Abstract
Mammalian telomeres are coated by the sequence-specific, DNA-binding protein, TRF1, a negative regulator of telomere length. Previous results showed that ADP-ribosylation of TRF1 by tankyrase 1 released TRF1 from telomeres and promoted telomere elongation. We now show that loss of TRF1 from telomeres results in ubiquitination and degradation of TRF1 by the proteasome and that degradation is required to keep TRF1 off telomeres. Ubiquitination of TRF1 is regulated by its telomere-binding status; only the telomere-unbound form of TRF1 is ubiquitinated. Our findings suggest a novel mechanism of sequential post translational modification of TRF1 (ADP-ribosylation and ubiquitination) for regulating access of telomerase to telomeres.
Keywords: Telomere, ubiquitin, TRF1, tankyrase 1, PARP
In most human tumor cells, telomere length is maintained by telomerase, a specialized reverse transcriptase that adds telomere repeats to chromosome ends (Greider and Blackburn 1985). Telomere length is tightly controlled in tumor cells, where despite high levels of telomerase, telomeres are maintained at a constant length setting (Counter et al. 1992). Cells employ a complex set of positive and negative regulatory mechanisms to modulate telomere length, including regulating access of telomerase to chromosome termini (Evans and Lundblad 2000).
Mammalian telomeres consist of tandem arrays of TTAGGG repeats bound by the sequence-specific, double-stranded, DNA-binding proteins TRF1 (Chong et al. 1995) and TRF2 (Bilaud et al. 1997; Broccoli et al. 1997). TRF2 is required to protect chromosome ends (van Steensel et al. 1998; de Lange 2002), possibly through its ability to assemble t-loops, higher-order structures at telomeres (Griffith et al. 1999). In addition, TRF2 can influence telomere length through a telomerase-independent mechanism (Ancelin et al. 2002; Karlseder et al. 2002). TRF1, on the other hand, is a negative regulator of telomerase-mediated telomere length, acting in cis at chromosome ends to repress telomere elongation (van Steensel and de Lange 1997; Ancelin et al. 2002).
Tankyrase 1 is a telomeric poly(ADP-ribose) polymerase (PARP) that binds and modifies TRF1 (Smith et al. 1998). Like other PARP family members (Smith 2001), tankyrase 1 uses NAD+ as a substrate to catalyze formation of ADP-ribose polymers onto specific protein acceptors, including itself and TRF1 (Smith et al. 1998; Rippmann et al. 2002). ADP-ribosylation of TRF1 by tankyrase 1 inhibits TRF1 binding to telomeric DNA in vitro (Smith et al. 1998). Overexpression of tankyrase 1 in human tumor cells releases TRF1 from telomeres and induces telomere elongation (Smith and de Lange 2000; Cook et al. 2002). Both tankyrase 1-induced activities (loss of TRF1 and telomere elongation) require the catalytic PARP activity of tankyrase 1 (Smith and de Lange 2000; Cook et al. 2002). These findings suggest that tankyrase 1-mediated removal of TRF1 from telomeres by ADP-ribosylation could allow access of telomerase to chromosome termini.
Here, we show that tankyrase 1 induces proteasome-mediated degradation of TRF1. We demonstrate that TRF1 is ubiquitinated in vivo and in vitro. We present evidence to suggest that it is not ADP-ribosylation per se, but rather, release of TRF1 from telomeres that serves as a signal for ubiquitination and subsequent degradation.
Results and Discussion
Tankyrase 1-mediated telomere elongation requires telomerase and correlates with loss of TRF1
Previous results indicated that tankyrase 1 could induce telomere elongation in telomerase-positive human tumor cells (HTC75), but not telomerase-negative primary cells (WI38; Smith and de Lange 2000; Cook et al. 2002). To determine if telomerase is required for tankyrase 1-induced telomere elongation, we rendered the WI38 cells telomerase-positive by infection with a retrovirus carrying the human telomerase reverse transcriptase (TERT). As expected, WI38 cells expressing TERT (Fig. 1A) displayed telomerase activity (Fig. 1B) and showed elongated telomeres (Fig. 1C). Coexpression of wild-type tankyrase 1 (FN-tankyrase1.WT) in these, now telomerase-positive WI38-TERT cells, resulted in additional telomere elongation (Fig. 1F), indicating that telomerase is required for tankyrase 1-mediated telomere elongation. FN-tankyrase1.WT had no effect on telomerase protein levels (Fig. 1D) or telomerase activity (Fig. 1E), consistent with an indirect effect on telomerase. However, FN-tankyrase1. WT expression resulted in a dramatic reduction in TRF1 levels (Fig. 1D). Expression of a PARP-dead tankyrase 1 mutant (FN-tankyrase1.HE/A; containing a double-point mutation in the catalytic PARP domain; Cook et al. 2002) had no effect on telomere length (Fig. 1F) or TRF1 levels (Fig. 1D) and was similar to the vector control. Together, these data demonstrate that tankyrase 1 regulates telomere elongation by telomerase, and as shown previously in other cell types (Smith and de Lange 2000; Cook et al. 2002), telomere lengthening correlates with loss of TRF1 and requires the PARP activity of tankyrase 1.
