Abstract
Bacteriophage λ Q-protein stably binds and modifies RNA polymerase (RNAP) to a termination-resistant form. We describe amino acid substitutions in RNAP that disrupt Q-mediated antitermination in vivo and in vitro. The positions of these substitutions in the modeled RNAP/DNA/RNA ternary elongation complex, and their biochemical properties, suggest that they do not define a binding site for Q in RNAP, but instead act by impairing interactions among core RNAP subunits and nucleic acids that are essential for Q modification. A specific conjecture is that Q modification stabilizes interactions of RNAP with the DNA/RNA hybrid and optimizes alignment of the nucleic acids in the catalytic site. Such changes would inhibit the activity of the RNA hairpin of an intrinsic terminator to disrupt the 5′-terminal bases of the hybrid and remove the RNA 3′ terminus from the active site.
Keywords: RNA polymerase, antitermination, termination, transcription, Q-protein, bacteriophage λ
Both specific and general elongation factors promote transcription through natural barriers in chromosomes. These include the discrete termination sites of prokaryotes, and less well-defined transcription barriers of eukaryotes: promoter-proximal pause sites, 3′-processing-linked terminators, and nucleosomes. General transcription elongation factors include NusA and NusG of bacteria and the highly conserved Spt4/Spt5/Spt6 proteins of eukaryotic Pol II; regulon-specific factors include the Q-protein and N-protein of the Escherichia coli bacteriophage λ and the TAT protein of HIV (Roberts 1988; Greenblatt et al. 1993; Laspia et al. 1993; Roberts et al. 1998). The recent atomic resolution structures of multisubunit prokaryotic and eukaryotic RNA polymerases (RNAP), including structures and models of elongation complexes, provide an opportunity to describe the activity of these factors in molecular detail (Zhang et al. 1999; Cramer et al. 2000; Ebright 2000; Korzheva et al. 2000; Gnatt et al. 2001; Korzheva and Mustaev 2001; Murakami et al. 2002a,b; Vassylyev et al. 2002). The polymerases share a strikingly similar structural core, suggesting that insights into regulatory regions, domains, and even individual residues may be universal.
The phage-λ-encoded Q-protein antitermination factor, and Q-proteins of related phages including φ21 and φ82, are stably incorporated into RNAP during a promoter-proximal σ70-dependent pause (Yarnell and Roberts 1992; Ring et al. 1996; Roberts et al. 1998). Q-modified complexes have fundamentally altered elongation properties, displaying increased elongation rate, diminished pausing, and resistance to all downstream intrinsic and ρ-dependent termination sites; these properties can be expressed in a defined minimal biochemical system containing RNAP, Q-protein, and the elongation factor NusA. Although the correlation of antipausing and antitermination originally suggested a kinetic basis of antitermination—namely, that RNAP might elongate faster than some rate-limiting step of termination—it is clear that Q induces a more fundamental stabilization of the transcription complex. Thus, static Q-modified elongation complexes stopped by a DNA cross-link at the release site of an intrinsic terminator in vitro are stabilized against the dissociation activity of the terminator (T.J. Santangelo and J.W. Roberts, unpubl.). Similarly, static Q-modified complexes are stabilized at any elongation site against attack by an oligonucleotide that simulates the activity of an intrinsic terminator (Yarnell and Roberts 1999). Finally, Q-modified elongation complexes resist the action of a strong intrinsic terminator when they are transcribing extremely slowly because of limiting NTP concentrations (Roberts et al. 1998).
To understand the molecular basis of Q modification, we isolated mutations altering the core subunits of RNAP that interfere with Q function but do not impair the basic enzymatic activity of RNAP. We used both Qλ and the analogously functioning Q82 from phage φ82. Although Qλ and Q82 both modify RNAP at a σ70-dependent promoter-proximal pause, they share no obvious sequence homology and act only on their own phage promoters with specific associated sequences (Yang et al. 1989). Presumably, they nonetheless act by the same basic mechanism and may share structural homology.
To interpret the phenotypes of the mutant RNAPs, we consider the distinctive nature of the interaction between Q and RNAP. Q can only productively bind and functionally modify RNAP in a paused elongation complex containing the initiating σ70 subunit; Q makes direct contact with a specific site in DNA, with RNAP core subunits, and, for Qλ at least, with region 4 of σ70 (Ko et al. 1998; Nickels et al. 2002). Importantly, after RNAP clears this pause, Q no longer can bind the unmodified complex. Productive engagement of Q releases RNAP from the pause site and Q becomes a firmly bound subunit, stable (for Q82 at least) to repeated washing of immobilized complexes in vitro; σ70 can (and might usually) dissociate at this stage.
Because of the site-specific and irreversible nature of this association, the only step in Q function that is plausibly sensitive to Q concentration is its initial engagement with RNAP at the pause site. In fact, most of the defects exhibited by the mutant RNAPs we identified are rescued partly or completely at increased concentrations of Q, implying that these defects are expressed at the pause site. Because formation of the paused complex itself is not impaired (data not shown), only two sorts of mutational change are likely to be rescued by a higher concentration of Q. First, amino acid substitutions could damage a site where Q makes direct contact with the enzyme. Second, a mutational change in RNAP could disrupt other molecular interactions required for the modification that stably incorporates Q at the pause site. The mutational sites that we identified and describe here form several discrete clusters in conserved and functionally important parts of RNAP that are largely exposed within the nucleic acid-binding, catalytic cleft, but likely are buried and inaccessible to an external protein in the elongation complex. Based on these locations, and the in vitro properties of the modified enzymes, we argue that our mutations act primarily to disrupt molecular interactions within the large core subunits and do not define a discrete binding site for Q on RNAP. We further conjecture that these loci identify molecular interactions required not only to incorporate the Q polypeptide, but also to mediate antitermination itself.
We report below in Results the locations of the amino acid substitutions and properties of the mutationally altered core RNAPs, and in the Discussion we propose structural interpretations and implications for Q function.
Results
Mutations affecting Q activity in vivo
Substitutions in the core subunits of RNAP that disrupt Q-mediated antitermination were isolated from mutagenized libraries using reporter constructs specific for each Q-protein (see Materials and Methods). Briefly, these reporters contained the λ late promoter upstream of two strong intrinsic terminators, followed by the lac operon, making β-galactosidase expression Q-dependent. Mutagenized plasmid libraries of rpoB and rpoC (encoding the β and β‘ subunits, respectively) were introduced to strains carrying the reporters and the appropriate Q source, and mutations limiting β-galactosidase expression were isolated. To generate a positive phenotype, the screens require that overproduced plasmid-encoded protein incorporate into and produce an active RNAP; the wild-type genomic copy of rpoB or rpoC was still expressed. RNAP preparations from cells grown under conditions used for the screens show that ∼85%–90% of RNAPs contained the plasmid-encoded His6-tagged subunit (data not shown).
