Abstract
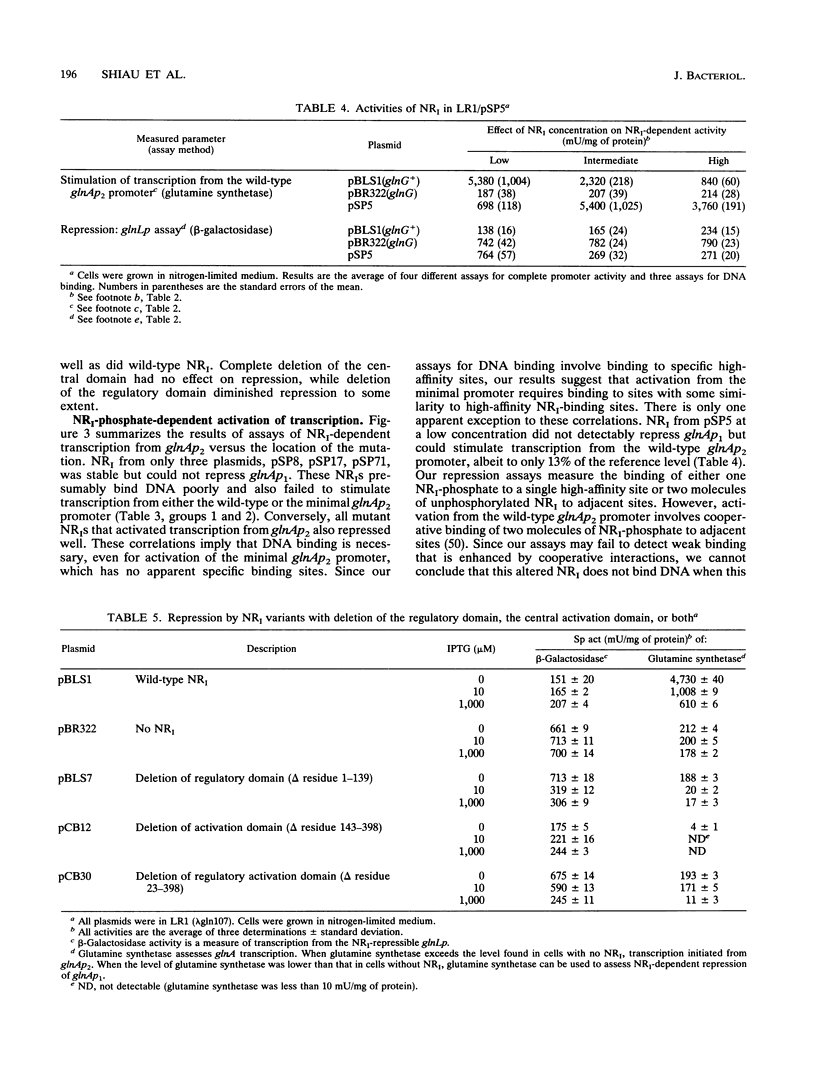
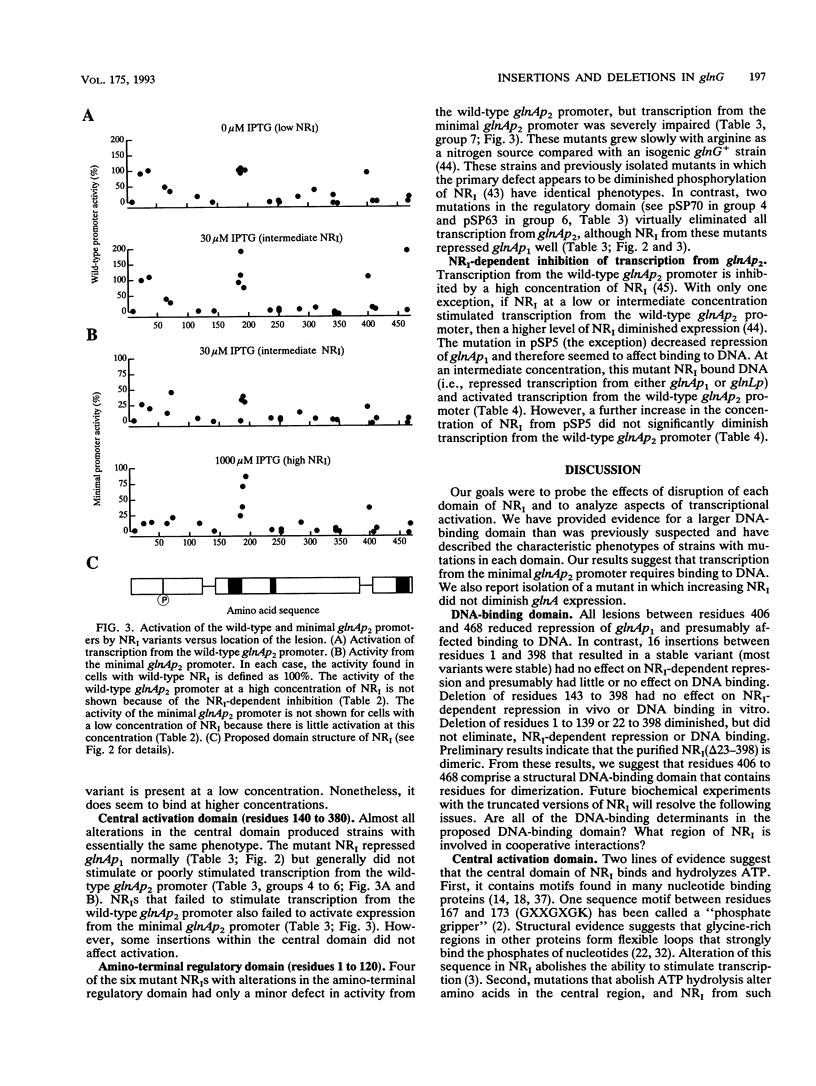
Phosphorylated nitrogen regulator I (NRI, also called NTRC), encoded by glnG (ntrC), stimulates transcription in Escherichia coli and other enteric bacteria from sites analogous to eukaryotic enhancers. We isolated 30 mutants, obtained without phenotypic selection, that have either a small insertion or deletion within glnG. Mutants were classified by the ability of NRI to repress the glnAp1 and glnL promoters and to activate two versions of the nitrogen-regulated glnAp2 promoter; each activity was measured in cells grown with three concentrations of NRI. The results were interpreted within the framework of the three-domain hypothesis of NRI structure. NRI is thought to contain a phosphorylated regulatory domain that controls binding of ATP, a central domain that hydrolyzes ATP and interacts with RNA polymerase, and a DNA-binding region of unknown extent. Our results suggest that the 70 amino acids from residue 400 to the carboxyl terminus constitute a DNA-binding domain that includes residues for specific contacts and dimerization. Our results also suggest that (i) transcription from glnAp2 without specific NRI-binding sites requires binding to sites with some similarity to the specific sites, and (ii) if an NRI variant can stimulate transcription, then increasing the concentration of NRI diminishes glnA expression for all mutants but one.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Atkins W. M., Stayton P. S., Villafranca J. J. Time-resolved fluorescence studies of genetically engineered Escherichia coli glutamine synthetase. Effects of ATP on the tryptophan-57 loop. Biochemistry. 1991 Apr 9;30(14):3406–3416. doi: 10.1021/bi00228a008. [DOI] [PubMed] [Google Scholar]
- Austin S., Kundrot C., Dixon R. Influence of a mutation in the putative nucleotide binding site of the nitrogen regulatory protein NTRC on its positive control function. Nucleic Acids Res. 1991 May 11;19(9):2281–2287. doi: 10.1093/nar/19.9.2281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barany F. Single-stranded hexameric linkers: a system for in-phase insertion mutagenesis and protein engineering. Gene. 1985;37(1-3):111–123. doi: 10.1016/0378-1119(85)90263-x. [DOI] [PubMed] [Google Scholar]
- Barany F. Two-codon insertion mutagenesis of plasmid genes by using single-stranded hexameric oligonucleotides. Proc Natl Acad Sci U S A. 1985 Jun;82(12):4202–4206. doi: 10.1073/pnas.82.12.4202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourret R. B., Borkovich K. A., Simon M. I. Signal transduction pathways involving protein phosphorylation in prokaryotes. Annu Rev Biochem. 1991;60:401–441. doi: 10.1146/annurev.bi.60.070191.002153. [DOI] [PubMed] [Google Scholar]
