Abstract
Ultraviolet (UV) irradiation of HeLa cells triggers an apoptotic response mediated by mitochondria. Biochemical analysis of this response revealed that the elimination of cytosolic inhibitors is required for mitochondrial release of cytochrome c and subsequent caspase activation. These inhibitors were found to be Mcl-1 and Bcl-xL, two antiapoptotic members of the Bcl-2 family. Following UV treatment, Mcl-1 protein synthesis is blocked, the existing pool of Mcl-1 protein is rapidly degraded by the proteasome, and cytosolic Bcl-xL translocates to the mitochondria. These events are sequential; the elimination of Mcl-1 is required for the translocation of Bcl-xL. The disappearance of Mcl-1 is also required for other mitochondrial apoptotic events including Bax translocation, cytochrome c release, and caspase activation.
Keywords: Apoptosis, cytochrome c, mitochondria, Mcl-1, Bcl-xL, proteasome
Apoptosis provides a powerful protection mechanism to eliminate harmful cells that have suffered a lethal dose of DNA damage. Failure to die results in the survival of cells harboring genetic mutations that may become cancerous (Johnstone et al. 2002). Therefore, molecular dissection of the cellular apoptotic response to DNA damage is of both theoretical and practical significance.
DNA-damaging reagents such as ultraviolet (UV) light and genotoxic chemotherapeutical agents induce apoptosis through a mitochondrial pathway (Kim et al. 1997; Kluck et al. 1997; Bossy-Wetzel et al. 1998). In response to these treatments, cytochrome c is released from the mitochondrial intermembrane space to the cytoplasm, where it binds to Apaf-1. Cytochrome c binding to Apaf-1 triggers the formation of the apoptosome, which activates procaspase-9 (Li et al. 1997; Acehan et al. 2002). Activated caspase-9 cleaves and activates caspase-3 and caspase-7, which subsequently cleave many intracellular substrates, leading to the characteristic morphological changes associated with apoptosis (Rodriguez and Lazebnik 1999; Hengartner 2000).
In addition to cytochrome c, several other apoptogenic proteins are also released from the mitochondrial intermembrane space. These include Smac/Diablo and Omi/HtrA2, which antagonize the caspase-inhibitory IAP proteins, and AIF and EndoG, which cause apoptotic changes independent of caspase activity (Susin et al. 1999; Du et al. 2000; Verhagen et al. 2000; Li et al. 2001; Suzuki et al. 2001; Hegde et al. 2002; Martins et al. 2002).
The balance between pro- and antiapoptotic Bcl-2 family members determines the mitochondrial response to apoptotic stimuli (Gross et al. 1999; Martinou and Green 2001). Antiapoptotic proteins such as Bcl-2, Bcl-xL, and Mcl-1 protect mitochondrial integrity, whereas the proapoptotic members of the family promote the release of apoptogenic proteins from mitochondria. The function of these proapoptotic proteins can be further divided into the BH3-only proteins including Bid, Bad, and Bim and their effectors Bak and Bax (Cheng et al. 2001; Wei et al. 2001; Zong et al. 2001). Activated BH3-only proteins induce the formation of mitochondrial oligomeric Bax/Bak complexes either directly or indirectly by binding and inactivating antiapoptotic Bcl-2 family members (Korsmeyer et al. 2000; Letai et al. 2002). These complexes may function as protein pores for cytochrome c and other proteins to pass through, or cause mitochondrial outer membrane destabilization (Vander Heiden et al. 1997; Kuwana et al. 2002).
The essential question of how the Bcl-2 family of proteins translates genotoxic stress into mitochondrial damage remains unaddressed. In the present report, we used classic biochemical fractionation and reconstitution to map a sequential signaling pathway composed of Bcl-2 family members that leads to cytochrome c release after UV treatment.
Results
Activation of a mitochondrial apoptotic pathway in response to UV irradiation
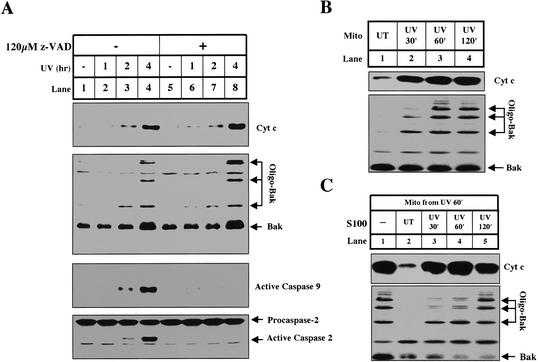
After receiving a strong dose of UV irradiation, cultured HeLa cells exhibit synchronized characteristic apoptotic changes beginning 2 h after irradiation. After 4 h, most cells die by apoptosis. As shown in Figure 1A (lanes 1-4), these changes include the oligomerization of Bak, the appearance of cytochrome c in the cytosol, and the activation of caspase-9 and caspase-2.
Figure 1.
UV induces cytochrome c release from HeLa cells in vivo and in vitro. (A) Mitochondria and S100 were harvested from HeLa cells treated with UV for different amounts of time as described in Materials and Methods. In lanes 5-8, we preincubated the cells in the presence of Z-VAD.fmk, a broad-spectrum caspase inhibitor as indicated. Cytochrome c, caspase-9, and caspase-2 were analyzed in S100. Bak oligomerization was analyzed in mitochondrial fractions as described (Wei et al. 2000). (B) Mitochondria from UV-treated cells release cytochrome c in vitro. Mitochondria (0.67 mg/mL) from untreated cells (lane 1) or cells that were UV-treated for 30 min (lane 2), 60 min (lane 3), or 120 min (lane 4) were incubated in vitro at 37°C for 30 min. Following incubation, the mitochondria were pelleted and analyzed as described in Materials and Methods. (C) S100 inhibits cytochrome c release from primed mitochondria. Mitochondria (0.67 mg/mL) from cells treated with UV for 1 h were coincubated (lanes 1-5) at 37°C for 30 min with only buffer (lane 1) or HeLa S100 (4 mg/mL) from untreated cells (lane 2) or cells that were UV-treated for 30 min (lane 3), 60 min (lane 4), or 120 min (lane 5). Mitochondria and supernatants were analyzed for Bak oligomerization and cytochrome c release, respectively.
When cells were irradiated in the presence of a pan-caspase inhibitor, z-VAD-fmk, caspase-9 activation was completely blocked (Fig. 1A, lanes 5-8). However, the oligomerization of Bak and release of cytochrome c remained intact, indicating that these two events are upstream of caspase activation. Caspase-2 activation was also blocked by zVAD-fmk, suggesting that caspase-2 activation is not required for UV-induced cytochrome c release in HeLa cells as it is in oncogene transformed human fibroblasts (Lassus et al. 2002).
The mitochondria from UV-irradiated cells were primed to release cytochrome c in vitro
In vivo, mitochondria from UV-irradiated cells start to release cytochrome c 120 min after UV irradiation; however, mitochondria isolated from cells just 30-60 min after UV irradiation readily formed oligomerized Bak and released cytochrome c when incubated in vitro (Fig. 1B, lanes 2-3). In contrast, mitochondria from untreated cells did not form oligomerized Bak or release cytochrome c under the same conditions (Fig. 1B, lane 1). Mitochondria from cells 30-60 min after UV are therefore “primed“ to release cytochrome c in vitro, at least 1 h before cytochrome c release can be detected in vivo.