We isolated 17 point mutations in rpoB or rpoC that significantly diminish Q reporter activity: 11 individual mutations, and 3 pairs for which each mutation alone has no phenotype (Fig. 1). Seven mutants were isolated with each Q reporter, excluding alternative changes at the same site. Of the 17 substitutions, 16 modify the β subunit of RNAP; most mutations cluster between or within conserved regions D, E, F, or G (Fig. 1B). The remaining mutation maps to conserved region G of the β‘ subunit. These sites are primarily in domains structurally conserved between Thermus aquaticus RNAP, Thermus thermophilus RNAP, and Saccharomyces cerevisiae RNA Pol II Δ4/Δ7 (Zhang et al. 1999; Cramer et al. 2000; Ebright 2000; Vassylyev et al. 2002), suggesting that they affect basic elements of the general transcription process, and could further define the role(s) of individual domains and residues for all multisubunit RNAPs.
Figure 1.
Mutant phenotypes. (A) Compilation of RNAP mutants affecting Q activity. Substitutions shown in pairs only diminish Q activity as pairs, and are shown twice to position each bold substitution in the correct spatial group. Mutant classes as discussed in the text are grouped and indicated by background color. β-Gal activity is the average of percent wild-type (% WT) activity at each arabinose concentration. Phage spotting efficiency is relative to the same strain carrying wild-type β or β‘. A value of 4 indicates growth identical to wild type; 0 represents no plaque growth. In vitro Q activity is indicated by two values: the ratio of Q concentrations (WT/mutant) required for half saturation of readthrough activity (1/2sat); and the fraction of wild-type readthrough obtained at the highest Q concentration (RTmax). (B) Maps of RpoB and RpoC with conserved segments in blue. The regions displaying essentially identical tertiary folds between T. aquaticus RNAP and yeast Pol II are shown beneath in green (Ebright 2000). The rifampicin-binding site is shown (Jin and Gross 1988); the active center is defined by hydroxyl radical cleavage mediated by an Fe2+ ion chelated in the active site (Mustaev et al. 1997). Substitutions isolated with the Qλ reporter are at top (black), and those isolated with the Q82 reporter are below (gray); the dot color indicates the mutant class. DR, dispensable region.
We tested each mutant, regardless of the reporter with which it was originally isolated, with both reporters (Figs. 1, 2). Each mutant decreases expression 2–10-fold with the reporter for which it was selected, and most reduce expression of both reporters, consistent with a common mechanism of action of both Q-proteins. However, some mutants are defective quite specifically for the reporter with which they were originally selected (e.g., β H673L for Q82). Furthermore, mutants obtained with the Qλ reporter tend to affect Q82 more strongly than mutants obtained with the Q82 reporter affect Qλ, suggesting some differences in the specific modification pathway of each Q-protein.
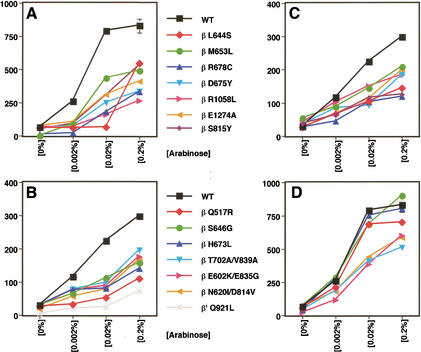
Figure 2.
β-Galactosidase activity of the mutant RNAPs. (A) Activity of the Qλ reporter with substitutions isolated with the Qλ reporter. (B) Activity of the Q82 reporter with substitutions isolated with the Q82 reporter. (C) Activity of the Qλ reporter with substitutions isolated with the Q82 reporter. (D) Activity of the Q82 reporter with substitutions isolated with the Qλ reporter. Activities of the Q82 and Qλ reporters were identical with wild-type β or wild-type β‘, and only one curve is shown for simplicity. Q-protein synthesis was induced with arabinose. Synthesis of the mutant polymerase subunit was induced with 1% lactose.
Strikingly, none of the mutations originally isolated against the Qλ reporter were found with the Q82 reporter, and vice versa, although in several cases substitutions selected with one reporter mapped within a few residues of substitutions selected for the other (Fig. 1). The Q82 screen was dominated by C-terminal rpoB deletions that inhibit Q82 activity, limiting the total number of mutations isolated, and therefore was not saturated (data not shown; see Materials and Methods).
The inhibitory effect and Q-protein specificity of each substitution was tested further by growing phage carrying different Q-proteins on lawns of bacteria expressing the mutant β or β‘ subunit (Figs. 1, 3). In addition to Qλ and Q82, we tested phage carrying the Q-protein from phage φ21 (Q21), which represents a third and distinct specificity, and has limited or no sequence homology with Qλ or Q82 (Guo et al. 1991). The strongest example of specificity, β Q517R, almost completely blocks growth of Q82 phage, whereas this same substitution does not significantly affect Qλ or Q21 phage. Several mutants, for example, β R678C, significantly inhibit growth of all phage. Some substitutions show overlapping deficiencies with Q21; for example, β‘ Q921L blocks growth of the Q82 and Q21 phages, but does not affect the growth of Qλ phage. The full results of the phage spotting are compiled in Figure 1. These results are in agreement with the specificity seen with the β-galactosidase assays, and in most cases show comparable effects.
Figure 3.
Phage spotting with mutant RNAPs. Tenfold serial dilutions of phage harboring different Q-proteins were spotted onto lawns of bacteria expressing the mutant RNAPs. Full results of the phage spotting are presented in Figure 1. Experimental plates contained 1% lactose to induce expression of the mutant RNAP subunit. Inducing expression of the wild-type subunits on pRL663 or pRL706 did not impair phage growth. Qλ phage are λ+ (left) and the clear mutant, λc17 (right); the Q21 phage is iλQ21; the Q82 phage are iλQ82 (left) and φ82 (right).