- Bourret R. B., Hess J. F., Simon M. I. Conserved aspartate residues and phosphorylation in signal transduction by the chemotaxis protein CheY. Proc Natl Acad Sci U S A. 1990 Jan;87(1):41–45. doi: 10.1073/pnas.87.1.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collado-Vides J., Magasanik B., Gralla J. D. Control site location and transcriptional regulation in Escherichia coli. Microbiol Rev. 1991 Sep;55(3):371–394. doi: 10.1128/mr.55.3.371-394.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Contreras A., Drummond M. The effect on the function of the transcriptional activator NtrC from Klebsiella pneumoniae of mutations in the DNA-recognition helix. Nucleic Acids Res. 1988 May 11;16(9):4025–4039. doi: 10.1093/nar/16.9.4025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon R., Eydmann T., Henderson N., Austin S. Substitutions at a single amino acid residue in the nitrogen-regulated activator protein NTRC differentially influence its activity in response to phosphorylation. Mol Microbiol. 1991 Jul;5(7):1657–1667. doi: 10.1111/j.1365-2958.1991.tb01913.x. [DOI] [PubMed] [Google Scholar]
- Drummond M. H., Contreras A., Mitchenall L. A. The function of isolated domains and chimaeric proteins constructed from the transcriptional activators NifA and NtrC of Klebsiella pneumoniae. Mol Microbiol. 1990 Jan;4(1):29–37. doi: 10.1111/j.1365-2958.1990.tb02012.x. [DOI] [PubMed] [Google Scholar]
- Drummond M., Whitty P., Wootton J. Sequence and domain relationships of ntrC and nifA from Klebsiella pneumoniae: homologies to other regulatory proteins. EMBO J. 1986 Feb;5(2):441–447. doi: 10.1002/j.1460-2075.1986.tb04230.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferro-Luzzi Ames G., Nikaido K. Nitrogen regulation in Salmonella typhimurium. Identification of an ntrC protein-binding site and definition of a consensus binding sequence. EMBO J. 1985 Feb;4(2):539–547. doi: 10.1002/j.1460-2075.1985.tb03662.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fry D. C., Kuby S. A., Mildvan A. S. ATP-binding site of adenylate kinase: mechanistic implications of its homology with ras-encoded p21, F1-ATPase, and other nucleotide-binding proteins. Proc Natl Acad Sci U S A. 1986 Feb;83(4):907–911. doi: 10.1073/pnas.83.4.907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirschman J., Wong P. K., Sei K., Keener J., Kustu S. Products of nitrogen regulatory genes ntrA and ntrC of enteric bacteria activate glnA transcription in vitro: evidence that the ntrA product is a sigma factor. Proc Natl Acad Sci U S A. 1985 Nov;82(22):7525–7529. doi: 10.1073/pnas.82.22.7525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huala E., Ausubel F. M. The central domain of Rhizobium meliloti NifA is sufficient to activate transcription from the R. meliloti nifH promoter. J Bacteriol. 