Inhibitors in normal cytosol prevent cytochrome c release from primed mitochondria
Our finding that cytochrome c release is delayed in vivo compared with in vitro is consistent with a previous report predicting that cytoplasm contains inhibitors of cytochrome c release (Duelli and Lazebnik 2000). To test this hypothesis in our system, primed mitochondria isolated from cells 60 min after UV irradiation were incubated with cytosol (S100) from untreated cells (naive), or cells 30-120 min after UV irradiation. As shown in Figure 1C, cytosol from naive cells efficiently inhibited cytochrome c release from UV-primed mitochondria (Fig. 1C, lane 2). The ability of cytosol to inhibit release was gradually lost with increasing time after UV irradiation (Fig. 1C, lanes 3-5).
Purification and identification of Mcl-1 and Bcl-xL as the cytosolic inhibitors of cytochrome c release
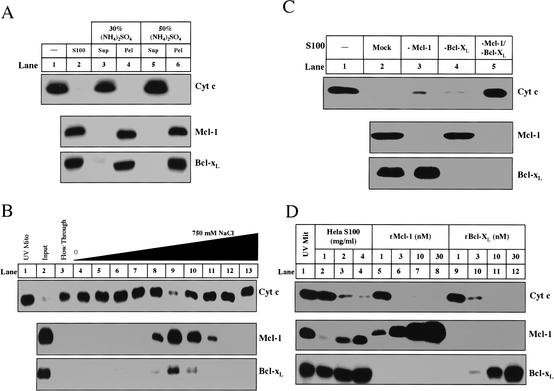
We used biochemical fractionation in concert with a candidate protein approach to identify the inhibitors present in the cytosol of naive cells. The cytosol from naive HeLa cells was fractionated by ammonium sulfate precipitation, ion exchange Mono Q column chromatography, gel-filtration column chromatography (data not shown), and hydroxyapatite chromatography (data not shown), and assayed for inhibition of cytochrome c release from UV-primed mitochondria. As shown in the upper panel of Figure 2A, 30% ammonium sulfate precipitated the activity (Fig. 2A, lanes 4,6). The precipitated activity was subsequently loaded onto a Mono Q column, and an activity peak was eluted from the column by ∼200 mM NaCl (Fig. 2B, upper panel, lane 9).
Figure 2.
Mcl-1 and Bcl-xL are necessary and sufficient for cytosolic inhibitory activity. (A) Mcl-1 and Bcl-xL correlate with inhibitory activity after ammonium sulfate fractionation. Ammonium sulfate was added to 1 mL of S100 (5 mg/mL) up to 30% (lanes 3,4) or 50% (lanes 5,6). Following incubation at 4°C for 1 h, the supernatant (Sup) and pellet (Pel) were separated by centrifugation (20,000g). The pellet was resuspended in 1 mL of Buffer A. Both the supernatants and pellets were dialyzed in Buffer A overnight. All fractions were assayed for inhibitory activity (as described in Materials and Methods) and evaluated for Mcl-1 and Bcl-xL. (B) HeLa S100 (36 mg) was first precipitated with 30% ammonium sulfate. The resulting pellet (7.5 mg) was resuspended and dialyzed in Buffer A and then loaded onto a 1-mL Hi-trap Q Sepharose column (Amersham) equilibrated in Buffer A. The protein was eluted with a gradient from 0 to 750 mM NaCl (in Buffer A) over 14 mL. Inhibitory activity was assayed for buffer alone (lane 1), input (lane 2), Q flow through (lane 3), and fractions eluting from Q sepharose (lanes 4-13). The amount of Mcl-1 and Bcl-xL in each sample and the mitochondria (lane 1) was measured by Western blot. (C) HeLa S100 was immunodepleted as described in Materials and Methods. Inhibitory activity was assayed in buffer alone (lane 1), S100 mock-immunodepleted (lane 2), depleted of Mcl-1 (lane 3), Bcl-xL (lane 4), or both (lane 5). The amount of Mcl-1 and Bcl-xL was determined by Western blot for each S100 sample. (D) Recombinant Mcl-1 (rMcl-1) and Bcl-xL (rBcl-xL) were prepared as described in Materials and Methods. S100, rMcl-1, and rBcl-xL were analyzed for inhibitory activity. The levels of Mcl-1 and Bcl-xL in recombinant fractions were compared with those in S100. Mitochondria solubilized in Buffer A with 1% NP-40 were analyzed for Mcl-1 and Bcl-xL to compare the levels of these proteins in mitochondria to those in the fractions.
Because antiapoptotic members of the Bcl-2 family of proteins are likely candidates for such an activity, we probed the column fractions using antibodies against these proteins. As shown in the middle and lower panels of Figure 2A and B, we found that Bcl-xL and Mcl-1 co-purified with the inhibitory activity.
We tested whether the inhibitory activity in naive cytosol was due to Mcl-1 and Bcl-xL by immunodepleting these two proteins either individually or in combination from naive S100. As shown in the middle and lower panels of Figure 2C, antibodies against Mcl-1 and Bcl-xL efficiently depleted these proteins from naive cytosol (Fig. 2C, lanes 3-5). Cytosol depleted of Mcl-1 (Fig. 2C, lane 3) or Bcl-xL (Fig. 2C, lane 4) lost some activity whereas cytosol depleted of both (Fig. 2C, lane 5) completely lost the ability to inhibit cytochrome c release, indicating that both Mcl-1 and Bcl-xL are necessary for full activity (Fig. 2C, upper panel).
To confirm that Bcl-xL and Mcl-1 are sufficient to inhibit cytochrome c release from UV-treated mitochondria, purified recombinant Bcl-xL or Mcl-1 was incubated with UV-primed mitochondria. As shown in the upper panel of Figure 2D, 3 nM Mcl-1 or 10 nM Bcl-xL was able to completely block cytochrome c when added directly to UV-primed mitochondria (Fig. 2D, lanes 6,11). The concentration of recombinant Bcl-xL and Mcl-1 in the reactions is comparable with that of Bcl-xL and Mcl-1 present on the mitochondria and in naive cytosol (Fig. 2D, middle and lower panels, lanes 1-4).
UV irradiation triggers a decrease in Mcl-1 levels and the translocation of Bcl-xL to the mitochondria
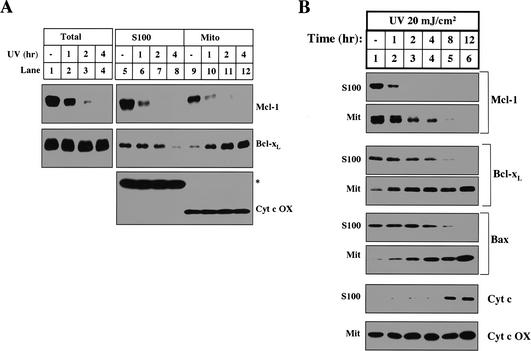
The inhibition of cytochrome c release in naive cytosol is removed by UV irradiation (Fig. 1C, lanes 3-5). We therefore examined the levels of Mcl-1 and Bcl-xL in cytosol and mitochondria isolated from naive and UV-treated cells. As shown in Figure 3A, the total amount of Mcl-1 was markedly decreased 1 h after UV treatment and completely disappeared after 2 h (Fig. 3A, lanes 1-4). On the other hand, the total amount of Bcl-xL in cells did not change even 4 h after UV irradiation. However, cytosolic Bcl-xL started to decrease 1 h after UV irradiation and disappeared by 4 h, when robust apoptosis is observed (Fig. 3A, middle panel, lanes 5-8). Unlike Mcl-1, there was a corresponding increase of Bcl-xL on mitochondria, indicating that Bcl-xL translocates from cytosol to mitochondria after UV irradiation (Fig. 3A, middle panel, lanes 9-12). As a loading control, the levels of cytochrome c oxidase, a mitochondrial inner membrane protein, and a cross-reactive cytosolic protein remained unchanged (Fig. 3A, lower panel). The translocation of Bcl-xL from the cytosol to the mitochondria was previously described in thymocytes undergoing apoptosis (Hsu et al. 1997). In that study, the authors also showed that Bax, a proapoptotic Bcl-2 family member, similarly translocated from the cytosol to the mitochondria. Bax translocation, along with cytochrome c release and caspase activation, is commonly used as a marker for apoptosis.