In vitro transcription with mutant RNAPs
We used in vitro transcription with purified mutant enzymes both to confirm that the defect in Q function is a property of the modified core subunit and to characterize the defect among the distinguishable steps of the in vitro reaction. All of the mutant RNAPs disrupt Q-mediated antitermination in vitro, with substantial conservation of the Q-protein specificity seen in vivo (Figs. 1, 4, 5). Furthermore, the magnitude of the defects seen in both the β-galactosidase and phage-plating assays correlates well with the magnitude of the in vitro defects. For example, β Q517R and β‘ Q921L show strong defects in supporting Q82-mediated antitermination in all three assays (β-gal assays, phage growth, and in vitro transcription), but mild effects on Qλ-mediated antitermination, particularly when antitermination activity is corrected to account for differences in basal level termination. In contrast to the Q-specificity shown by β Q517R, both β R678C and β N620I/β D814V display significant defects supporting antitermination by both Q-proteins both in vivo and in vitro, although the effects are greater for Qλ.
Figure 4.
Gel analysis of in vitro transcription with purified mutant RNAPs. (A) A gel image of transcription with Qλ protein, at 0, 0.1, 1, 5, 10, 100, and 200 nM; all reactions contained 150 nM NusA. The positions of the abortive products (AP), +16/+17 (the σ70-dependent pause), terminated (T), and readthrough product (RT) are shown to the left. (B) Molar percent readthrough of terminator in experiment of A. (C) The gel image of abortive products (AP) and cleavage products (CP) made by β R678C RNAP. A single-round reaction was sampled at 0.25, 0.5, 1, 2, 3, 5, 8, and 12 min. Note that the absence of abortive products >8 nt in β R678C reveals a short-lived paused species at +11, +12, and +13.
Figure 5.
Compilation of mutant RNAP activity in vitro. (A) Transcription is as in Figure 4 with a sample of RNAPs that were isolated against either Q-reporter. The right panel shows the level of Qλ-mediated antitermination supported by these substitutions; the left panel shows the Q82-mediated antitermination supported by these substitutions; all reactions contained 150 nM NusA. (B) Defect of β S646G in NusA-stimulated Q-mediated antitermination. (C) Termination defects of β E1274A and β‘ Q921L in the absence of NusA. The addition of NusA to transcription reactions (150 nM) eliminates much of the termination defect. Both mutant RNAPs are defective in response to Q, although the large backgrounds make the defect difficult to quantify.
The concentration dependence of Q function with mutant enzymes reveals two patterns of activity: some mutant RNAPs achieve wild-type levels of antitermination but require higher concentrations (e.g., β‘ Q921L), whereas others reach only partial activity at the highest concentrations (e.g., β Q517R and β R678C with both Q-proteins). Thus, in Figure 1 we report both maximal activity and the concentration of Q that gives half-maximal activity.
In general, a mutation could affect either the initial engagement of Q at the pause site or Q antitermination activity during downstream transcription. As argued above, the Q-concentration-dependent rescue of antitermination activity implies that the mutants affect primarily the engagement step. Note in Figure 4 that chase of the +16/+17 pause by Q is inefficient in the mutant β R678C, as is true for all of the mutants (data not shown), consistent with a defect at the site of modification. Although the downstream antitermination activity might be affected for mutant enzymes not rescued completely by higher concentrations of Q, in most such cases the activity curves are shifted to higher concentrations (e.g., cf. WT and β R678C in Fig. 5), which again supports the argument for an effect at the engagement site.
We consider three ways that the engagement step might be impaired. First, a mutation might disrupt σ70–core interactions or other elements that maintain RNAPs in the promoter-proximal pause site required for Q engagement. However, no substitution altered the efficiency of pausing or the half-life of this σ70-dependent pause (data not shown). Second, a substitution might impair the ability of the host elongation factor NusA to stimulate Q-mediated antitermination; however, with one exception (β S646G), the substitutions do not solely affect NusA-stimulation of Q-mediated antitermination (see below; Fig. 5B). Finally, a substitution might prevent or impair the conversion of core by Q-protein to a termination-resistant form, either by altering a recognition site or by changing the ability of core to respond to Q. The positions of the substitutions are inconsistent with a simple binding site for Q and we suggest therefore that these substitutions disrupt other elements of RNAP required for its conversion to a termination-resistant form at the pause site.
β S646G is specifically defective in allowing NusA to stimulate antitermination at low Q concentrations, showing little to no effect on Q activity in the absence of NusA (Fig. 5B). Furthermore, its effect is substantially specific to Q82, although the deficiency is overcome completely at higher Q concentrations. NusA acts at least in part by stabilizing the binding of Q to the paused complex (Yarnell and Roberts 1992), and β S646G could impair this interaction.
There are two other phenotypes of these mutants, independent of antitermination, that could offer insight into their mechanism. Substitutions that modify residues within the secondary channel (β 673, 675, 678; see Discussion), and, perhaps surprisingly, residues on the surface of β (644, 653), show a loss of 3′-end cleavage products, which are generated from cleavage of an internal phosphodiester linkage in a backtracked complex (Fig. 4C; Orlova et al. 1995). These same substitutions also alter the amount and maximum size of abortive products made from the λ late promoter (Fig. 4C). The mechanism underlying abortive initiation is unknown, although the process likely is akin to backtracking, in which the RNA 3′ terminus exits the active site and enters the secondary channel. The relevance of these phenotypes remains to be understood.
It is noteworthy that all of the mutant RNAPs show some change in termination efficiency in the absence of Q, although for most RNAPs this change is modest. For any given mutant RNAP, termination efficiency is increased at some terminators but decreased at other terminators (data not shown), making a simple interpretation of the underlying mechanism impossible. However, this effect implies that the mutations influence enzymatic states that determine both the ability of Q-proteins to mediate antitermination and the general efficiency of intrinsic termination. Two mutant RNAPs (β E1274A and β‘ Q921L), which map near the active site, display particularly strong termination anomalies in the absence of Q (Fig. 5C), leading to dramatic increases or decreases in termination efficiency at different terminators. These termination defects can be almost completely suppressed by the addition of NusA to transcription reactions (Fig. 5C).
Discussion
To consider how these substitutions might inhibit Q function, we localize them in the ternary complex model of the bacterial RNAP of Korzheva et al. (2000), with reference also to the yeast Pol II elongation complex crystal structure (Gnatt et al. 2001) and the T. thermophilus RNAP structure (Vassylyev et al. 2002). Figure 6 shows the modeled elongation complex, highlighting the proposed path of the DNA template and RNA transcript through the enzyme. The nucleic acids bind into a channel comprised entirely of the β and β‘ subunits, which form a claw-like structure; the active site resides deep within the channel at the base of the claw (Fig. 7).