1989 Jun;171(6):3354–3365. doi: 10.1128/jb.171.6.3354-3365.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunt T. P., Magasanik B. Transcription of glnA by purified Escherichia coli components: core RNA polymerase and the products of glnF, glnG, and glnL. Proc Natl Acad Sci U S A. 1985 Dec;82(24):8453–8457. doi: 10.1073/pnas.82.24.8453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Husain I., Van Houten B., Thomas D. C., Sancar A. Sequences of Escherichia coli uvrA gene and protein reveal two potential ATP binding sites. J Biol Chem. 1986 Apr 15;261(11):4895–4901. [PubMed] [Google Scholar]
- Keener J., Kustu S. Protein kinase and phosphoprotein phosphatase activities of nitrogen regulatory proteins NTRB and NTRC of enteric bacteria: roles of the conserved amino-terminal domain of NTRC. Proc Natl Acad Sci U S A. 1988 Jul;85(14):4976–4980. doi: 10.1073/pnas.85.14.4976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kustu S., Santero E., Keener J., Popham D., Weiss D. Expression of sigma 54 (ntrA)-dependent genes is probably united by a common mechanism. Microbiol Rev. 1989 Sep;53(3):367–376. doi: 10.1128/mr.53.3.367-376.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LOWRY O. H., ROSEBROUGH N. J., FARR A. L., RANDALL R. J. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951 Nov;193(1):265–275. [PubMed] [Google Scholar]
- Lowry D. F., Cool R. H., Redfield A. G., Parmeggiani A. NMR study of the phosphate-binding elements of Escherichia coli elongation factor Tu catalytic domain. Biochemistry. 1991 Nov 12;30(45):10872–10877. doi: 10.1021/bi00109a010. [DOI] [PubMed] [Google Scholar]
- Magasanik B. Reversible phosphorylation of an enhancer binding protein regulates the transcription of bacterial nitrogen utilization genes. Trends Biochem Sci. 1988 Dec;13(12):475–479. doi: 10.1016/0968-0004(88)90234-4. [DOI] [PubMed] [Google Scholar]
- Minchin S. D., Austin S., Dixon R. A. The role of activator binding sites in transcriptional control of the divergently transcribed nifF and nifLA promoters from Klebsiella pneumoniae. Mol Microbiol. 1988 Jul;2(4):433–442. doi: 10.1111/j.1365-2958.1988.tb00049.x. [DOI] [PubMed] [Google Scholar]