Figure 3.
UV treatment induces the disappearance of Mcl-1 and the translocation of Bcl-xL. (A) HeLa cells left untreated or treated with UV for different amounts of time were either harvested for 1% CHAPS total cell lysate or fractionated into mitochondria (Mito) and S100. The levels of Mcl-1, Bcl-xL, and cytochrome c oxidase were determined in total cell lysate (lanes 1-4), S100 (lanes 5-8), and Mito (lanes 9-12). (B) Human fibroblast cells were treated (as described in Materials and Methods) with UV at different time points before harvest. Mcl-1, Bcl-xL, Bax, and cytochrome c levels were compared in the S100. Mcl-1, Bcl-xL, Bax, and cytochrome c oxidase were compared in the mitochondria.
To test whether the disappearance of Mcl-1 and translocation of Bcl-xL is specific to HeLa cells, we treated human fibroblasts with UV and tracked the levels of Mcl-1, Bcl-xL, and Bax in the mitochondria and cytosol. Following UV treatment, both the disappearance of Mcl-1 and the translocation of Bax and Bcl-xL precede cytochrome c release (Fig. 3B). These data demonstrate that following UV treatment the disappearance of Mcl-1 and translocation of Bcl-xL are not restricted to HeLa cells. Additionally, this experiment demonstrates that the translocation of Bax and Bcl-xL occurs with similar kinetics.
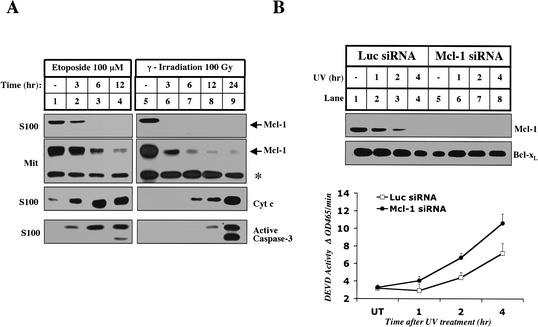
To determine if the disappearance of Mcl-1 is a general response to genotoxic stress, we analyzed the levels of Mcl-1 in cells treated with etoposide and γ-irradiation. In both cases, Mcl-1 disappears from the cytosol and mitochondria well before cytochrome c is released and caspases are activated (Fig. 4A). To evaluate the importance of the disappearance of Mcl-1 during UV-induced apoptosis, we used RNA interference (RNAi) to specifically knock down the levels of Mcl-1 (Fig. 4B; Elbashir et al. 2001). The levels of Mcl-1 were decreased in cells transfected with Mcl-1 siRNA, whereas the levels of Bcl-xL remained unchanged (Fig. 4B, lane 5). When Mcl-1 was eliminated by siRNA treatment, UV-induced caspase-3 activation was accelerated, suggesting that the disappearance of Mcl-1 is an important early event in apoptosis (Fig. 4B, lower panel). However, Mcl-1 siRNA-treated cells did not activate caspase-3 in the absence of UV, indicating that the elimination of Mcl-1 is not sufficient to activate caspase-3.
Figure 4.
Mcl-1 disappears in the early stages of apoptosis induced by other DNA-damaging agents. (A) Mitochondria and S100 were isolated from HeLa cells that were left untreated (lanes 1,5), treated with 100 μM Etoposide (Sigma; lanes 2-4), or treated with 100 Gy of γ-irradiation (lanes 6-9) and harvested for cytosol and mitochondria at the indicated times. The levels of cleaved caspase-3, cytochrome c and Mcl-1 were measured in S100. Mcl-1 mitochondrial levels were compared between the samples. The asterisk denotes a cross-reactive band that indicates equal loading. (B) Elimination of Mcl-1 by RNAi accelerates UV-induced apoptosis. HeLa cells were pretreated with Luciferase or Mcl-1 siRNA as described in Materials and Methods. Triplicate samples were harvested without treatment (UT) or at different time points after UV treatment. Whole-cell lysate prepared with 0.5% CHAPS was used to determine the levels of Mcl-1 and Bcl-xL by Western blot and caspase-3 activity by a fluorogenic assay as described in Materials and Methods.
Mcl-1 elimination is caused by a lack of synthesis
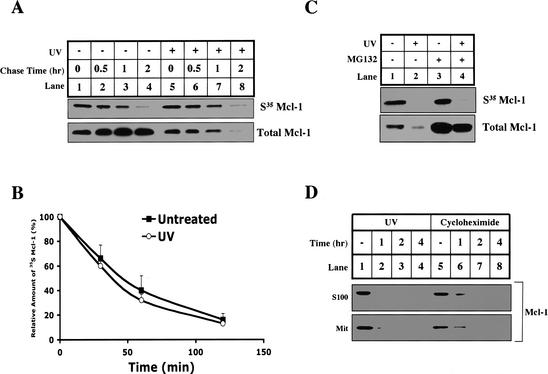
UV-induced Mcl-1 elimination could be caused by accelerated protein degradation or inhibition of synthesis. To distinguish between these possibilities, we pulse-labeled cells with 35S methionine and chased with or without UV irradiation. Newly synthesized Mcl-1 was analyzed by immunoprecipitation. As shown in Figure 5A and B, Mcl-1 is a short-lived protein with a half-life of ∼40 min. Interestingly, the half-life of Mcl-1 was the same with or without UV irradiation, even though the total amount of Mcl-1 protein was dramatically decreased after UV irradiation (Fig. 5A, lower panel).
Figure 5.
The synthesis of Mcl-1 is blocked by UV, and the protein half-life is unchanged. (A,B) HeLa cells were incubated in methionine starvation medium for 30 min before adding [35S]methionine to pulse for 1 h. Following the pulse, cells were either UV-treated (lanes 5-8) or untreated (lanes 1-4), and then immediately chased by complete medium for 0 (lanes 1,5), 30 min (lanes 2,6), 60 min (lanes 3,7), or 120 min (lanes 4,8). Mcl-1 was immunoprecipitated and analyzed by SDS-PAGE. (B) The amount of 35S-labeled Mcl-1 was quantified by PhosphorImager analysis and plotted with respect to time. Values are representative of three independent experiments. (C) HeLa cells were either left untreated (lanes 1,3) or UV-treated (lanes 2,4), and pretreated with DMSO (lanes 1,2) or 10 μM MG132 (lanes 3,4), and then methionine-starved for 30 min followed by 1 h of labeling with [35S]methionine. After labeling, the synthesis of new Mcl-1 was measured by Mcl-1 immunoprecipitation, and total Mcl-1 levels were determined by Western blot. (D) HeLa cells were left untreated (lanes 1,5), UV-treated (lanes 2-4), or treated with 20 μM cycloheximide (lanes 6-8). At the indicated times after treatment, the cells were harvested for S100 and mitochondria. The levels of Mcl-1 were determined by Western blotting both fractions.