Figure 6.
Structural model of the prokaryotic TEC, highlighting the position of substitutions. (A) A model of T. aquaticus TEC was used to map the substitutions onto the three-dimensional structure of RNAP (Korzheva et al. 2000). Equivalent positions in the T. aquaticus structure are highlighted and labeled with E. coli residue numbers. The left and center panels are equivalent except that the β subunit has been turned to a wire frame to expose the path of the nucleic acids through the enzyme. RNAP in this orientation is moving from left to right. The colors are as follows: α1 NTD, yellow; α2 NTD, green; β, cyan; β‘, violet; nontemplate strand, dark gray; template strand, light gray; RNA transcript, orange; β‘ f-helix, purple; β helix 673–687, dark blue; active-site Mg2+, dark green. Substitutions are labeled, space-filled, and colored as in Figure 1. Residue 1274 is hidden in this view, as indicated by the broken line. (B) The surface patch required for efficient Q activity, shown in two views of T. aquaticus RNAP. The left view is essentially down the secondary channel, and the right is as in A.
Figure 7.
Substitutions within the active site and hybrid region of RNAP. A stereo view looking directly into the main channel of RNAP (RNAP is moving right to left in this view). Substitutions are colored and labeled equivalently to Figure 6 (none of the surface patch substitutions are seen in this view). The top image is essentially identical to that on the lower one, which contains the nucleic acids modeled into the complex with the template strand and RNA transcript shown in ball and stick (white and orange, respectively). The nontemplate strand was reduced to wire frame to clarify the image (gray). The colors are as follows: β, cyan; β‘, violet; both α NTDs, gray, for clarity only; β flap, dark blue, partially visible on extreme right side of both images; β‘ f-helix and rudder, purple; β helix from 673 to 687, dark blue.
Our substitutions cluster into four distinct groups, three of which are in the conserved core of RNAP and surface-exposed within the main nucleic acid-binding cleft, and thus effectively internal in the clamped elongation complex. The substitutions change residues: I—along the path of the DNA/RNA hybrid; II—within the secondary channel; III—near the active site and the template-primer end; and IV—in an external surface-exposed patch, which we argue nevertheless may change protein–nucleic acid interactions in the nucleic-acid-binding channel.
Although we have no direct evidence of their mechanism of action, we consider it unlikely that most of these substitutions reflect sites of interaction between Q and the RNAP core. Their locales suggest, instead, roles in Q-dependent function of the core elements, as we discuss specifically for each class.
Class I. Substitutions within the hybrid binding region/β flap domain (β 517, β 835, β 839, and β 1058)
These substitutions, colored dark green in Figures 6A and 7, change residues in the main channel of RNAP immediately adjacent to the DNA/RNA hybrid, from positions −4 to −8. The elongation complex of Pol II, which is likely to be very similar to that of E. coli, lacks protein–nucleic acid interactions with the emerging transcript beyond position −4. Gnatt et al. (2001) suggest that RNA beyond −4 is held in place solely by base-pairing to the very tightly held template strand. The locations of the four residues we identified suggest that they could extend the region of intimate protein–hybrid contacts in a reconfigured, termination-resistant complex. A specific suggestion is that Q modification of RNAP repositions β 517, β 835, β 839, and β 1058 and/or neighboring residues to make direct contacts with the hybrid, thereby stabilizing the ternary elongation complex (TEC) against the dissociation by an intrinsic terminator hairpin; in this model, the substitutions we isolated would impair these contacts. This proposal implies that these residues are not involved in contacts with the hybrid in the absence of Q modification, and it is consistent that the mutant enzymes display no significant differences in overall activity compared with wild-type (WT) RNAP. It is noteworthy that Class I mutations are in or near the binding pocket of the antibiotic rifampicin (Campbell et al. 2001). Some mutations that confer rifampicin resistance, such as the well-studied alleles rpoB2 and rpoB8, elicit strong effects on elongation rate, pausing, and termination efficiency (Jin et al. 1988; McDowell et al. 1994; Yarnell and Roberts 1999; Campbell et al. 2001).
Classes II and III. Substitutions surrounding the catalytic center: the secondary channel (β 673, β 675, β 678) and active site or bridge helix (β 814, β 815, β 1274, β‘ 921)
We consider together these two sets of mutations that cluster around the active center and that could affect catalysis.
The main channel is divided by an extended helical segment, the β‘ f-helix or “bridge” helix (shown in purple in Figs. 6A, 7), forming the “secondary channel” that may serve as the entry site of NTP substrates (Zhang et al. (1999). The three substitutions β 673, β 675, and β 678 (colored yellow in Figs. 6A, 7) are all surface-exposed in the secondary channel near its junction with the main channel and active site. They also cluster near one end of an amphipathic α-helix (β 673–687, shown as a blue ribbon in Figs. 6A, 7), that runs perpendicular to the β‘ f-helix and traverses the secondary and main channels, and is adjacent to sites of Class I mutations at its other end. Direct evidence that the substitution β R678C affects the active site is its inhibition of 3′-transcript cleavage (Fig. 4C); the effect could be manifested through the role of β R678 in helping chelate the second metal ion in the active site via a water contact (Vassylyev et al. 2002). Alternatively, β R678C might inhibit extrusion of the 3′ end of the transcript through the secondary channel, as is believed to occur to create a backtracked complex. A further connection of this cluster to the active center involves contact of β 675 with the β‘ f-helix, movements of which have been proposed to mediate translocation (Gnatt et al. 2001; Bushnell et al. 2002; Epshtein et al. 2002; Vassylyev et al. 2002). Another potential effect of Class II mutations results from the close approach of the other end of the β 673–687 helix to the Class I mutations; by subtly changing the position of this helix, substitutions at β 673, β 675, and β 678 could interfere with protein–RNA contacts in the hybrid, as proposed for Class I mutations.
Class III substitutions at β 814 and β 815 (colored red in Figs. 6A, 7) are at the base of the secondary channel, in direct proximity to the catalytic site. Substitution of these residues may result in direct changes in the arrangement of the active-site metals. The T. thermophilus RNAP structure shows a network of interactions coordinating two Mg2+ ions in the active site (Vassylyev et al. 2002); β D814 participates in chelating a second metal ion in the active site of RNAP, via two water contacts. Mutation of β D814 to valine removes the charge required to help chelate this metal ion without impairing RNAP function, implying that this contact is not strictly required, although the substitution could well perturb the activity of the catalytic center.