- Ninfa A. J., Magasanik B. Covalent modification of the glnG product, NRI, by the glnL product, NRII, regulates the transcription of the glnALG operon in Escherichia coli. Proc Natl Acad Sci U S A. 1986 Aug;83(16):5909–5913. doi: 10.1073/pnas.83.16.5909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ninfa A. J., Reitzer L. J., Magasanik B. Initiation of transcription at the bacterial glnAp2 promoter by purified E. coli components is facilitated by enhancers. Cell. 1987 Sep 25;50(7):1039–1046. doi: 10.1016/0092-8674(87)90170-x. [DOI] [PubMed] [Google Scholar]
- Parker R. C., Watson R. M., Vinograd J. Mapping of closed circular DNAs by cleavage with restriction endonucleases and calibration by agarose gel electrophoresis. Proc Natl Acad Sci U S A. 1977 Mar;74(3):851–855. doi: 10.1073/pnas.74.3.851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popham D. L., Szeto D., Keener J., Kustu S. Function of a bacterial activator protein that binds to transcriptional enhancers. Science. 1989 Feb 3;243(4891):629–635. doi: 10.1126/science.2563595. [DOI] [PubMed] [Google Scholar]
- Redfield A. G., Papastavros M. Z. NMR study of the phosphoryl binding loop in purine nucleotide proteins: evidence for strong hydrogen bonding in human N-ras p21. Biochemistry. 1990 Apr 10;29(14):3509–3514. doi: 10.1021/bi00466a013. [DOI] [PubMed] [Google Scholar]
- Reitzer L. J., Magasanik B. Expression of glnA in Escherichia coli is regulated at tandem promoters. Proc Natl Acad Sci U S A. 1985 Apr;82(7):1979–1983. doi: 10.1073/pnas.82.7.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reitzer L. J., Magasanik B. Transcription of glnA in E. coli is stimulated by activator bound to sites far from the promoter. Cell. 1986 Jun 20;45(6):785–792. doi: 10.1016/0092-8674(86)90553-2. [DOI] [PubMed] [Google Scholar]
- Reitzer L. J., Movsas B., Magasanik B. Activation of glnA transcription by nitrogen regulator I (NRI)-phosphate in Escherichia coli: evidence for a long-range physical interaction between NRI-phosphate and RNA polymerase. J Bacteriol. 1989 Oct;171(10):5512–5522. doi: 10.1128/jb.171.10.5512-5522.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richet E., Raibaud O. Purification and properties of the MalT protein, the transcription activator of the Escherichia coli maltose regulon. J Biol Chem. 1987 Sep 15;262(26):12647–12653. [PubMed] [Google Scholar]
- Ronson C. W., Astwood P. M., Nixon B. T., Ausubel F. M. Deduced products of C4-dicarboxylate transport regulatory genes of Rhizobium leguminosarum are homologous to nitrogen regulatory gene products. Nucleic Acids Res. 1987 Oct 12;15(19):7921–7934. doi: 10.1093/nar/15.19.7921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothstein D. M., Pahel G., Tyler B., Magasanik B. Regulation of expression from the glnA promoter of Escherichia coli in the absence of glutamine synthetase. Proc Natl Acad Sci U S A. 1980 Dec;77(12):7372–7376. doi: 10.1073/pnas.77.12.7372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders D. A., Gillece-Castro B. L., Burlingame A. L., Koshland D. E., Jr Phosphorylation site of NtrC, a protein phosphatase whose covalent intermediate activates transcription. J Bacteriol. 