The above experiment suggests that Mcl-1 protein synthesis must be blocked after UV irradiation. To demonstrate that directly, we pulse-labeled the cells with 35S methionine at the same time as UV irradiation (Fig. 5C, lanes 1,2). After a 60-min pulse, newly synthesized Mcl-1 was dramatically decreased if cells were exposed to UV irradiation. This decrease was not affected by blocking degradation with the proteasome inhibitor, MG132 (Fig. 5C, lanes 3,4; Palombella et al. 1994). To verify that the disappearance of Mcl-1 protein is consistent with the inhibition of its synthesis, we treated HeLa cells with an inhibitor of protein synthesis, cycloheximide, and checked the fate of Mcl-1 protein in the same time course as UV irradiation. As shown in Figure 5D, Mcl-1 disappeared after UV or cycloheximide treatment at a similar rate.
Proteasome-mediated degradation of Mcl-1 is required for UV-induced apoptosis
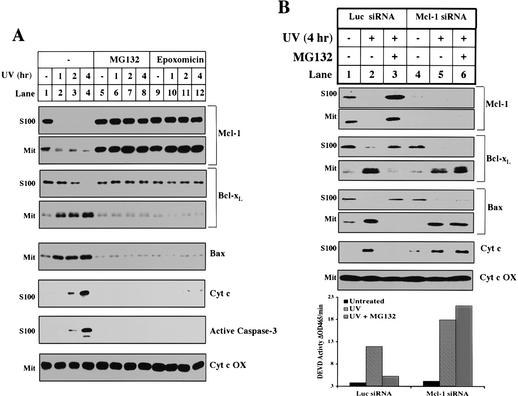
The short half-life of Mcl-1 is due to constitutive polyubiquitination and subsequent degradation by the proteasome (data not shown). Hence, to test whether the elimination of Mcl-1 is necessary for apoptosis, we incubated UV-treated cells in the presence of MG132 or another structurally unrelated proteasome inhibitor, epoxomicin (Meng et al. 1999). As shown in Figure 6A, both inhibitors efficiently block the degradation of Mcl-1 (Fig. 6A, lanes 5-12). Concomitantly, they also block other biochemical markers of apoptosis including Bcl-xL and Bax translocation, Bak oligomerization (data not shown), cytochrome c release, and caspase-3 activation.
Figure 6.
Proteasome inhibitors block the decrease of Mcl-1, the translocation of Bcl-xL, and other apoptotic events after UV treatment. (A) Mitochondria and S100 were fractionated from HeLa cells treated with UV for different amounts of time and pretreated for 1 h with DMSO (lanes 1-4), 10 μM MG132 (lanes 5-8), or 10 μM epoxomicin (lanes 9-12). The levels of Mcl-1, Bcl-xL, Bax, and cytochrome c oxidase were measured in the mitochondria. The levels of Mcl-1, Bcl-xL, cytochrome c, and cleaved caspase-3 were measured in the cytosol. (B) Six 6-well dishes of HeLa cells were transfected with Luciferase or Mcl-1 siRNA as described in Materials and Methods. For each sample, two dishes were pretreated (30 min) 24 h later with DMSO (lanes 1,2,4,5) or 10 μM MG132 (lanes 3,6). Cells were either left untreated (lanes 1,4) or UV-treated (lanes 2,3,5,6) and harvested 4 h later. A small aliquot of the cell pellet was used to make whole cell lysate in 0.5% CHAPS for the caspase assay. The remaining cells were fractionated for mitochondria and S100. The levels of Mcl-1, Bcl-xL, Bax, and cytochrome c oxidase were measured in the mitochondria. The levels of Mcl-1, Bcl-xL, Bax, and cytochrome c were measured in the cytosol.
To ensure that the antiapoptotic effect of the proteasome inhibitors was due specifically to the accumulation of Mcl-1, we first eliminated Mcl-1 using RNAi and then tested the effect of the proteasome inhibitors. As shown in Figure 6B, pretreatment of HeLa cells with Mcl-1 siRNA completely blocked the ability of MG132 or epoxomicin (data not shown) to prevent Bcl-xL and Bax translocation, cytochrome c release, and caspase-3 activation (Fig. 6B, lanes 4-6, and bar graph). However, in control siRNA-treated cells, proteasome inhibitors efficiently blocked all of these events (Fig. 6B, lanes 1-3, and bar graph).
Mcl-1 is upstream of Bcl-xL and Bax translocation
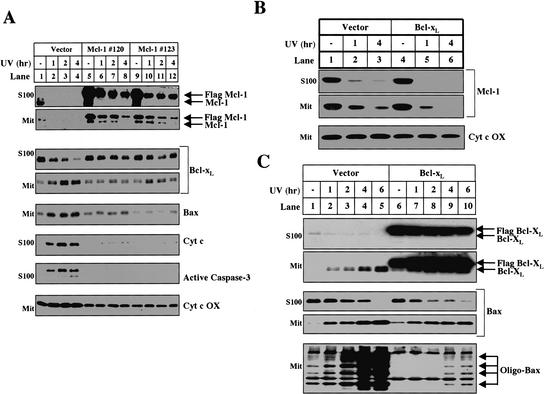
The ability of proteasome inhibitors to block Bcl-xL/Bax translocation, Bak oligomerization, cytochrome c release, and caspase-3 activation indicates that proteasome-mediated degradation of Mcl-1 is an upstream step in this apoptotic pathway. This hypothesis predicts that if the levels of Mcl-1 are artificially elevated, downstream events such as Bax and Bcl-xL translocation will be delayed. We tested this hypothesis by developing two independent HeLa cell lines that stably express Flag-tagged Mcl-1 under a CMV promoter. In response to UV irradiation, both the levels of endogenous Mcl-1 and the tagged Mcl-1 expressed from the transgene decreased (Fig. 7A, lanes 5-12). Nevertheless, 4 h after UV irradiation, there was still some Flag-tagged Mcl-1 protein in the cytosol and mitochondria (Fig. 7A, lanes 8,12). When the levels of Mcl-1 are artificially elevated in Mcl-1 transgenic cells, cytochrome c, Bcl-xL, and Bax do not translocate 4 h after UV irradiation in contrast to a control HeLa cell line that expresses the Neo vector (Fig. 7A, lanes 1-4), indicating that the elimination of Mcl-1 is a required upstream event for Bcl-xL and Bax translocation.
Figure 7.
The disappearance of Mcl-1 is a required upstream event for UV-induced apoptosis. (A) After UV treatment, mitochondria and S100 were isolated from vector control (lanes 1-4) and two independent Flag-Mcl-1-overexpressing lines: line 120 (lanes 5-8) and line 123 (lanes 9-12). The levels of Mcl-1, Bcl-xL, Bax, and cytochrome c oxidase were measured in the mitochondria. The levels of Mcl-1, Bcl-xL, cytochrome c, and cleaved caspase-3 were measured in the cytosol. (B) Mcl-1 levels were analyzed in mitochondria and S100 fractionated from Vector (lanes 1-3) or Flag-Bcl-xL-overexpressing (lanes 4-6) cells treated with UV for 1 h (lanes 2,5) or 4 h (lanes 3,6). (C) HeLa cells that stably express the Neo vector (lanes 1-5) or Flag-Bcl-xL (lanes 6-10) were left untreated or UV-treated and harvested between 1 and 6 h later for mitochondria and S100. The levels of Flag-Bcl-xL, Bcl-xL, and Bax were determined in the S100 and mitochondria. Mitochondria were also used to evaluate Bax oligomerization.