The configuration of the active site could be altered by the other two Class III substitutions, at β E1274 and β‘ Q921 (colored black in Figs. 6A, 7). β E1274 is directly adjacent to the template strand at positions −1 and +1, and participates in a network of interactions that contact and position the nucleic acids in the active site. Gnatt et al. (2001) identified a specific contact between the adjacent residue, β M1273, and the phosphate backbone of the template strand at −1/+1. The position and orientation of β M1273 likely is directly influenced by β E1274, and therefore changing β E1274 to a small residue (as was the case in all three substitutions described here) could alter contacts with the phosphate backbone of the template strand at this critical location. β E1274 also forms a salt bridge with conserved residue β‘ R425. Gnatt et al. (2001) identified a specific contact between β‘ R425 and the phosphate backbone of the RNA at −1/+1; therefore, changing β E1274 to an uncharged residue incapable of forming a salt bridge is likely to influence the positioning of the 3′-RNA terminus in the active site. β‘ Q921 is somewhat removed from the active-site residues, although it is near the base of the β‘ f-helix and could influence movements of this helix, such as those proposed to drive the translocation of RNAP (Gnatt et al. 2001; Epshtein et al. 2002; Vassylyev et al. 2002).
In summary, both Class II and Class III substitutions are positioned to alter the orientation of the active site and the template-primer end. We suggest that these mutations identify contacts normally made in Q-modified complexes that act to stabilize the active configuration of the catalytic site; in particular, contacts affected by Class II and III mutations could underlie the antipausing effect of the Q modification. The screens used here require that the mutant RNAP retains catalytic activity, thereby eliminating substitutions that grossly disrupt the catalytic center; presumably many more residues around the active site are critical to support Q modification of RNAP.
Class IV. Substitutions that define a surface-exposed patch on β (β 602, β 620, β 644, β 646, β 653, β 702)
Six substitutions (colored light green in Fig. 6B) are clustered and largely exposed on the surface of the β subunit in the elongation complex model, well away from the main or secondary channel. This external patch of mutations is fully 65–70 Å from the specific DNA-binding site for Q in the modeled elongation complex. These substitutions are in regions that border strongly conserved domains, although they are less conserved among even bacterial RNAPs than the other classes of substitutions; their positions in E. coli RNAP might not be modeled accurately by the T. aquaticus structure. Furthermore, the exact positions of these residues differ between the T. aquaticus and T. thermophilus structures, possibly because of errors in tracing the chain in the T. aquaticus structure (Vassylyev et al. 2002); however, in both structures the general position of these exposed loops on the surface of RNAP is clear. No specific functions or interactions have been ascribed to this region, although the mutationally defined surface patch is linked to the main channel of RNAP by both genetic and biochemical data. β S643R was isolated as a site-specific suppressor of the termination-altering properties of rpoB2 (β H526Y), demonstrating that substitutions within this surface patch can alter the configuration of the rifampicin-binding pocket, and therefore presumably alter the contacts made between the β subunit and the hybrid (R. Landick, unpubl.).
Our study defines three pairs of substitutions, which display defects only as pairs, that link this surface patch to the main channel of RNAP. β 620/814, β 602/835, and β 702/839 all have one residue in the main channel and one residue in the surface-exposed patch. We also found biochemical evidence linking substitutions in this surface patch to the active site, as both β L644S and β M653L display the same loss of cleavage products and abortive phenotype as β R678C (Fig. 4C; data not shown). Possibly alterations in the surface patch could be transmitted to either the active site or the hybrid binding cleft to optimize either reactive alignments or hybrid–RNAP stability.
Mechanism of Q-mediated antitermination
Several models for the mechanism of termination have been proposed (Landick 1997; Gusarov and Nudler 1999; Yarnell and Roberts 1999; Toulokhonov et al. 2001; Komissarova et al. 2002). Although their details differ, the models commonly propose that an initial step of intrinsic termination is the disruption of the upstream portion of the hybrid induced by hairpin formation; hairpin formation and hybrid disruption are linked. We envision two major, and not mutually exclusive, models to explain how Q modification of RNAP could counteract these linked reactions. First, a Q-modified complex might physically block hairpin formation, for example, by preventing nucleation of hairpin annealing at the upstream end of the downstream hairpin segment. Second, Q modification might stabilize interactions between RNAP and the hybrid, preventing the disruption of the upstream portion of the hybrid required for the concerted formation of the full hairpin.
The locations and behavior of our mutations are consistent with the second model. We argue that the phenotypes mostly are not direct defects in antitermination, but instead reflect impaired Q incorporation into the transcription complex at the pause site; for most mutants, a higher concentration of Q overcomes the defect. However, it seems unlikely that the mutations damage a simple binding site for Q; Classes I, II, and III are internal to the elongation complex, and even the external Class IV substitutions are implausibly distant from the DNA site to which Q binds in the elongation complex. Instead, we conjecture that the substitutions affect internal molecular interactions required for Q to functionally modify the complex, presumably after an initial binding event. Furthermore, we imagine that these alterations are also the molecular changes that define the antiterminating state itself. Specifically, we suggest that, in concert with incorporation of Q into the TEC, regions defined by our mutations undergo rearrangements to further clamp down on the nucleic acids within the TEC, both strengthening interactions with the hybrid and maintaining an optimal alignment of the active site; as we describe, the sites are plausibly located for this role. Such reconfiguration would inhibit termination by countering the disruptive force of hairpin formation, and reduce pausing by stabilizing the elongation competent configuration of the active site. Clearly, biochemical and structural analysis are required to test this model.
Materials and methods
Strains and plasmids
TOM100 is a derivative of SG22182A (Δara, Δlac, malP∷lacIq) with modifications made with standard molecular biology techniques: integration of the JG200 Qλ reporter (Simons et al. 1987; Ko et al. 1998; Guo 1999) and introduction of recA− [recA∷KanR via P1vir transduction from NT440 p226; a gift of S. Gottesman (Bethseda, MD)]. Single-copy integration of the reporter was confirmed via PCR (Powell et al. 1994). The recA− phenotype was confirmed by increased sensitivity to UV light (data not shown). TOM101 is like TOM100, except that it carries an integrated reporter of Q82 activity, JG205 (Guo 1999).
The JG200 reporter carries λ sequence containing the late promoter, λpR‘, to +49, followed by the intrinsic terminators t82 and to upstream of an otherwise promoterless lac operon (lacZYA). The JG205 reporter is identical to JG200 except it carries φ82 sequence containing the φ82 late promoter, p82a, to +51 in place of λpR‘.