Figure 1.
Tankyrase 1-induced telomere elongation is telomerase-dependent. (A–C) Generation of a stable WI38 cell line expressing telomerase. WI38 cells at PD 4 expressing vector control (V) or telomerase (TERT) were analyzed for telomerase expression by immunoblotting whole-cell extracts with anti-TERT 374 antibody (A), or for telomerase activity by TRAP (telomere repeat amplification protocol; B) or for telomere length by Southern blot analysis (C). (D–F) Analysis of stable WI38 cells expressing telomerase (WI38-TERT) and tankyrase 1 alleles. WI38-TERT cells expressing vector control (V), FN-tankyrase1.WT (WT), or FN-tankyrase1.HE/A (HE/A) were analyzed at PD 9 by immunoblotting whole-cell extracts with the antibodies anti-tankyrase 1 376 (TNKS1), anti-poly(ADP-ribose) (PAR), anti-telomerase (TERT), or anti-TRF1 415 (D); or at PD 11 for telomerase activity by TRAP (E); or at the indicated PDs for telomere length by Southern blot analysis (F).
TRF1 is ubiquitinated in vivo and degraded by the proteasome
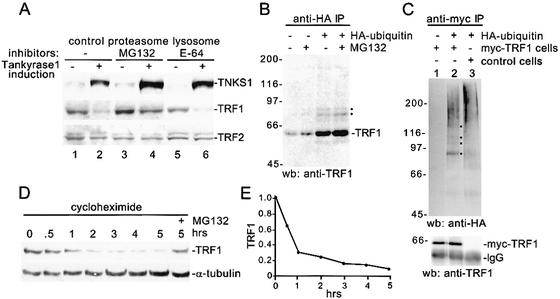
TRF1 loss could be due to regulated degradation of the protein. To address this question, we used a previously characterized HTC75 cell line (FN-30) expressing an inducible allele of tankyrase 1 (Smith and de Lange 2000). As shown in Figure 2A, immunoblot analysis indicates that upon induction of tankyrase 1, TRF1 is lost (cf. lanes 1 and 2). Incubation of cells with MG132 (a specific inhibitor of the proteasome) rescued the tankyrase 1-induced loss of TRF1 (Fig. 2A, cf. lanes 2 and 4). Whereas, incubation of cells with E-64 (an inhibitor of lysosomal proteolysis) had no effect (Fig. 2A, cf. lanes 2 and 6). TRF2 was unaffected by tankyrase 1 overexpression or proteasome inhibition. These results indicate that the tankyrase 1-induced loss of TRF1 is due to TRF1 degradation and that this degradation is mediated by the proteasome pathway.
Figure 2.
TRF1 degradation by the ubiquitin-proteasome pathway. (A) TRF1 is degraded by the proteasome in vivo. Immunoblot analysis of cell extracts from FN30 cells grown in the absence (-) or presence (+) of tankyrase 1 induction. Prior to harvesting, cells were treated without (-) or with (+) proteasome inhibitor (12.5 μM MG132) or with (+) lysosomal inhibitor (12.5 μM E-64) for 10 h. (B,C) TRF1 is ubiquitinated in vivo. (B) HelaI.2.11 cells were transfected with (+) or without (-) a plasmid expressing HA-ubiquitin for 24 h. Four hours prior to harvest, cells were treated with (+) or without (-) 12.5 μM MG132, lysed, and incubated with anti-HA affinity matrix. (C) HTC75 cells expressing myc-TRF1 or a vector control were transfected with (+) or without (-) HA-ubiquitin for 24 h, lysed and incubated with anti-myc affinity matrix. (A–C) Proteins were fractionated on SDS-PAGE and analyzed by Western blotting (wb) with anti-tankyrase 1 376 (TNKS1), anti-TRF1 415, anti-TRF2, or anti-HA antibodies. (B,C) Black dot indicates ubiquitinated TRF1. (D) TRF1 has a short half-life. HT1080 cells were incubated with 100 μg/mL cycloheximide for 0–5 h or for 5 h with 100 μg/mL cycloheximide and 12.5 μM MG132 (+). Cells were harvested, lysed, and cell extracts were fractionated by SDS-PAGE and analyzed by immunoblotting with anti-TRF1 415 or anti-α-tubulin antibodies. (E) Graphical representation of relative TRF1 levels normalized against the α-tubulin loading control using Image Quant version 1.2. The graph represents an average of four experiments. Bars represent the average ± standard error calculated from four independent experiments.