To verify that Mcl-1 degradation and Bax/Bcl-xL translocation are sequential events and not merely a product of antiapoptotic protein overexpression, we examined a Bcl-xL-overexpressing HeLa cell line, which also does not release cytochrome c after UV treatment (data not shown). After UV treatment, despite extremely high levels of exogenous Bcl-xL expression, Mcl-1 is eliminated at the same rate (Fig. 7B, lanes 4-6). Moreover, in contrast to Mcl-1 overexpression, exogenous expression of Bcl-xL does not inhibit the translocation of Bax or Bcl-xL to the mitochondria (Fig. 7C, lanes 6-10). Bcl-xL overexpression does, however, block the oligomerization of Bax on the mitochondria (Fig. 7C, bottom panel) suggesting the following sequence of events: Mcl-1 degradation, Bcl-xL/Bax translocation to the mitochondria, Bax oligomerization, and cytochrome c release.
Discussion
Mcl-1 and Bcl-xL function at different steps in a UV-induced apoptotic pathway
The Bcl-2 family is composed of both pro- and antiapoptotic proteins that share sequence and structural homology (Cory and Adams 2002). Most of the initial studies of the Bcl-2 family used overexpression as a means of characterizing the apoptotic nature of the proteins (Vaux et al. 1988; Oltvai et al. 1993). Although effective at separating pro- and antiapoptotic characteristics, overexpression studies do not differentiate specific functions within the pro- or antiapoptotic groups.
Recently, genetic experiments separated the proapoptotic members of the Bcl-2 family into two functional classes: Bax/Bak and BH3-only proteins. BH3-only proteins are upstream of Bax/Bak because BH3-only proteins fail to trigger cytochrome c release in Bax/Bak-deficient cells (Cheng et al. 2001; Wei et al. 2001; Zong et al. 2001). On the other hand, mouse knock'out models have proven less effective at sorting out the functions of the antiapoptotic Bcl-2 family members. Mcl-1-null embryos have a preimplantation lethal phenotype, revealing only that Mcl-1 has an important and early role in development (Rinkenberger et al. 2000). The knockout of Bcl-xL was also early embryonic lethal; the embryo displayed widespread apoptosis, indicating that this gene also functions early in development (Motoyama et al. 1995).
Using a biochemical approach, this study demonstrates that the antiapoptotic members of the Bcl-2 family also function at distinct steps. Mcl-1 is at the apical point in the pathway. The disappearance of Mcl-1 is required for downstream events including Bcl-xL and Bax translocation, Bak and Bax oligomerization, cytochrome c release, and caspase activation. Mcl-1 may regulate downstream events by sequestering proapoptotic BH3 proteins that activate downstream events such as Bax/Bcl-xL translocation and Bax/Bak oligomerization (Letai et al. 2002).
The cytosol-to-membrane translocation of both proapoptotic Bax and antiapoptotic Bcl-xL is counterintuitive and seemingly counterproductive. However, because the levels of Bax are in excess of Bcl-xL in HeLa cells (M. Fang and X. Wang, unpubl.), continuous translocation of Bax may eventually result in excess free Bax on the mitochondria, leading to oligomerization. Consistent with this model, Bcl-xL overexpression blocks oligomerization but does not affect Bcl-xL or Bax translocation (Fig. 7C). The effect of Bcl-2 overexpression on Bax translocation when apoptosis is induced by growth factor deprivation is variable. Bcl-2 overexpression blocks Bax translocation in B-cells after IL-3 withdrawal but not in primary neurons after NGF withdrawal (Gross et al. 1998; Putcha et al. 1999).
Mcl-1 is an apical sensor for apoptotic stimuli
The elimination of Mcl-1 is not limited to UV irradiation. Genotoxic stress induced by etoposide or γ-irradiation also efficiently induces Mcl-1 elimination (Fig. 4A), suggesting that the disappearance of Mcl-1 may be initiated through a common pathway. Previous studies have shown that UV irradiation, γ-irradiation, and etoposide treatment induce an arrest in protein synthesis (Clemens et al. 2000). UV light treatment triggers the ubiquitination and subsequent degradation of RNA polymerase II and the GCN2-mediated phosphorylation of eIF2α to arrest transcription and translation, respectively (Bregman et al. 1996; Deng et al. 2002). Because Mcl-1 protein and mRNA both have short half-lives (Yang et al. 1996), Mcl-1 can serve as a sensor of acute changes in the rate of mRNA and protein synthesis after genotoxic stress. On the other hand, increased Mcl-1 synthesis may enhance the general well being of the cell. Growth factor pathways that promote cell survival through the AKT/PI3 kinase pathway up-regulate Mcl-1 mRNA levels (Wang et al. 1999; Huang et al. 2000; Liu et al. 2001). Our study predicts that this up-regulation of Mcl-1 may explain how growth factors render cells less sensitive to apoptotic stimuli (Datta et al. 1999).
Although the elimination of Mcl-1 by an arrest in synthesis is required, this event is not sufficient for activation of the mitochondrial apoptosis pathway. The elimination of Mcl-1 by RNAi did not induce Bax/Bcl-xL translocation, Bax/Bak oligomerization (data not shown), cytochrome c release, and caspase activation (Figs. 4B, 6B). Therefore, another UV-induced event in addition to the elimination of Mcl-1 must be required to promote this pathway. In other systems, it appears that Mcl-1 disappearance is both necessary and sufficient for apoptosis. For example, it is possible to induce apoptosis in a multiple myeloma cell line merely by treating it with inhibitors of protein synthesis or by specifically removing Mcl-1 with anti-sense oligonucleotides (Zhang et al. 2002). In this system, depletion of Mcl-1 is the only hit needed for apoptosis.
Stress-activated pathways have also been implicated in UV-induced cytochrome c release. Fibroblasts deficient in JNK1/JNK2 kinases fail to release cytochrome c in response to UV treatment (Tournier et al. 2000). JNK kinases might activate the Mcl-1 pathway by directly phosphorylating and inactivating Mcl-1 as they do after oxidative stress (Inoshita et al. 2002). Alternatively, JNK activity might regulate pathways leading to a stop in Mcl-1 synthesis or initiate a separate required pathway. All of these possibilities can now be tested in JNK-deficient cells.
The quest to delineate the apoptosis pathway upstream of cytochrome c release is partly motivated by data suggesting that mitochondrial damage from proapoptotic Bcl-2 family members is toxic to cells even in the absence of caspase activation (Xiang et al. 1996). Hence, potential therapies that inhibit apoptosis before the onset of mitochondrial dysfunction are likely to be more effective than direct caspase inhibitors. The sequential nature of the apoptotic signal transduction pathway revealed by this study offers several points for potential intervention. Novel therapies that block Mcl-1 degradation may effectively prevent apoptosis and cell death.
Materials and methods
Reagents
The following antibodies were used for Western blots: monoclonal Bak Ab-1 (Oncogene), monoclonal cytochrome c (Pharmingen), polyclonal Bcl-xL (Cell signaling), monoclonal Mcl-1 (Pharmingen), monoclonal caspase-2 (Pharmingen), polyclonal caspase-3 (cell signaling), monoclonal caspase-9 (Cell Signaling), and polyclonal Bax N-20 (Santa Cruz). Immunoprecipitations or immunodepletions for Mcl-1 and Bcl-xL were performed with polyclonal antibodies to Mcl-1 (Pharmingen) and Bcl-xL (Cell Signaling). Z-VAD.fmk, cycloheximide, MG132, epoxomicin, caspase-3 fluorogenic II substrate, phosphatase inhibitor cocktail II, and λ Protein Phosphatase were all obtained from Calbiochem.