All plasmids were made with standard molecular biology techniques. pBAD33-Qλ places Qλ under control of the arabinose-dependent pBAD promoter from pBAD33 (Guzman et al. 1995). pBAD33-Q82 is like pBAD33-Qλ except that it contains Q82. pRL663 (Wang et al. 1995) and pRL706 (Severinov et al. 1997) encode C-terminal His6-tagged versions of the rpoC and rpoB genes, respectively, under control of a lac-repressor-regulated trc promoter. The ORFs of β and β‘ were divided using flanking and internal unique restriction endonuclease sites into three or five sections, respectively (internal ClaI and BspEI sites for rpoB; SnaBI, SphI, SalI, and BspEI sites for rpoC). Each section was amplified via error-prone PCR (Caldwell and Joyee 1992), ligated into the original backbone containing unmutagenized flanking segments, and transformed into JM109 to yield >104 individual colonies. Plasmid DNA was prepared from the pooled colonies without further growth to yield eight independent PCR mutagenized libraries: five individual libraries of rpoC and three libraries for rpoB. Random isolates of the N-terminal rpoC library were sequenced and found to contain on average one to two randomly distributed base substitutions. After retransformation and replica plating to rich medium containing 1 mM IPTG and 50 μg Rif/mL, the middle segment rpoB library yielded approximately the frequency of rifampicin-resistant colonies expected, assuming one to two substitutions per plasmid and 40 possible bases that could be mutated to give Rif-resistance (Campbell et al. 2001).
Screen for mutations that affect Q activity in vivo
TOM100 carrying pBAD33-Qλ or TOM101 carrying pBAD33-Q82 was transformed with each library of pRL663 or pRL706 individually. Transformants were grown in LB plus appropriate antibiotics for 3 h, washed, and plated onto MacConkey indicator agar with 1% lactose, appropriate antibiotics, and 0.002% arabinose (13 μM). White colonies were picked and rescreened on identical plates. Total plasmid DNA was prepared from positive colonies and transformed into DH5α; AmpR, CmS colonies were selected. The AmpR library plasmid was purified from DH5α and rescreened in TOM100 pBAD33-Qλ or TOM101 pBAD33-Q82 on MacConkey indicator plates. The mutagenized interval was cloned into a wild-type background, and its phenotype was confirmed and sequenced; multiple mutations were separated via subcloning, or were produced individually via Quikchange (Stratagene), until individual mutations responsible for the positive phenotype could be established.
Sequence analysis of the mutagenized segments of the selected β and β‘ mutants showed an overall mutation frequency of ∼1 in 5000. A total of 3.5 × 105 and 1.7 × 105 colonies from the libraries of β and β‘ were screened against the Qλ and Q82 reporters, respectively. A sufficient number of colonies were screened against the Qλ reporter to pick up most mutations more then once (in each case, individual isolates were shown not to be sister colonies because of additional sequence differences). Difficulties with the Q82 screen, in which a particular class of mutations dominated the phenotypic positives, limited the number of mutants isolated.
A similar screen for substitutions in the remaining core subunit of RNAP (α, the product of rpoA) revealed several mutations within the CTD of α that diminish Q activity in vivo, which will be described elsewhere (data not shown).
β-Galactosidase assays
Late log phase cultures of TOM100 pBAD33-Qλ or TOM101 pBAD33-Q82 harboring the library plasmids were used to inoculate (1:100) LB with appropriate antibiotics, 1% lactose, and varied concentrations of arabinose. Cultures were grown at 37°C for 3 h (reaching an OD600 of ∼0.5), and standard β-galactosidase assays were performed. All values listed are the average of three independent cultures (each done in triplicate) for each arabinose concentration.
Phage spotting tests
TOM100, or TOM101, carrying mutant pRL663 or pRL706 was grown in LB with appropriate antibiotics to an OD600 of 0.4. Cultures were top-plated onto LB or LB plus 1% lactose with appropriate antibiotics and equivalent top agar. In this, 5-μL drops of phage stocks (10-fold serial dilutions) were spotted directly onto the top agar, and allowed to adsorb. Plates were incubated at 37°C overnight, visually inspected, and compared to cultures harboring wild-type pRL663 or pRL706 and to equivalent cultures spotted on LB without lactose. Qλ phage are λ+ (far left in Fig. 3) and the variant λc17 as a second source of Qλ; the Q21 phage is iλQ21; the Q82 phage are iλQ82 (left, Fig. 3) and φ82 (right, Fig. 3). The phage carrying immλ are essentially isogenic except for the QSR region of each.
RNAP purification
RNAP was prepared using a combination of standard techniques (Burgess and Jendrisak 1975), and affinity chromatography. RNAP was purified using a Hi-trap heparin agarose column (Pharmacia), a Sephacryl S-300 sizing column (Pharmacia), and an Ni2+-charged chelating column (Pharmacia). Purified RNAP was ∼50% σ70-saturated. Purified σ70 was added to fully saturate each RNAP preparation; σ70 was prepared as described (Marr and Roberts 1997). RNAP containing a His6 tag on the C terminus of β or β‘ showed no differences in the percentage of active RNAP, general elongation rate, response to NusA, Qλ, Q82, GreA, or GreB when compared with untagged RNAP (data not shown).
Purification of Q-proteins
Qλ was prepared as described (Marr and Roberts 2000), except that all buffers contained 100 mM DTT. Q82 was prepared as described (Goliger and Roberts 1989)
In vitro transcription
For in vitro transcription, 20 nM RNAP holoenzyme was added to 1–2 nM linear templates in transcription buffer (20 mM Tris-HCl at pH 8.0; 0.1 mM EDTA; 50 mM KCl; 1 mM DTT; 100 μg/mL BSA-acetylated; 200 μM ATP, CTP, and GTP; 50 μM UTP; 0.2–1.0 μC/μL 32P-α-UTP) and allowed to form open complex at 37°C for 5–10 min. When added, NusA was present at 150 nM. Single round transcription was initiated by simultaneous addition of MgCl2 [4 mM]final and rifampicin [10 μg/mL]final. Qλ-protein was diluted into 10 mM Tris-HCl (pH 7.5), 500 μg/mL BSA-acetylated, 100 mM DTT, 10% glycerol, and 50 mM K-glutamate immediately before use. Q82-protein was diluted into 10 mM Tris-HCl (pH 8.0), 5% glycerol, 0.1 mM EDTA, 0.1 mM DTT, and 50 mM KCl immediately before use and added to transcription reactions 30 sec before the addition of Mg2+/rifampicin. Reactions were stopped by addition of 5 volumes of 1.2× STOP buffer (0.6 M Tris-HCl at pH 8.0, 12.0 mM EDTA, 80 μg/mL tRNA). Equal volume phenol/chloroform/isoamyl alcohol (25:24:1) extractions were performed, and the aqueous phase was precipitated by addition of 2.6 volumes of 100% EtOH. RNA products were resolved through denaturing polyacrylamide gels, exposed, and quantitated using ImageQuant software (Molecular Dynamics, Storm Model 840). Percent readthrough = {(moles of readthrough RNA)/[(moles of readthrough RNA) + (moles of terminated RNA)]} × 100.