Degradation of proteins by the proteasome depends upon conjugation of ubiquitin to the target protein. To determine if TRF1 is ubiquitinated in vivo, HeLa cells were transfected with a HA-tagged ubiquitin construct (HA-ubiquitin) and ubiquitinated proteins were isolated on an anti-HA antibody affinity matrix. As shown in Figure 2B, slower migrating ubiquitin conjugates of TRF1 could be detected specifically in the HA-ubiquitin transfected cells. This modification was slightly stimulated by proteasome inhibitor. In an alternative approach, HTC75 cells stably expressing a mycpitope tagged allele of TRF1 (myc-TRF1) were transfected with HA-ubiquitin, isolated on an anti-myc antibody affinity matrix, and immunoblotted with anti-HA antibodies. As shown in Figure 2C, lane 2, slower migrating ubiquitin conjugates were detected specifically in the myc-TRF1, HA-ubiquitin transfected cells. Notably, the presence of multiple slower migrating forms indicates that TRF1 is polyubiquitinated in vivo.
Finally, a common feature of proteins targeted for degradation by ubiquitin-mediated proteolysis is a relatively short half-life. To determine the half-life of TRF1, cells were incubated with cycloheximide and analyzed by immunoblotting with anti-TRF1 antibody. As shown in Figure 2D, and graphically in Figure 2E, TRF1 (in contrast to the long-lived α-tubulin) was turned over with a half-life of <1 h, indicating that TRF1 is a short-lived protein.
TRF1 is ubiquitinated in vitro, independent of ADP-ribosylation
To establish a cell-free system for ubiquitination of TRF1, 35S-methionine-labeled TRF1 was generated by coupled in vitro transcription/translation (Fig. 3A, lane 1) and then diluted into a ubiquitination reaction mix [retic(+)] containing reticulocyte lysate supplemented with methyl ubiquitin, ubiquitin aldehyde, and an energy source. As shown in Figure 3A, lane 2, upon incubation with retic(+), TRF1 distributed to slower migrating forms, which likely represent TRF1 conjugated with one to several ubiquitin chains. Immunoblot analysis with anti-TRF1 antibodies confirmed that these slower migrating bands comprise TRF1 (Fig. 3B, lane 2). To confirm that these bands represented ubiquitinated forms of TRF1, exogenous excess ubiquitin was added to the reaction. As shown in Figure 3A, lane 3, addition of ubiquitin (which dilutes out the chain limiting methyl ubiquitin in the reaction) shifted the bands to much higher molecular weight species, indicating longer ubiquitin chains.
Figure 3.
In vitro ubiquitination of TRF1 is not dependent on ADP-ribosylation. (A-B) TRF1 is ubiquitinated in vitro. 35S-labeled TRF1 was generated by coupled in vitro transcription/translation and then diluted into a ubiquitination reaction mix containing reticulocyte lysate supplemented with methyl ubiquitin, ubiquitin aldehyde, and an energy source [retic(+)] without (-) or with (+) exogenous ubiquitin. (C) TRF1 interaction with tankyrase 1 is not required for ubiquitination. 35S-labeled TRF1 (C) or Δacidic TRF1 (Δ) was incubated without (-) or with (+) retic(+) ubiquitination reaction mix. (D) TRF1 is ubiquitinated in the presence of the PARP inhibitor 3AB. 35S-labeled in vitro translated TRF1 generated in the presence of 0, 1, or 10 mM 3AB was incubated without (-) or with (+) retic(+) ubiquitination reaction mix containing 0, 1, or 10 mM 3AB. Following incubations proteins were fractionated on SDS-PAGE and visualized by fluorography (A,C,D) or analyzed by immunoblotting (B) with anti-TRF1 415 antibody.
As shown in Figure 1D and previously (Cook et al. 2002), tankyrase 1-induced loss of TRF1 requires a catalytically active form of tankyrase 1, suggesting that ADP-ribosylation of TRF1 may be required for ubiquitination and subsequent degradation. Since immunoblot analysis indicated that the reticulocyte lysate used in the in vitro reactions contained tankyrase 1 (data not shown), we sought to determine if tankyrase 1 was required for ubiquitination. To address this question, we first asked if interaction between TRF1 and tankyrase 1 was required for ubiquitination of TRF1. A TRF1 construct lacking the N-terminal, tankyrase 1-binding (Smith et al. 1998), acidic domain of TRF1 was generated. As shown in Figure 3C, Δacidic TRF1 was efficiently ubiquitinated in vitro, although there was a shift in the distribution of ubiquitinated species from low to higher molecular weight forms in wild type versus Δacidic TRF1. These results indicate that TRF1 binding to tankyrase 1 is not required for ubiquitination of TRF1 in vitro.
In a second approach, we asked if PARP activity was required for ubiquitination of TRF1. In vitro translation and subsequent ubiquitination of TRF1 was performed in the presence of the PARP inhibitor 3-aminobenzamide (3AB). Previous results indicated that 1 mM 3AB was sufficient to inhibit ADP ribosylation of TRF1 by tankyrase 1 in vitro (Smith et al. 1998). As shown in Figure 3D, inclusion of 1 or 10 mM 3AB in the reactions did not inhibit ubiquitination of TRF1, indicating that ADP-ribosylation of TRF1 is not required for ubiquitination in vitro.