UV treatment and cellular fractionation
HeLa cells were plated at a density of 2 × 107 cells in a 15-cm dish with DMEM medium supplemented with 10% FBS. Several hours before treatment, each dish was replaced with 13 mL of fresh medium. The cover of each dish was removed before the cells were treated in a Stratagene stratalinker with 200 mJ/cm2 of UV irradiation (254 nm). At the indicated times after treatment, cells were scraped, collected, and washed once in PBS. The cell pellet was resuspended in 5 times the volume of Buffer A (20 mM HEPES, 10 mM KCl, 1.5 mM MgCl2, 1 mM EDTA, 1 mM EGTA, 1 mM DTT, 0.1 mM PMSF, and Complete Protease Inhibitor; Roche) supplemented with 250 mM sucrose. The resuspended cell pellet was incubated on ice for 15 min before the cells were broken by passing them through a 22-gauge needle 25 times. The resulting broken cell mixture was centrifuged in three sequential steps: 1000g, 10,000g, and 100,000g. The 10,000g pellet was considered the “mitochondrial“ fraction and the 100,000g supernatant (S100) the cytosol. Mitochondrial pellets were lysed in SDS loading buffer, and equal volumes were analyzed by SDS-PAGE. S100 protein concentrations were normalized using the Bio-Rad Bradford reagent.
Recombinant Mcl-1 and Bcl-xL
The cDNA of full-length human Mcl-1 was subcloned into NdeI and SapI sites of the pTYB1 vector (New England Biolabs). Full-length Bcl-xL cDNA was cloned into NdeI and XhoI sites of the pet15B vector (Novagen). BL21 (DE3) cells were transformed with these constructs, and the recombinant proteins were purified according to the manufacturer's protocol (New England Biolabs and QIAGEN, respectively). The eluted recombinant Mcl-1 protein was purified in two additional steps: Hitrap Q sepharose and Superdex 200 size exclusion chromatography. The eluted recombinant Bcl-xL was further purified on a Hitrap Q sepharose column.
In vitro assays for mitochondrial priming and inhibitory activity
Mitochondria (0.67 mg/mL) alone or with S100 (4 mg/mL) were incubated in Buffer A with 250 mM sucrose and 150 mM NaCl at 37°C for 15 min. Following incubation, the mitochondria were pelleted, and the supernatant was tested for cytochrome c release. The mitochondria pellet was tested for Bak oligomerization (Wei et al. 2000).
During biochemical fractionation, to increase our yield we made S100 from HeLa cells using Buffer A without sucrose. Hypotonic buffers cause mitochondrial rupture and cytochrome c contamination in the S100. Contaminating cytochrome c was removed from the S100 by incubating with SP-sepharose XL resin (Amersham) at 4°C for 1 h. Cytochrome c binds SP resin, but our activity does not (data not shown).
Mcl-1 RNA interference
Annealed, purified, and desalted double-stranded siRNA, Luciferase (AACGUACGCGGAAUACUUCGA), and Mcl-1 (AAGAAACGCGGUAAUCGGACU) were ordered from Dharmacon Research. Then 1.5 × 105 HeLa cells were plated in a 6-well dish on day 0. On day 1, cells were transfected with 200 nM siRNA in Opti-MEM medium (Invitrogen) without FBS using Oligofectamine reagent (Invitrogen) according to the manufacturer's transfection protocol. After 4 h, FBS was added to a final concentration of 10%. On day 2, the medium over the cells was adjusted to 1 mL before treatment.
Fluorogenic assay for caspase-3activity
In a 20-μL system, 10 μg of whole cell lysate was incubated in Buffer A containing 0.125% CHAPS and 10 μM fluorogenic DEVD substrate. Samples were applied to a 384-well micro-plate, and the reaction was carried out at 30°C. Every 10 min, the samples were excited at 360 nM and the OD465 nM was measured by the SpectraFluor4 spectrometry reader (TECAN).
Mcl-1 pulse-chase experiments
HeLa suspension cells were plated at a density of 5 × 106 cells per 10-cm dish. For the pulse chase, the cells were washed with phosphate-buffered saline (PBS), starved in methionine/cysteine-free DMEM (GIBCO-BRL) at 37°C for 30 min, then labeled with 200-500 μCi of [35S]cysteine/methionine (ICN) per plate at 37°C for 60 min. After labeling, the cells were chased with complete DMEM containing 10% FBS and 2 M cold methionine at 37°C for the indicated amount of time. A total of 107 cells were collected at each time point. Cells were subsequently subjected to immunoprecipitation analysis as described below.
Immunoprecipitation
For immunodepletion and Mcl-1 immunoprecipitation experiments, samples were incubated with beads precoupled to antibody. Protein A agarose beads (Santa Cruz; 0.5-mL bed volume) were incubated in 1 mL of PBS with 1 mg/mL of BSA at 4°C alone overnight, with 100 μg of polyclonal antibodies to Mcl-1 or Bcl-xL. After incubation, the beads were pelleted by centrifugation and washed at least 5 times with Buffer A. For immunodepletion reactions, 50 μL of beads were incubated with 500 μL of S100 (5 mg/mL) with rotation at 4°C overnight. After incubation, the beads were pelleted, and the supernatant was collected as immunodepleted S100.
For pulse-chase experiments, radiolabeled cells were lysed in cold RIPA lysis buffer (10 mM Tris-HCl at pH 7.4, 1% NP-40, 1 mM EDTA, 0.1% SDS, 150 mM NaCl) with fresh proteinase inhibitors, followed by breaking through a 25-gauge syringe 20 times. Equivalent amounts of cell extract were adjusted to equal volumes with RIPA buffer, precleared with protein A agarose beads, and clarified by centrifugation. Lysates were then incubated with polyclonal antibodies to Mcl-1 at 4°C overnight, and the immune complexes were precipitated with protein A agarose beads for 4-6 h. The immunoprecipitates were washed five times with RIPA buffer and incubated at 95°C for 5 min in SDS sample buffer. Samples were subjected to (SDS-PAGE) followed by autoradiography.
Mcl-1- and Bcl-xL-overexpressing stable cell lines
Full-length Mcl-1 cDNA was cloned into HindIII and BamHI sites of p3XFLAG-CMV-10 (Sigma). The resulting fusion protein was full-length Mcl-1 with three Flag epitopes at the N terminus followed by a two-amino-acid linker. The full-length Bcl-xL was cloned into XhoI and NdeI sites of pCDNA 3.1(-) (Invitrogen) with a single Flag tag at the N terminus followed by a short linker sequence. Empty vector or subcloned plasmids were transfected into 5 × 105 attached HeLa cells grown in DMEM with 10% FBS using Lipofectamine Plus transfection reagent (Invitrogen) according to the manufacturer's protocol. Then, 2 d later, the cells were transferred to 20 100-mm dishes in DMEM with 10% FBS and 0.5 mg/mL of G418. After several weeks, individual clones were lifted and tested for expression of the transgene.
Acknowledgments
We thank X. Luo for Bcl-xL expression constructs. X. Jiang and I. Alibhai gave us critical insight and helpful advice. Michael Brown, Joseph Goldstein, and A. Shulman offered helpful advice in the preparation of our manuscript. R. Harold provided valuable technical assistance. This work was also supported by NIH (GMR01-57158) and the Welch foundation (2-1412). D.N. and E.T. are medical scientist training fellows supported by the Perot Family foundation.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement“ in accordance with 18 USCsection 1734 solely to indicate this fact.