1992 Aug;174(15):5117–5122. doi: 10.1128/jb.174.15.5117-5122.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasse-Dwight S., Gralla J. D. Probing the Escherichia coli glnALG upstream activation mechanism in vivo. Proc Natl Acad Sci U S A. 1988 Dec;85(23):8934–8938. doi: 10.1073/pnas.85.23.8934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider B. L., Shiau S. P., Reitzer L. J. Role of multiple environmental stimuli in control of transcription from a nitrogen-regulated promoter in Escherichia coli with weak or no activator-binding sites. J Bacteriol. 1991 Oct;173(20):6355–6363. doi: 10.1128/jb.173.20.6355-6363.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiau S. P., Schneider B. L., Gu W., Reitzer L. J. Role of nitrogen regulator I (NtrC), the transcriptional activator of glnA in enteric bacteria, in reducing expression of glnA during nitrogen-limited growth. J Bacteriol. 1992 Jan;174(1):179–185. doi: 10.1128/jb.174.1.179-185.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stock J. B., Ninfa A. J., Stock A. M. Protein phosphorylation and regulation of adaptive responses in bacteria. Microbiol Rev. 1989 Dec;53(4):450–490. doi: 10.1128/mr.53.4.450-490.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su W., Porter S., Kustu S., Echols H. DNA-looping and enhancer activity: association between DNA-bound NtrC activator and RNA polymerase at the bacterial glnA promoter. Proc Natl Acad Sci U S A. 1990 Jul;87(14):5504–5508. doi: 10.1073/pnas.87.14.5504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wedel A., Weiss D. S., Popham D., Dröge P., Kustu S. A bacterial enhancer functions to tether a transcriptional activator near a promoter. Science. 1990 Apr 27;248(4954):486–490. doi: 10.1126/science.1970441. [DOI] [PubMed] [Google Scholar]
- Weiss D. S., Batut J., Klose K. E., Keener J., Kustu S. The phosphorylated form of the enhancer-binding protein NTRC has an ATPase activity that is essential for activation of transcription. Cell. 1991 Oct 4;67(1):155–167. doi: 10.1016/0092-8674(91)90579-n. [DOI] [PubMed] [Google Scholar]
- Weiss V., Claverie-Martin F., Magasanik B. Phosphorylation of nitrogen regulator I of Escherichia coli induces strong cooperative binding to DNA essential for activation of transcription. Proc Natl Acad Sci U S A. 1992 Jun 1;89(11):5088–5092. doi: 10.1073/pnas.89.11.5088. [DOI] [PMC free article] [PubMed] [Google Scholar]