Telomere-bound TRF1 is protected from ubiquitination
Our studies thus far indicate that while catalytic activity of tankyrase 1 is required for TRF1 degradation in vivo, ADP-ribosylation of TRF1, per se, is not a prerequisite for ubiquitination of TRF1 in vitro. What then is the role of ADP-ribosylation? One possibility is that this modification is required solely to remove TRF1 from telomeres in order to render TRF1 accessible to the ubiquitination machinery; that is, perhaps telomere bound TRF1 is protected from ubiquitination. To address this question, increasing amounts of double-stranded TTAGGG repeat DNA were included in the in vitro reaction. As shown in Figure 4A, ubiquitination of TRF1 was inhibited by double-stranded TTAGGG repeats, but not by nonvertebrate, double-stranded telomeric TTAGGC repeats nor by single-stranded vertebrate telomeric repeats TTAGGG or CCCTAA.
Figure 4.
Telomeric TRF1 is protected from ubiquitination. (A) Ubiquitination of TRF1 is specifically inhibited by binding to double-stranded telomeric TTAGGG repeats. In vitro ubiquitination reactions containing 35S-labeled TRF1 were carried out in the absence (-) or presence of double-stranded (DS; 0, 0.01, 0.05, 0.25, or 1 μg TTAGGG, or 0.25 or 1 μg TTAGGC) repeat DNA or single-stranded [SS; 1 μg TTAGGG (G) or AATCCC (C)] repeat DNA. (B) Ubiquitination of a TRF1 DNA-binding mutant is not inhibited by telomeric DNA. 35S-labeled TRF1.RV was incubated without (-) or with [+; retic(+)] ubiquitination reaction mix in the absence (-) or presence of double-stranded (0.25 or 1 μg TTAGGG) repeat DNA. (C) Ubiquitination of TRF1 lacking the myb domain is greatly reduced. 35S-labeled TRF1 (C) or ΔmybTRF1 (Δmyb) was incubated without (-) or with (+) retic(+) ubiquitination reaction mix. (A–C) Following incubations, proteins were fractionated on SDS-PAGE and visualized by fluorography. (D,E) Proteasome inhibition attenuates tankyrase 1-induced loss of telomeric TRF1. HelaI.2.11 cells were transiently transfected with FN-tankyrase1.WT for 24 h. Eight hours prior to harvest, cells were incubated without (-; D) or with (+; E) 25 μM MG132 and analyzed by indirect immunofluorescence. Paraformaldehyde-fixed cells were stained with anti-FLAG (green) or anti-TRF1 415 (red) antibodies. DAPI staining of DNA is shown in blue. Bar, 5 μm.
To rule out the possibility that the TTAGGG DNA had a nonspecific effect on ubiquitination, we generated a point mutation in TRF1 (conversion of R to V at position 425 in the myb domain), which abolishes DNA binding (Fairall et al. 2001). As shown in Figure 4B, TRF1.RV (like TRF1) was efficiently ubiquitinated; however, ubiquitination of TRF1.RV (unlike that of TRF1) was not inhibited by double-stranded TTAGGG telomeric repeat DNA. These data show that TRF1 is protected from ubiquitination when bound to telomeric DNA and suggest the possibility that the DNA-binding, myb domain of TRF1 might serve as a site for recognition or modification by the ubiquitinating machinery. Indeed, as shown in Figure 4C, ubiquitination of TRF1 lacking the myb domain (ΔmybTRF1) was dramatically reduced compared to wild-type TRF1.
Our findings are consistent with the notion that TRF1 is subjected to a set of sequential, posttranslational modifications. First, tankyrase 1 ADP-ribosylates TRF1 to release it from telomeres. Second, the telomere-dissociated form of TRF1 is then ubiquitinated and targeted for degradation. We wondered why degradation of TRF1 was necessary; that is, was it not sufficient to release TRF1 from telomeres? Studies indicate that because of rapid hydrolysis by the glycohydrolase PARG, the intracellular half-life of ADP-ribose polymers is ∼1 min (Davidovic et al. 2001). Thus, one possibility is that while tankyrase 1-mediated ADP-ribosylation of TRF1 is sufficient to release TRF1 from telomeres, rapid degradation of the polymer by PARG would allow immediate reassociation of TRF1 with telomeres. Indeed, the abundant repetitive sequences at telomeres could act as a sink for rapid rebinding of TRF1 to telomeres. Thus, degradation of TRF1 may be necessary to keep TRF1 off telomeres for an extended period of time. This question was addressed by determining the fate of TRF1 localization when it is released from telomeres by ADP-ribosylation, but not degraded by the proteasome. As shown in Figure 4D, and previously (Smith and de Lange 2000; Cook et al. 2002), overexpression of tankyrase 1 in the nucleus results in release of TRF1 from telomeres. However, when proteasome-mediated degradation is inhibited, TRF1 is found relocalized to telomeres (Fig. 4E). These results suggest that tankyrase 1-mediated ADP-ribosylation of TRF1 is not sufficient to keep TRF1 off telomeres; that is, if TRF1 is not degraded it reassociates with telomeres, even in the presence of excess tankyrase 1.