Acknowledgments
We thank S. Gottesman for strains, Erika Woodbury for assistance with the screen, S. Darst for providing coordinates of the modeled transcription elongation complex, and A. Hochschild and members of the laboratory for criticism of the manuscript. This work was supported by NIH grants GM 21941 (to J.W.R.) and GM 38660 (to R.L.).
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL jwr7@cornell.edu; FAX (607) 255-2428.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.1082103.
References
- Burgess RR, Jendrisak JJ. A procedure for the rapid, large-scale purification of Escherichia coli DNA-dependent RNA polymerase involving Polymin P precipitation and DNA-cellulose chromatography. Biochemistry. 1975;14:4634–4638. doi: 10.1021/bi00692a011. [DOI] [PubMed] [Google Scholar]
- Bushnell DA, Cramer P, Kornberg RD. Structural basis of transcription: α-Amanitin–RNA polymerase II cocrystal at 2.8 Å resolution. Proc Natl Acad Sci. 2002;99:1218–1222. doi: 10.1073/pnas.251664698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caldwell RC, Joyee GF. Randomization of genes by PCR mutagenesis. PCR Methods Applic. 1992;2:28–33. doi: 10.1101/gr.2.1.28. [DOI] [PubMed] [Google Scholar]
- Campbell EA, Korzheva N, Mustaev A, Murakami K, Nair S, Goldfarb A, Darst SA. Structural mechanism for rifampicin inhibition of bacterial RNA polymerase. Cell. 2001;104:901–912. doi: 10.1016/s0092-8674(01)00286-0. [DOI] [PubMed] [Google Scholar]
- Cramer P, Bushnell DA, Fu J, Gnatt AL, Maier-Davis B, Thompson NE, Burgess RR, Edwards AM, David PR, Kornberg RD. Architecture of RNA polymerase II and implications for the transcription mechanism. Science. 2000;288:640–649. doi: 10.1126/science.288.5466.640. [DOI] [PubMed] [Google Scholar]
- Ebright RH. RNA polymerase: Structural similarities between bacterial RNA polymerase and eukaryotic RNA polymerase II. J Mol Biol. 2000;304:687–698. doi: 10.1006/jmbi.2000.4309. [DOI] [PubMed] [Google Scholar]
- Epshtein V, Mustaev A, Markovtsov V, Bereshchenko O, Nikiforov V, Goldfarb A. Swing-gate model of nucleotide entry into the RNA polymerase active center. Mol Cell. 2002;10:623–634. doi: 10.1016/s1097-2765(02)00640-8. [DOI] [PubMed] [Google Scholar]
- Gnatt AL, Cramer P, Fu J, Bushnell DA, Kornberg RD. Structural basis of transcription: An RNA polymerase II elongation complex at 3.3 Å resolution. Science. 2001;292:1876–1882. doi: 10.1126/science.1059495. [DOI] [PubMed] [Google Scholar]
- Goliger JA, Roberts JW. Sequences required for antitermination by phage 82 Q protein. J Mol Biol. 1989;210:461–471. doi: 10.1016/0022-2836(89)90123-x. [DOI] [PubMed] [Google Scholar]
- Greenblatt J, Nodwell JR, Mason SW. Transcriptional antitermination. Nature. 1993;364:401–406. doi: 10.1038/364401a0. [DOI] [PubMed] [Google Scholar]
- Guo HC, Kainz M, Roberts JW. Characterization of the late-gene regulatory region of phage 21. J Bacteriol. 1991;173:1554–1560. doi: 10.1128/jb.173.4.1554-1560.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo J. “Study of the λQ functional domain.” Ph.D. thesis. Ithaca, NY: Cornell University; 1999. [Google Scholar]
- Gusarov I, Nudler E. The mechanism of intrinsic transcription termination. Mol Cell. 1999;3:495–504. doi: 10.1016/s1097-2765(00)80477-3. [DOI] [PubMed] [Google Scholar]
- Guzman LM, Belin D, Carson MJ, Beckwith J. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J Bacteriol. 1995;177:4121–4130. doi: 10.1128/jb.177.14.4121-4130.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin DJ, Gross CA. Mapping and sequencing of mutations in the Escherichia coli rpoB gene that lead to rifampicin resistance. J Mol Biol. 1988;202:45–58. doi: 10.1016/0022-2836(88)90517-7. [DOI] [PubMed] [Google Scholar]
- Jin DJ, Walter WA, Gross CA. Characterization of the termination phenotypes of rifampicin-resistant mutants. J Mol Biol. 1988;202:245–253. doi: 10.1016/0022-2836(88)90455-x. [DOI] [PubMed] [Google Scholar]
- Ko DC, Marr MT, Guo J, Roberts JW. A surface of Escherichia coli σ70 required for promoter function and antitermination by phage λ Q protein. Genes & Dev. 1998;12:3276–3285. doi: 10.1101/gad.12.20.3276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komissarova N, Becker J, Solter S, Kireeva M, Kashlev M. Shortening of RNA:DNA hybrid in the elongation complex of RNA polymerase is a prerequisite for transcription termination. Mol Cell. 2002;10:1151–1162. doi: 10.1016/s1097-2765(02)00738-4. [DOI] [PubMed] [Google Scholar]
- Korzheva N, Mustaev A. Transcription elongation complex: Structure and function. Curr Opin Microbiol. 2001;4:119–125. doi: 10.1016/s1369-5274(00)00176-4. [DOI] [PubMed] [Google Scholar]
- Korzheva N, Mustaev A, Kozlov M, Malhotra A, Nikiforov V, Goldfarb A, Darst SA. A structural model of transcription elongation. Science. 2000;289:619–625. doi: 10.1126/science.289.5479.619. [DOI] [PubMed] [Google Scholar]
- Landick R. RNA polymerase slides home: Pause and termination site recognition. Cell. 1997;88:741–744. doi: 10.1016/s0092-8674(00)81919-4. [DOI] [PubMed] [Google Scholar]
