix–loop–helix proteins with a two-hybrid system. Mol Cell Biol. 1995;15:3813–3822. doi: 10.1128/mcb.15.7.3813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikegawa S, Isomura M, Koshizuka Y, Nakamura Y. Cloning and characterization of ASH2L and Ash2l, human and mouse homologs of the Drosophila ash2 gene. Cytogenet Cell Genet. 1999;84:167–172. doi: 10.1159/000015248. [DOI] [PubMed] [Google Scholar]
- Jenuwein T, Allis CD. Translating the histone code. Science. 2001;293:1074–1080. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- Kennison JA. The polycomb and trithorax group proteins of Drosophila: Trans-regulators of homeotic gene function. Annu Rev Genet. 1995;29:289–303. doi: 10.1146/annurev.ge.29.120195.001445. [DOI] [PubMed] [Google Scholar]
- Knoepfler PS, Eisenman RN. Sin meets NuRD and other tails of repression. Cell. 1999;99:447–450. doi: 10.1016/s0092-8674(00)81531-7. [DOI] [PubMed] [Google Scholar]
- Kouzarides T. Histone methylation in transcriptional control. Curr Opin Genet Dev. 2002;12:198–209. doi: 10.1016/s0959-437x(02)00287-3. [DOI] [PubMed] [Google Scholar]
- Kristie TM. The mouse homologue of the human transcription factor C1 (host cell factor). Conservation of forms and function. J Biol Chem. 1997;272:26749–26755. doi: 10.1074/jbc.272.42.26749. [DOI] [PubMed] [Google Scholar]
- Kristie T, Sharp P. Interactions of the Oct-1 POU subdomains with specific DNA sequences and the HSV a trans-activator protein. Genes & Dev. 1990;4:2383–2396. doi: 10.1101/gad.4.12b.2383. [DOI] [PubMed] [Google Scholar]
- Kristie TM, Pomerantz JL, Twomwey TC, Parent SA, Sharp PA. The cellular C1 factor of the herpes simplex virus enhancer complex is a family of polypeptides. J Biol Chem. 1995;270:4387–4394. doi: 10.1074/jbc.270.9.4387. [DOI] [PubMed] [Google Scholar]
- Krogan NJ, Dover J, Khorrami S, Greenblatt JF, Schneider J, Johnston M, Shilatifard A. COMPASS, a histone H3 (lysine 4) methyltransferase required for telomeric silencing of gene expression. J Biol Chem. 2002;277:10753–10755. doi: 10.1074/jbc.C200023200. [DOI] [PubMed] [Google Scholar]
- Kurdistani SK, Robyr D, Tavazoie S, Grunstein M. Genome-wide binding map of the histone deacetylase Rpd3 in yeast. Nat Genet. 2002;31:248–254. doi: 10.1038/ng907. [DOI] [PubMed] [Google Scholar]
- Kuzmichev A, Nishioka K, Erdjument-Bromage H, Tempst P, Reinberg D. Histone methyltransferase activity associated with a human multiprotein complex containing the Enhancer of Zeste protein. Genes & Dev. 2002;16:2893–2905. doi: 10.1101/gad.1035902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaBoissière S, Walker S, O'Hare P. Concerted activity of host cell factor subregions in promoting stable VP16 complex assembly and preventing interference by the acidic activation domain. Mol Cell Biol. 1997;17:7108–7118. doi: 10.1128/mcb.17.12.7108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laherty CD, Billin AN, Lavinsky RM, Yochum GS, Bush AC, Sun JM, Mullen TM, Davie JR, Rose DW, Glass CK, et al. SAP30, a component of the mSin3 corepressor complex involved in N-CoR-mediated repression by specific transcription factors. Mol Cell. 1998;2:33–42. doi: 10.1016/s1097-2765(00)80111-2. [DOI] [PubMed] [Google Scholar]
- Lai JS, Herr W. Ethidium bromide provides a simple tool for identifying genuine DNA-independent protein associations. Proc Natl Acad Sci. 1992;89:6958–6962. doi: 10.1073/pnas.89.15.6958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaJeunesse D, Shearn A. Trans-regulation of thoracic homeotic selector genes of the antennapedia and bithorax complexes by the trithorax group genes: Absent, small, and homeotic discs 1 and 2. Mech Dev. 1995;53:123–139. doi: 10.1016/0925-4773(95)00430-0. [DOI] [PubMed] [Google Scholar]
- Leinbach SS, Summers WC. The structure of herpes simplex virus type 1 DNA as probed by micrococcal nuclease digestion. J Gen Virol. 1980;51:45–59. doi: 10.1099/0022-1317-51-1-45. [DOI] [PubMed] [Google Scholar]