A model for telomere length regulation
Our results are presented in terms of a model for telomere length regulation. As shown in Figure 5, TRF1-bound telomeres are in a configuration that blocks access to telomerase. Poly(ADP-ribosyl)ation of TRF1 by tankyrase 1 releases TRF1 from telomeres. The telomere-unbound form of TRF1 is then ubiquitinated and degraded by the proteasome, thereby preventing its rapid reassociation with telomeres. Telomerase can then gain access to the TRF1-free telomere to add telomeric repeats. The telomere is reassembled using newly synthesized TRF1 into a configuration that once again blocks access to telomerase.
Figure 5.
Model: sequential posttranslational modification of TRF1 regulates access of telomerase to telomeres. TRF1-bound telomeres are inaccessible to telomerase. Tankyrase 1 ADP-ribosylates TRF1, removing it from telomeres. Once TRF1 is off telomeres, it is ubiquitinated and degraded by the proteasome. TRF1-free telomeres are elongated by telomerase and then reassembled into a telomerase-inaccessible state using newly synthesized TRF1.
This model offers a number of interesting features. First, it provides a rapid mechanism for release of TRF1 from telomeres. Since tankyrase 1 contains five binding sites for TRF1 (Seimiya and Smith 2002), it has a large capacity to bind, modify, and remove TRF1 from telomeres. Second, since poly(ADP-ribosyl)ation reactions are tightly controlled in vivo, activation of tankyrase 1's PARP activity could be regulated to coordinate release of TRF1 and access to telomerase with the cell cycle. And third, because of the rapid turnover of TRF1, reassembly of telomeres with TRF1 into a telomerase-inaccessible state would occur with newly synthesized TRF1. This could provide an opportunity for TRF1 to recruit regulatory proteins to chromosome termini. Indeed, previous results indicate that TRF1 can recruit tankyrase 1 to telomeres when the proteins are coexpressed (Smith and de Lange 1999).
How does loss of TRF1 change telomere configuration to allow telomerase access? Telomeres exist in a protective configuration that requires TRF2 and likely involves a higher-order structure, such as the t-loop (de Lange 2002). Studies indicate that when TRF1 is lost from telomeres (either through a dominant-negative allele of TRF1 or through ADP-ribosylation by tankyrase 1) TRF2 remains on telomeres and telomeres remain protected (as indicated by unimpeded cell growth), yet they undergo telomere elongation (van Steensel and de Lange 1997; Smith and de Lange 2000; Cook et al. 2002). Thus, loss of TRF1 may induce an intermediate state where telomeres remain in a protected configuration, but still allow access to telomerase for telomere elongation. Whether TRF1 loss influences higher-order structure at telomeres and/or the proteins that bind and recruit telomerase, will be important questions for the future.
While poly(ADP-ribosyl)ation of TRF1 is a very dramatic reaction, sufficient to release TRF1 from telomeres, because of its transient nature, a second modification (ubiquitination and subsequent degradation) appears to be required to keep TRF1 off telomeres. The ubiquitin-proteasome degradation pathway consists of a series of enzymatic reactions involving a ubiquitin-activating enzyme (E1), a ubiquitin-conjugating enzyme (E2) and ubiquitin ligase (E3; Hershko and Ciechanover 1998). Of particular interest are the E3s, a growing family of proteins that determine the timing and specificity of ubiquitination. Undoubtedly, identification of the E3 that recognizes TRF1 will be informative. Our studies suggest that the putative E3 does not recognize telomere-bound TRF1. E3 recognition may occur through the myb-type DNA-binding domain of TRF1. Consistent with this notion, in vitro ubiquitination of TRF1 lacking a myb domain is severely reduced (Fig. 4C). Thus, when TRF1 is bound to telomeres, the myb domain may not be accessible to the E3 and/or lysine residues (that serve as ubiquitination sites) may be masked by DNA interactions.
Although a role for ubiquitination in telomere function has not been directly demonstrated, a number of studies suggest the possibility. Mutations in Drosophila UbcD1, a ubiquitin conjugating (E2) enzyme, induce transient, resolvable telomere-telomere associations in mitosis and meiosis, suggesting that a telomere-associated protein could be a target for ubiquitination (Cenci et al. 1997). More recently, the fission yeast F-box protein Pof3 was found to be required for genomic integrity and telomere function (Katayama et al. 2002). F-box proteins are members of a large family of proteins that provide substrate specificity for the SCF ubiquitin ligase (E3) complexes (Kipreos and Pagano 2000). Yeast cells lacking Pof3 displayed shortened telomeres and were defective in telomeric silencing, suggesting again that a telomere-associated protein could be a target for ubiquitination. In this report, we have identified a telomeric target for ubiquitination, TRF1. While we have yet to identify the ubiquitin machinery responsible for this reaction in human cells, our studies, along with those in other divergent organisms, suggest the possibility of a conserved role for ubiquitin-mediated proteolysis in telomere function.