Corresponding author.
Article published online ahead of print. Article and publication date are at http://www.genesdev.org/cgi/doi/10.1101/gad.1093903.
References
- Acehan D., Jiang, X., Morgan, D.G., Heuser, J.E., Wang, X., and Akey, C.W. 2002. Three-dimensional structure of the apoptosome: Implications for assembly, procaspase-9 binding, and activation. Mol. Cell 9: 423–432. [DOI] [PubMed] [Google Scholar]
- Bossy-Wetzel E., Newmeyer, D.D., and Green, D.R. 1998. Mitochondrial cytochrome c release in apoptosis occurs upstream of DEVD-specific caspase activation and independently of mitochondrial transmembrane depolarization. EMBO J. 17: 37–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bregman D.B., Halaban, R., van Gool, A.J., Henning, K.A., Friedberg, E.C., and Warren, S.L. 1996. UV-induced ubiquitination of RNA polymerase II: A novel modification deficient in Cockayne syndrome cells. Proc. Natl. Acad. Sci. 93: 11586–11590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng E.H., Wei, M.C., Weiler, S., Flavell, R.A., Mak, T.W., Lindsten, T., and Korsmeyer, S.J. 2001. BCL-2, BCL-XL sequester BH3 domain-only molecules preventing BAX- and BAK-mediated mitochondrial apoptosis. Mol. Cell 8: 705–711. [DOI] [PubMed] [Google Scholar]
- Clemens M.J., Bushell, M., Jeffrey, I.W., Pain, V.M., and Morley, S.J. 2000. Translation initiation factor modifications and the regulation of protein synthesis in apoptotic cells. Cell Death Differ. 7: 603–615. [DOI] [PubMed] [Google Scholar]
- Cory S. and Adams, J.M. 2002. The Bcl2 family: Regulators of the cellular life-or-death switch. Nat. Rev. Cancer 2: 647–656. [DOI] [PubMed] [Google Scholar]
- Datta S.R., Brunet, A., and Greenberg, M.E. 1999. Cellular survival: A play in three Akts. Genes & Dev. 13: 2905–2927. [DOI] [PubMed] [Google Scholar]
- Deng J., Harding, H.P., Raught, B., Gingras, A.C., Berlanga, J.J., Scheuner, D., Kaufman, R.J., Ron, D., and Sonenberg, N. 2002. Activation of GCN2 in UV-irradiated cells inhibits translation. Curr. Biol. 12: 1279–1286. [DOI] [PubMed] [Google Scholar]
- Du C., Fang, M., Li, Y., Li, L., and Wang, X. 2000. Smac, a mitochondrial protein that promotes cytochrome c-dependent caspase activation by eliminating IAP inhibition. Cell 102: 33–42. [DOI] [PubMed] [Google Scholar]
- Duelli D.M. and Lazebnik, Y.A. 2000. Primary cells suppress oncogene-dependent apoptosis. Nat. Cell Biol. 2: 859–862. [DOI] [PubMed] [Google Scholar]
- Elbashir S.M., Harborth, J., Lendeckel, W., Yalcin, A., Weber, K., and Tuschl, T. 2001. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature 411: 494–498. [DOI] [PubMed] [Google Scholar]
- Gross A., Jockel, J., Wei, M.C., and Korsmeyer, S.J. 1998. Enforced dimerization of BAX results in its translocation, mitochondrial dysfunction and apoptosis. EMBO J. 17: 3878–3885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross A., McDonnell, J.M., and Korsmeyer, S.J. 1999. BCL-2 family members and the mitochondria in apoptosis. Genes & Dev. 13: 1899–1911. [DOI] [PubMed] [Google Scholar]
- Hegde R., Srinivasula, S.M., Zhang, Z., Wassell, R., Mukattash, R., Cilenti, L., DuBois, G., Lazebnik, Y., Zervos, A.S., Fernandes-Alnemri, T., et al. 2002. Identification of Omi/HtrA2 as a mitochondrial apoptotic serine protease that disrupts inhibitor of apoptosis protein-caspase interaction. J. Biol. Chem. 277: 432–438. [DOI] [PubMed] [Google Scholar]
- Hengartner M.O. 2000. The biochemistry of apoptosis. Nature 407: 770–776. [DOI] [PubMed] [Google Scholar]
- Hsu Y.T., Wolter, K.G., and Youle, R.J. 1997. Cytosol-to-membrane redistribution of Bax and Bcl-XL during apoptosis. Proc. Natl. Acad. Sci. 94: 3668–3672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang H.M., Huang, C.J., and Yen, J.J. 2000. Mcl-1 is a common target of stem cell factor and interleukin-5 for apoptosis prevention activity via MEK/MAPK and PI-3K/Akt pathways. Blood 96: 1764–1771. [PubMed] [Google Scholar]
- Inoshita S., Takeda, K., Hatai, T., Terada, Y., Sano, M., Hata, J., Umezawa, A., and Ichijo, H. 2002. Phosphorylation and inactivation of myeloid cell leukemia 1 by JNK in response to oxidative stress. J. Biol. Chem. 277: 43730–43734. [DOI] [PubMed] [Google Scholar]
- Johnstone R.W., Ruefli, A.A., and Lowe, S.W. 2002. Apoptosis: A link between cancer genetics and chemotherapy. Cell 108: 153–164. [DOI] [PubMed] [Google Scholar]
- Kim C.N., Wang, X., Huang, Y., Ibrado, A.M., Liu, L., Fang, G., and Bhalla, K. 1997. Overexpression of Bcl-XL inhibits Ara-C-induced mitochondrial loss of cytochrome c and other perturbations that activate the molecular cascade of apoptosis. Cancer Res. 57: 3115–3120. [PubMed] [Google Scholar]
- Kluck R.M., Bossy-Wetzel, E., Green, D.R., and Newmeyer, D.D. 1997. The release of cytochrome c from mitochondria: A primary site for Bcl-2 regulation of apoptosis. Science 275: 1132–1136. [DOI] [PubMed] [Google Scholar]
- Korsmeyer S.J., Wei, M.C., Saito, M., Weiler, S., Oh, K.J., and Schlesinger, P.H. 2000. Pro-apoptotic cascade activates BID, which oligomerizes BAK or BAX into pores that result in the release of cytochrome c. Cell Death Differ. 7: 1166–1173. [DOI] [PubMed] [Google Scholar]
- Kuwana T., Mackey, M.R., Perkins, G., Ellisman, M.H., Latterich, M., Schneiter, R., and Newmeyer, D.D. 2002. Bid, Bax, and lipids cooperate to form supramolecular openings in the outer mitochondrial membrane. Cell 111: 331–342. [DOI] [PubMed] [Google Scholar]
- Lassus P., Opitz-Araya, X., and Lazebnik, Y. 2002. Requirement for caspase-2 in stress-induced apoptosis before mitochondrial permeabilization. Science 297: 1352–1354. [DOI] [PubMed] [Google Scholar]
- Letai A., Bassik, M.C., Walensky, L.D., Sorcinelli, M.D., Weiler, S., and Korsmeyer, S.J. 2002. Distinct BH3 domains either sensitize or activate mitochondrial apoptosis, serving as prototype cancer therapeutics. Cancer Cell 2: 183–192. [DOI] [PubMed] [Google Scholar]
- Li L.Y., Luo, X., and Wang, X. 2001. Endonuclease G is an apoptotic DNase when released from mitochondria. Nature 412: 95–99. [DOI] [PubMed] [Google Scholar]