- Litt MD, Simpson M, Gaszner M, Allis CD, Felsenfeld G. Correlation between histone lysine methylation and developmental changes at the chicken β-globin locus. Science. 2001;293:2453–2455. doi: 10.1126/science.1064413. [DOI] [PubMed] [Google Scholar]
- Lu R, Yang P, O'Hare P, Misra V. Luman, a new member of the CREB/ATF family, binds to herpes simplex virus VP16-associated host cellular factor. Mol Cell Biol. 1997;17:5117–5126. doi: 10.1128/mcb.17.9.5117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu R, Yang P, Padmakumar S, Misra V. The herpesvirus transactivator VP16 mimics a human basic domain leucine zipper protein, Luman, in its interaction with HCF. J Virol. 1998;72:6291–6297. doi: 10.1128/jvi.72.8.6291-6297.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luciano RL, Wilson AC. An activation domain in the C-terminal subunit of HCF-1 is important for transactivation by VP16 and LZIP. Proc Natl Acad Sci. 2002;99:13403–13408. doi: 10.1073/pnas.202200399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller T, Krogan NJ, Dover J, Erdjument-Bromage H, Tempst P, Johnston M, Greenblatt JF, Shilatifard A. COMPASS: A complex of proteins associated with a trithorax-related SET domain protein. Proc Natl Acad Sci. 2001;98:12902–12907. doi: 10.1073/pnas.231473398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagy L, Kao HY, Chakravarti D, Lin RJ, Hassig CA, Ayer DE, Schreiber SL, Evans RM. Nuclear receptor repression mediated by a complex containing SMRT, mSin3A, and histone deacetylase. Cell. 1997;89:373–380. doi: 10.1016/s0092-8674(00)80218-4. [DOI] [PubMed] [Google Scholar]
- Nagy PL, Griesenbeck J, Kornberg RD, Cleary ML. A trithorax-group complex purified from Saccharomyces cerevisiae is required for methylation of histone H3. Proc Natl Acad Sci. 2002;99:90–94. doi: 10.1073/pnas.221596698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng HH, Bird A. Histone deacetylases: Silencers for hire. Trends Biochem Sci. 2000;25:121–126. doi: 10.1016/s0968-0004(00)01551-6. [DOI] [PubMed] [Google Scholar]
- Noma K, Grewal SI. Histone H3 lysine 4 methylation is mediated by Set1 and promotes maintenance of active chromatin states in fission yeast. Proc Natl Acad Sci. 2002;99 Suppl 4:16438–16445. doi: 10.1073/pnas.182436399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noma K, Allis CD, Grewal SI. Transitions in distinct histone H3 methylation patterns at the heterochromatin domain boundaries. Science. 2001;293:1150–1155. doi: 10.1126/science.1064150. [DOI] [PubMed] [Google Scholar]
- O'Hare P. The virion transactivator of herpes simplex virus. Semin Virol. 1993;4:145–155. [Google Scholar]
- Papamichos-Chronakis M, Petrakis T, Ktistaki E, Topalidou I, Tzamarias D. Cti6, a PHD domain protein, bridges the Cyc8-Tup1 corepressor and the SAGA coactivator to overcome repression at GAL1. Mol Cell. 2002;9:1297–1305. doi: 10.1016/s1097-2765(02)00545-2. [DOI] [PubMed] [Google Scholar]
- Perez-Canadillas JM, Varani G. Recent advances in RNA–protein recognition. Curr Opin Struct Biol. 2001;11:53–58. doi: 10.1016/s0959-440x(00)00164-0. [DOI] [PubMed] [Google Scholar]
- Proft M, Struhl K. Hog1 kinase converts the Sko1–Cyc8–Tup1 repressor complex into an activator that recruits SAGA and SWI/SNF in response to osmotic stress. Mol Cell. 2002;9:1307–1317. doi: 10.1016/s1097-2765(02)00557-9. [DOI] [PubMed] [Google Scholar]
- Rea S, Eisenhaber F, O'Carroll D, Strahl BD, Sun Z-W, Schmid M, Opravil S, Mechtler K, Ponting CP, Allis CD, et al. Regulation of chromatin structure by site-specific histone H3 methyltransferases. Nature. 2000;406:593–598. doi: 10.1038/35020506. [DOI] [PubMed] [Google Scholar]
- Reilly PT, Herr W. Spontaneous reversion of tsBN67 cell proliferation and cytokinesis defects in the absence of HCF-1 function. Exp Cell Res. 2002;277:119–130. doi: 10.1006/excr.2002.5551. [DOI] [PubMed] [Google Scholar]