Materials and methods
Plasmids
TRF1 constructs were cloned into the retroviral vector pLPC (Serrano et al. 1997) and contain an N-terminal myc-epitope tag followed by amino acids 2–439 (pLPCTRF1), amino acids 66–439 (ΔacidicTRF1), or amino acids 2–378 (ΔmybTRF1). pLPC-TRF1.RV was generated using the Stratagene quickchange site directed mutagenesis kit. FN-tankyrase1.WT and HE/A contain full-length tankyrase 1 (amino acids 2–1327) with an N-terminal FLAG-epitope tag and nuclear localization signal in pLPC (Cook et al. 2002).
Retroviruses and cell lines
Retroviruses were generated and used to infect cells as described previously (Cook et al. 2002). WI38 cells (ATCC), human primary fibroblasts at population doubling (PD) 30 were infected with pBABE-hygro or pBABE-hygro-TERT (Counter et al. 1998) and selected in 90 μg/mL hygromycin. WI38-TERT cells at PD 5 were infected with pLPC, pLPC-FN-Tankyrase1.WT, or pLPC-FN-Tankyrase1.HE/A and selected with 2 μg/mL puromycin. On day 3 of retroviral infection, cells were subcultured 1:2 and upon confluence designated PD 0.
HT1080 (ATCC) is a human fibrosarcoma cell line. HTC75 is a HT1080-derived clonal cell line that stably expresses the tetracycline(tet)-controlled transactivator (van Steensel and de Lange 1997). FN30 is a HTC75-derived clonal cell line that stably expresses doxycylin-inducible FN-tankyrase1.WT (Smith and de Lange 2000). Stable HTC75 cell lines expressing myc-TRF1 or vector control were generated by retroviral infection using pLPCTRF1 or pLPC as described (Cook et al. 2002).
Genomic blotting and TRAP assays
Southern blotting for telomere-length analysis was performed as described previously (Cook et al. 2002). TRAP assays (Kim et al. 1994) contained 1 μg CHAPS (Pierce) extract with or without 10 μg/mL RNase A.
Immunoblotting
Immunoblots were incubated with the following primary antibodies: rabbit anti-poly(ADP-ribose) serum (1:1000; Alexis Biochemicals), rabbit anti-TRF1 415 (0.2 μg/mL; Cook et al. 2002), rabbit anti-tankyrase 1 376 (0.1 μg/mL; Cook et al. 2002), mouse anti-α-tubulin ascites (1:500,000; Sigma), rabbit anti-TERT 374 (0.8 μg/mL; raised and affinity purified against Escherichia coli-derived fusion protein containing hTERT amino acids 561–698), or mouse monoclonal anti-TRF2 (1.0 μg/mL; Imgenex Clone 4A794), followed by horseradish peroxidase-conjugated donkey anti-rabbit or anti-mouse IgG (Amersham; 1:2500). Bound antibody was detected using the Enhanced Chemiluminescence (Amersham), Super-Signal West Dura, or Femto (Pierce) kits.
Cell extracts and immunoprecipitation
For immunoblot analysis, whole-cell extracts were prepared as described (Cook et al. 2002) and 25 μg was fractionated by SDS-PAGE.
For immunoprecipitation, HA-ubiquitin transfected cell extracts were prepared in buffer C [20 mM Hepes-KOH at pH 7.9, 420 mM KCl, 25% glycerol, 0.1 mM EDTA, 5 mM MgCl2, 0.2% NP-40, 1 mM dithiothreitol, and 2.5% protease inhibitor cocktail (Sigma)] containing 10 mM N-ethylmaleimide (Sigma), and then incubated with anti-HA (Roche) or anti-myc (Sigma) affinity matrix for 3 h with shaking at 4°C. HA-matrix-bound proteins were washed three times in buffer D (20 mM Hepes at pH 7.9, 100 mM KCl, 20% glycerol, 0.2 mM EDTA, 0.2 mM EGTA) and myc-matrix bound proteins were washed four times in buffer C (without NP40).
Transient transfections and indirect immunofluorescence
HelaI.2.11 cells (van Steensel et al. 1998) were transfected for 24 h with pMT123 (encoding HA-ubiquitin; Treier et al. 1994) or pcDNA3-FN-tankyrase1.WT, full-length tankyrase 1 containing an N-terminal FLAG tag and nuclear localization signal in the expression vector pcDNA3 (Invitrogen; B. Houghtaling and S. Smith, unpubl.), using Lipofectamine 2000 regeant (Invitrogen). Cells were processed for immunofluorescence as described previously (Cook et al. 2002) using mouse monoclonal anti-FLAG M2 (1 μg/mL; Sigma) and rabbit anti-TRF1 415 (0.1 μg/mL; Cook et al. 2002) as primary antibodies.