- Laspia MF, Wendel P, Mathews MB. HIV-1 Tat overcomes inefficient transcriptional elongation in vitro. J Mol Biol. 1993;232:732–746. doi: 10.1006/jmbi.1993.1427. [DOI] [PubMed] [Google Scholar]
- Marr MT, Roberts JW. Promoter recognition as measured by binding of polymerase to nontemplate strand oligonucleotide. Science. 1997;276:1258–1260. doi: 10.1126/science.276.5316.1258. [DOI] [PubMed] [Google Scholar]
- ————— Function of transcription cleavage factors GreA and GreB at a regulatory pause site. Mol Cell. 2000;6:1275–1285. doi: 10.1016/s1097-2765(00)00126-x. [DOI] [PubMed] [Google Scholar]
- McDowell JC, Roberts JW, Jin DJ, Gross C. Determination of intrinsic transcription termination efficiency by RNA polymerase elongation rate. Science. 1994;266:822–825. doi: 10.1126/science.7526463. [DOI] [PubMed] [Google Scholar]
- Murakami KS, Masuda S, Campbell EA, Muzzin O, Darst SA. Structural basis of transcription initiation: An RNA polymerase holoenzyme–DNA complex. Science. 2002a;296:1285–1290. doi: 10.1126/science.1069595. [DOI] [PubMed] [Google Scholar]
- Murakami KS, Masuda S, Darst SA. Structural basis of transcription initiation: RNA polymerase holoenzyme at 4 Å resolution. Science. 2002b;296:1280–1284. doi: 10.1126/science.1069594. [DOI] [PubMed] [Google Scholar]
- Mustaev A, Kozlov M, Markovtsov V, Zaychikov E, Denissova L, Goldfarb A. Modular organization of the catalytic center of RNA polymerase. Proc Natl Acad Sci. 1997;94:6641–6645. doi: 10.1073/pnas.94.13.6641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nickels BE, Roberts CW, Sun H, Roberts JW, Hochschild A. The σ70 subunit of RNA polymerase is contacted by the Qλ antiterminator during early elongation. Mol Cell. 2002;10:611–622. doi: 10.1016/s1097-2765(02)00648-2. [DOI] [PubMed] [Google Scholar]
- Orlova M, Newlands J, Das A, Goldfarb A, Borukhov S. Intrinsic transcript cleavage activity of RNA polymerase. Proc Natl Acad Sci. 1995;92:4596–4600. doi: 10.1073/pnas.92.10.4596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powell BS, Rivas MP, Court DL, Nakamura Y, Turnbough CL., Jr Rapid confirmation of single copy λ prophage integration by PCR. Nucleic Acids Res. 1994;22:5765–5766. doi: 10.1093/nar/22.25.5765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ring BZ, Yarnell WS, Roberts JW. Function of E. coli RNA polymerase sigma factor σ70 in promoter-proximal pausing. Cell. 1996;86:485–493. doi: 10.1016/s0092-8674(00)80121-x. [DOI] [PubMed] [Google Scholar]
- Roberts JW. Phage λ and the regulation of transcription termination. Cell. 1988;52:5–6. doi: 10.1016/0092-8674(88)90523-5. [DOI] [PubMed] [Google Scholar]
- Roberts JW, Yarnell W, Bartlett E, Guo J, Marr M, Ko DC, Sun H, Roberts CW. Antitermination by bacteriophage λ Q protein. Cold Spring Harb Symp Quant Biol. 1998;63:319–325. doi: 10.1101/sqb.1998.63.319. [DOI] [PubMed] [Google Scholar]
- Severinov K, Mooney R, Darst SA, Landick R. Tethering of the large subunits of Escherichia coli RNA polymerase. J Biol Chem. 1997;272:24137–24140. doi: 10.1074/jbc.272.39.24137. [DOI] [PubMed] [Google Scholar]
- Simons RW, Houman F, Kleckner N. Improved single and multicopy lac-based cloning vectors for protein and operon fusions. Gene. 1987;53:85–96. doi: 10.1016/0378-1119(87)90095-3. [DOI] [PubMed] [Google Scholar]
- Toulokhonov I, Artsimovitch I, Landick R. Allosteric control of RNA polymerase by a site that contacts nascent RNA hairpins. Science. 2001;292:730–733. doi: 10.1126/science.1057738. [DOI] [PubMed] [Google Scholar]
- Vassylyev DG, Sekine S, Laptenko O, Lee J, Vassylyeva MN, Borukhov S, Yokoyama S. Crystal structure of a bacterial RNA polymerase holoenzyme at 2.6 Å resolution. Nature. 2002;417:712–719. doi: 10.1038/nature752. [DOI] [PubMed] [Google Scholar]
- Wang D, Meier TI, Chan CL, Feng G, Lee DN, Landick R. Discontinuous movements of DNA and RNA in RNA polymerase accompany formation of a paused transcription complex. Cell. 1995;81:341–350. doi: 10.1016/0092-8674(95)90387-9. [DOI] [PubMed] [Google Scholar]
- Yang XJ, Goliger JA, Roberts JW. Specificity and mechanism of antitermination by Q proteins of bacteriophages λ and 82. J Mol Biol. 1989;210:453–460. doi: 10.1016/0022-2836(89)90122-8. [DOI] [PubMed] [Google Scholar]
- Yarnell WS, Roberts JW. The phage λ gene Q transcription antiterminator binds DNA in the late gene promoter as it modifies RNA polymerase. Cell. 1992;69:1181–1189. doi: 10.1016/0092-8674(92)90639-t. [DOI] [PubMed] [Google Scholar]
- ————— Mechanism of intrinsic transcription termination and antitermination. Science. 1999;284:611–615. doi: 10.1126/science.284.5414.611. [DOI] [PubMed] [Google Scholar]
- Zhang G, Campbell EA, Minakhin L, Richter C, Severinov K, Darst SA. Crystal structure of Thermus aquaticus core RNA polymerase at 3.3 Å resolution. Cell. 1999;98:811–824. doi: 10.1016/s0092-8674(00)81515-9. [DOI] [PubMed] [Google Scholar]