- Reilly PT, Wysocka J, Herr W. Inactivation of the retinoblastoma protein family can bypass the HCF-1 defect in tsBN67 cell proliferation and cytokinesis. Mol Cell Biol. 2002;22:6767–6778. doi: 10.1128/MCB.22.19.6767-6778.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roguev A, Schaft D, Shevchenko A, Pijnappel WW, Wilm M, Aasland R, Stewart AF. The Saccharomyces cerevisiae Set1 complex includes an Ash2 homologue and methylates histone 3 lysine 4. EMBO J. 2001;20:7137–7148. doi: 10.1093/emboj/20.24.7137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos-Rosa H, Schneider R, Bannister AJ, Sherriff J, Bernstein BE, Emre NCT, Schreiber SL, Mellor J, Kouzarides T. Active genes are tri-methylated at K4 of histone H3. Nature. 2002;419:407–411. doi: 10.1038/nature01080. [DOI] [PubMed] [Google Scholar]
- Serrano M, Lin AW, McCurrach ME, Beach D, Lowe SW. Oncogenic ras provokes premature cell senescence associated with accumulation of p53 and p16INK4a. Cell. 1997;88:593–602. doi: 10.1016/s0092-8674(00)81902-9. [DOI] [PubMed] [Google Scholar]
- Strahl BD, Ohba R, Cook RG, Allis CD. Methylation of histone H3 at lysine 4 is highly conserved and correlates with transcriptionally active nuclei in Tetrahymena. Proc Natl Acad Sci. 1999;96:14967–14972. doi: 10.1073/pnas.96.26.14967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel JL, Kristie TM. The novel coactivator C1 (HCF) coordinates multiprotein enhancer formation and mediates transcription activation by GABP. EMBO J. 2000;19:683–690. doi: 10.1093/emboj/19.4.683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vojtek AB, Hollenberg SM, Cooper JA. Mammalian Ras interacts directly with the serine/threonine kinase Raf. Cell. 1993;74:205–214. doi: 10.1016/0092-8674(93)90307-c. [DOI] [PubMed] [Google Scholar]
- Wang J, Zhou Y, Yin B, Du G, Huang X, Li G, Shen Y, Yuan J, Qiang B. ASH2L: Alternative splicing and downregulation during induced megakaryocytic differentiation of multipotential leukemia cell lines. J Mol Med. 2001;79:399–405. doi: 10.1007/s001090100222. [DOI] [PubMed] [Google Scholar]
- Wells L, Vosseller K, Hart GW. Glycosylation of nucleocytoplasmic proteins: Signal transduction and O-GlcNAc. Science. 2001;291:2376–2378. doi: 10.1126/science.1058714. [DOI] [PubMed] [Google Scholar]
- Wilson AC, LaMarco K, Peterson MG, Herr W. The VP16 accessory protein HCF-1 is a family of polypeptides processed from a large precursor protein. Cell. 1993a;74:115–125. doi: 10.1016/0092-8674(93)90299-6. [DOI] [PubMed] [Google Scholar]
- Wilson AC, Cleary MA, Lai J-S, LaMarco K, Peterson MG, Herr W. Combinatorial control of transcription: The herpes simplex virus VP16-induced complex. Cold Spring Harbor Symp Quant Biol. 1993b;18:167–178. doi: 10.1101/sqb.1993.058.01.021. [DOI] [PubMed] [Google Scholar]
- Wilson AC, Peterson MG, Herr W. The HCF repeat is an unusual proteolytic cleavage signal. Genes & Dev. 1995;9:2445–2458. doi: 10.1101/gad.9.20.2445. [DOI] [PubMed] [Google Scholar]
- Wilson AC, Freiman RN, Goto H, Nishimoto T, Herr W. VP16 targets an amino-terminal domain of HCF involved in cell cycle progression. Mol Cell Biol. 1997;17:6139–6146. doi: 10.1128/mcb.17.10.6139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wysocka J, Reilly PT, Herr W. Loss of HCF-1 chromatin association precedes temperature-induced growth arrest of tsBN67 cells. Mol Cell Biol. 2001;21:3820–3829. doi: 10.1128/MCB.21.11.3820-3829.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X, Zhang F, Kudlow JE. Recruitment of O-GlcNAc transferase to promoters by corepressor mSin3A: Coupling protein O-GlcNAcylation to transcriptional repression. Cell. 2002;110:69–80. doi: 10.1016/s0092-8674(02)00810-3. [DOI] [PubMed] [Google Scholar]
- Zhang W, Chait BT. ProFound: An expert system for protein identification using mass spectrometric peptide mapping information. Anal Chem. 2000;72:2482–2489. doi: 10.1021/ac991363o. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Reinberg D. Transcription regulation by histone methylation: Interplay between different covalent modifications of the core histone tails. Genes & Dev. 2001;15:2343–2360. doi: 10.1101/gad.927301. [DOI] [PubMed] [Google Scholar]