In vitro ubiquitination reactions
The ubiquitination reaction mix [retic(+)] contained 5 μL rabbit reticulocyte lysate (Promega), 1 μM ubiquitin aldehyde (Boston Biochem), 121 μM methyl ubiquitin (Boston Biochem), 1× energy solution (Boston Biochem), and 150 μM additional ubiquitin (Boston Biochem) in 10 μL total reaction mix. The reaction indicated in Figure 3A and B, lane 2, lacked additional ubiquitin. The ubiquitin reaction mix was preincubated at 37°C for 5 min to allow ubiquitin aldehyde to inhibit deubiquitinating enzymes. To generate 35S-labeled TRF1, pLPC-TRF1, pLPC-TRF1.RV, or pLPC-Δacidic TRF1, plasmids were in vitro transcribed and translated with S35 methionine in a 12.5 μL reaction using Promega TNT coupled-reticulocyte lysate system. Then, 2.5 μL was incubated with the ubiquitination reaction mix at 37°C for 30 min. Reactions were stopped by addition of 2× sample buffer.
For telomere inhibition studies, oligonucleotides containing (TTAGGG)6 and (CCCTAA)6 for DS (TTAGGG) or (TTAGGC)6 and (GCCTAA)6 for DS (TTAGGC) or (TTAGGG)6 for SS (G) or (CCCTAA)6 for SS (C) were used. In vitro translated TRF1 was incubated with DS or SS oligonucleotides at 37°C for 30 min prior to addition to the ubiquitination reaction.
Acknowledgments
We thank Robert Weinberg for the pBABEhygro-TERT plasmid and Dirk Bohmann for the MT123 plasmid. We are grateful to Michelle Pagano, Tom Meier, and members of the Smith lab for comments on the manuscript. This work was supported by grants from the Edward Mallinckrodt Jr. Foundation, the New York City Council Speaker's Fund for Biomedical Research, the Kimmel Scholar Award, and the NIH (RO1 CA95099-01).
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Corresponding author.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.1077103
References
- Ancelin K., Brunori, M., Bauwens, S., Koering, C.E., Brun, C., Ricoul, M., Pommier, J.P., Sabatier, L., and Gilson, E. 2002. Targeting assay to study the cis functions of human telomeric proteins: Evidence for inhibition of telomerase by TRF1 and for activation of telomere degradation by TRF2. Mol. Cell. Biol. 22: 3474–3487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bilaud T., Brun, C., Ancelin, K., Koering, C.E., Laroche, T., and Gilson, E. 1997. Telomeric localization of TRF2, a novel human telobox protein. Nat. Genet. 17: 236–239. [DOI] [PubMed] [Google Scholar]
- Broccoli D., Smogorzewska, A., Chong, L., and de Lange, T. 1997. Human telomeres contain two distinct Myb-related proteins, TRF1 and TRF2. Nat. Genet. 17: 231–235. [DOI] [PubMed] [Google Scholar]
- Cenci G., Rawson, R.B., Belloni, G., Castrillon, D.H., Tudor, M., Petrucci, R., Goldberg, M.L., Wasserman, S.A., and Gatti, M. 1997. UbcD1, a Drosophila ubiquitin-conjugating enzyme required for proper telomere behavior. Genes & Dev. 11: 863–875. [DOI] [PubMed] [Google Scholar]
- Chong L., van Steensel, B., Broccoli, D., Erdjument-Bromage, H., Hanish, J., Tempst, P., and de Lange, T. 1995. A human telomeric protein. Science 270: 1663–1667. [DOI] [PubMed] [Google Scholar]
- Cook B.D., Dynek, J.N., Chang, W., Shostak, G., and Smith, S. 2002. Role for the related poly(ADP-Ribose) polymerases tankyrase 1 and 2 at human telomeres. Mol. Cell. Biol. 22: 332–342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Counter C.M., Avilion, A.A., LeFeuvre, C.E., Stewart, N.G., Greider, C.W., Harley, C.B., and Bacchetti, S. 1992. Telomere shortening associated with chromosome instability is arrested in immortal cells which express telomerase activity. EMBO J. 11: 1921–1929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Counter C.M., Hahn, W.C., Wei, W., Caddle, S.D., Beijersbergen, R.L., Lansdorp, P.M., Sedivy, J.M., and Weinberg, R.A. 1998. Dissociation among in vitro telomerase activity, telomere maintenance, and cellular immortalization. Proc. Natl. Acad. Sci. 95: 14723–14728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidovic L., Vodenicharov, M., Affar, E.B., and Poirier, G.G. 2001. Importance of poly(ADP-ribose) glycohydrolase in the control of poly-(ADP-ribose) metabolism. Exp. Cell Res. 268: 7–13. [DOI] [PubMed] [Google Scholar]