- Li P., Nijhawan, D., Budihardjo, I., Srinivasula, S.M., Ahmad, M., Alnemri, E.S., and Wang, X. 1997. Cytochrome c and dATP-dependent formation of Apaf-1/caspase-9 complex initiates an apoptotic protease cascade. Cell 91: 479–489. [DOI] [PubMed] [Google Scholar]
- Liu H., Perlman, H., Pagliari, L.J., and Pope, R.M. 2001. Constitutively activated Akt-1 is vital for the survival of human monocyte-differentiated macrophages. Role of Mcl-1, independent of nuclear factor (NF)-κB, Bad, or caspase activation. J. Exp. Med. 194: 113–126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinou J.C. and Green, D.R. 2001. Breaking the mitochondrial barrier. Nat. Rev. Mol. Cell. Biol. 2: 63–67. [DOI] [PubMed] [Google Scholar]
- Martins L.M., Iaccarino, I., Tenev, T., Gschmeissner, S., Totty, N.F., Lemoine, N.R., Savopoulos, J., Gray, C.W., Creasy, C.L., Dingwall, C., et al. 2002. The serine protease Omi/HtrA2 regulates apoptosis by binding XIAP through a reaper-like motif. J. Biol. Chem. 277: 439–444. [DOI] [PubMed] [Google Scholar]
- Meng L., Mohan, R., Kwok, B.H., Elofsson, M., Sin, N., and Crews, C.M. 1999. Epoxomicin, a potent and selective proteasome inhibitor, exhibits in vivo antiinflammatory activity. Proc. Natl. Acad. Sci. 96: 10403–10408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motoyama N., Wang, F., Roth, K.A., Sawa, H., Nakayama, K., Nakayama, K., Negishi, I., Senju, S., Zhang, Q., Fujii, S., et al. 1995. Massive cell death of immature hematopoietic cells and neurons in Bcl-x-deficient mice. Science 267: 1506–1510. [DOI] [PubMed] [Google Scholar]
- Oltvai Z.N., Milliman, C.L., and Korsmeyer, S.J. 1993. Bcl-2 heterodimerizes in vivo with a conserved homolog, Bax, that accelerates programmed cell death. Cell 74: 609–619. [DOI] [PubMed] [Google Scholar]
- Palombella V.J., Rando, O.J., Goldberg, A.L., and Maniatis, T. 1994. The ubiquitin-proteasome pathway is required for processing the NF-κ B1 precursor protein and the activation of NF-κ B. Cell 78: 773–785. [DOI] [PubMed] [Google Scholar]
- Putcha G.V., Deshmukh, M., and Johnson Jr., E.M. 1999. BAX translocation is a critical event in neuronal apoptosis: Regulation by neuroprotectants, BCL-2, and caspases. J. Neurosci. 19: 7476–7485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinkenberger J.L., Horning, S., Klocke, B., Roth, K., and Korsmeyer, S.J. 2000. Mcl-1 deficiency results in peri-implantation embryonic lethality. Genes & Dev. 14: 23–27. [PMC free article] [PubMed] [Google Scholar]
- Rodriguez J. and Lazebnik, Y. 1999. Caspase-9 and APAF-1 form an active holoenzyme. Genes & Dev. 13: 3179–3184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Susin S.A., Lorenzo, H.K., Zamzami, N., Marzo, I., Snow, B.E., Brothers, G.M., Mangion, J., Jacotot, E., Costantini, P., Loeffler, M., et al. 1999. Molecular characterization of mitochondrial apoptosis-inducing factor. Nature 397: 441–446. [DOI] [PubMed] [Google Scholar]
- Suzuki Y., Imai, Y., Nakayama, H., Takahashi, K., Takio, K., and Takahashi, R. 2001. A serine protease, HtrA2, is released from the mitochondria and interacts with XIAP, inducing cell death. Mol. Cell 8: 613–621. [DOI] [PubMed] [Google Scholar]
- Tournier C., Hess, P., Yang, D.D., Xu, J., Turner, T.K., Nimnual, A., Bar-Sagi, D., Jones, S.N., Flavell, R.A., and Davis, R.J. 2000. Requirement of JNK for stress-induced activation of the cytochrome c-mediated death pathway. Science 288: 870–874. [DOI] [PubMed] [Google Scholar]
- Vander Heiden M.G., Chandel, N.S., Williamson, E.K., Schumacker, P.T., and Thompson, C.B. 1997. Bcl-xL regulates the membrane potential and volume homeostasis of mitochondria. Cell 91: 627–637. [DOI] [PubMed] [Google Scholar]
- Vaux D.L., Cory, S., and Adams, J.M. 1988. Bcl-2 gene promotes haemopoietic cell survival and cooperates with c-myc to immortalize pre-B cells. Nature 335: 440–442. [DOI] [PubMed] [Google Scholar]
- Verhagen A.M., Ekert, P.G., Pakusch, M., Silke, J., Connolly, L.M., Reid, G.E., Moritz, R.L., Simpson, R.J., and Vaux, D.L. 2000. Identification of DIABLO, a mammalian protein that promotes apoptosis by binding to and antagonizing IAP proteins. Cell 102: 43–53. [DOI] [PubMed] [Google Scholar]
- Wang J.M., Chao, J.R., Chen, W., Kuo, M.L., Yen, J.J., and Yang-Yen, H.F. 1999. The antiapoptotic gene mcl-1 is up-regulated by the phosphatidylinositol 3-kinase/Akt signaling pathway through a transcription factor complex containing CREB. Mol. Cell. Biol. 19: 6195–6206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei M.C., Lindsten, T., Mootha, V.K., Weiler, S., Gross, A., Ashiya, M., Thompson, C.B., and Korsmeyer, S.J. 2000. tBID, a membrane-targeted death ligand, oligomerizes BAK to release cytochrome c. Genes & Dev. 14: 2060–2071. [PMC free article] [PubMed] [Google Scholar]
- Wei M.C., Zong, W.X., Cheng, E.H., Lindsten, T., Panoutsakopoulou, V., Ross, A.J., Roth, K.A., MacGregor, G.R., Thompson, C.B., and Korsmeyer, S.J. 2001. Proapoptotic BAX and BAK: A requisite gateway to mitochondrial dysfunction and death. Science 292: 727–730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang J., Chao, D.T., and Korsmeyer, S.J. 1996. BAX-induced cell death may not require interleukin 1 β-converting enzyme-like proteases. Proc. Natl. Acad. Sci. 93: 14559–14563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang T., Buchan, H.L., Townsend, K.J., and Craig, R.W. 1996. MCL-1, a member of the BLC-2 family, is induced rapidly in response to signals for cell differentiation or death, but not to signals for cell proliferation. J. Cell Physiol. 166: 523–536. [DOI] [PubMed] [Google Scholar]
- Zhang B., Gojo, I., and Fenton, R.G. 2002. Myeloid cell factor-1 is a critical survival factor for multiple myeloma. Blood 99: 1885–1893. [DOI] [PubMed] [Google Scholar]
- Zong W.X., Lindsten, T., Ross, A.J., MacGregor, G.R., and Thompson, C.B. 2001. BH3-only proteins that bind pro-survival Bcl-2 family members fail to induce apoptosis in the absence of Bax and Bak. Genes & Dev. 15: 1481–1486. [DOI] [PMC free article] [PubMed] [Google Scholar]