- de Lange T. 2002. Protection of mammalian telomeres. Oncogene 21: 532–540. [DOI] [PubMed] [Google Scholar]
- Evans S.K. and Lundblad, V. 2000. Positive and negative regulation of telomerase access to the telomere. J. Cell Sci. 113: 3357–3364. [DOI] [PubMed] [Google Scholar]
- Fairall L., Chapman, L., Moss, H., de Lange, T., and Rhodes, D. 2001. Structure of the TRFH dimerization domain of the human telomeric proteins TRF1 and TRF2. Mol. Cell 8: 351–361. [DOI] [PubMed] [Google Scholar]
- Greider C.W. and Blackburn, E.H. 1985. Identification of a specific telomere terminal transferase activity in Tetrahymena extracts. Cell 43: 405–413. [DOI] [PubMed] [Google Scholar]
- Griffith J.D., Comeau, L., Rosenfield, S., Stansel, R.M., Bianchi, A., Moss, H., and de Lange, T. 1999. Mammalian telomeres end in a large duplex loop. Cell 97: 503–514. [DOI] [PubMed] [Google Scholar]
- Hershko A. and Ciechanover, A. 1998. The ubiquitin system. Annu. Rev. Biochem. 67: 425–479. [DOI] [PubMed] [Google Scholar]
- Karlseder J., Smogorzewska, A., and de Lange, T. 2002. Senescence induced by altered telomere state, not telomere loss. Science 295: 2446–2449. [DOI] [PubMed] [Google Scholar]
- Katayama S., Kitamura, K., Lehmann, A., Nikaido, O., and Toda, T. 2002. Fission yeast F-box protein Pof3 is required for genome integrity and telomere function. Mol. Biol. Cell 13: 211–224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim N.W., Piatyszek, M.A., Prowse, K.R., Harley, C.B., West, M.D., Ho, P.L., Coviello, G.M., Wright, W.E., Weinrich, S.L., and Shay, J.W. 1994. Specific association of human telomerase activity with immortal cells and cancer. Science 266: 2011–2015. [DOI] [PubMed] [Google Scholar]
- Kipreos E.T. and Pagano, M. 2000. The F-box protein family. Genome Biol. 1: REVIEWS3002.1–3002.7 [DOI] [PMC free article] [PubMed]
- Rippmann J.F., Damm, K., and Schnapp, A. 2002. Functional characterization of the poly(ADP-ribose) polymerase activity of tankyrase 1, a potential regulator of telomere length. J. Mol. Biol. 323: 217–224. [DOI] [PubMed] [Google Scholar]
- Seimiya H. and Smith, S. 2002. The telomeric poly(ADP-ribose) polymerase, tankyrase 1, contains multiple binding sites for telomeric repeat binding factor 1 (TRF1) and a novel acceptor, 182-kDa tankyrase-binding protein (TAB182). J. Biol. Chem. 277: 14116–14126. [DOI] [PubMed] [Google Scholar]
- Serrano M., Lin, A.W., McCurrach, M.E., Beach, D., and Lowe, S.W. 1997. Oncogenic ras provokes premature cell senescence associated with accumulation of p53 and p16INK4a. Cell 88: 593–602. [DOI] [PubMed] [Google Scholar]
- Smith S. 2001. The world according to PARP. Trends Biochem. Sci. 26: 174–179. [DOI] [PubMed] [Google Scholar]
- Smith S. and de Lange, T. 1999. Cell cycle dependent localization of the telomeric PARP, tankyrase, to nuclear pore complexes and centrosomes. J. Cell Sci. 112: 3649–3656. [DOI] [PubMed] [Google Scholar]
- ____ 2000. Tankyrase promotes telomere elongation in human cells. Curr. Biol. 10: 1299–1302. [DOI] [PubMed] [Google Scholar]
- Smith S., Giriat, I., Schmitt, A., and de Lange, T. 1998. Tankyrase, a poly(ADP-ribose) polymerase at human telomeres. Science 282: 1484–1487. [DOI] [PubMed] [Google Scholar]
- Treier M., Staszewski, L.M., and Bohmann, D. 1994. Ubiquitin-dependent c-Jun degradation in vivo is mediated by the δ domain. Cell 78: 787–798. [DOI] [PubMed] [Google Scholar]
- van Steensel B. and de Lange, T. 1997. Control of telomere length by the human telomeric protein TRF1. Nature 385: 740–743. [DOI] [PubMed] [Google Scholar]
- van Steensel B., Smogorzewska, A., and de Lange, T. 1998. TRF2 protects human telomeres from end-to-end fusions. Cell 92: 401–413. [DOI] [PubMed] [Google Scholar]