Abstract
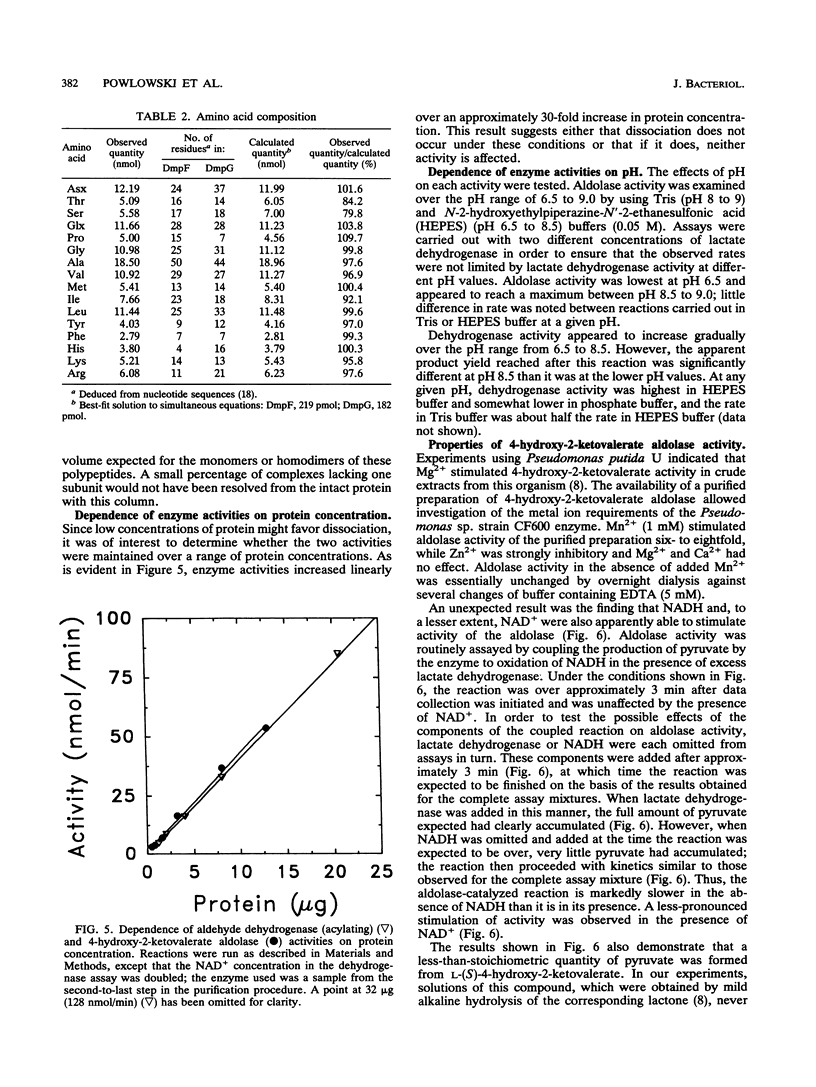
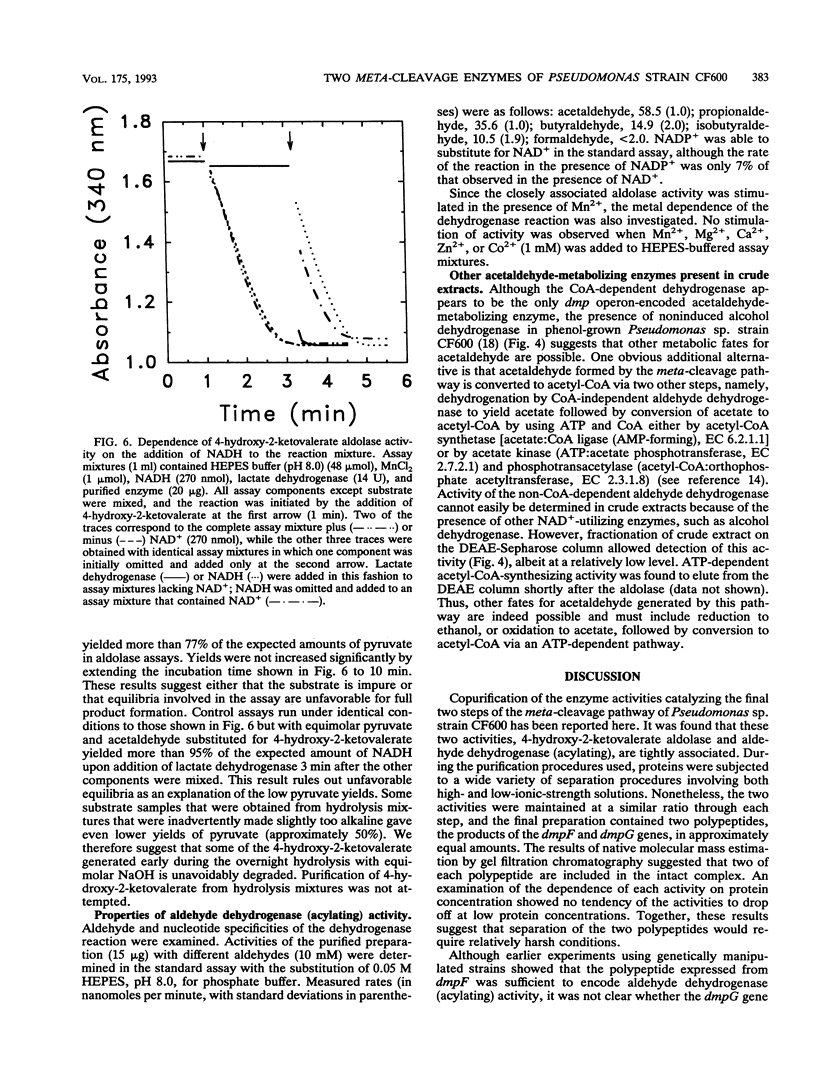
The final two steps in the dmp operon-encoded meta-cleavage pathway for phenol degradation in Pseudomonas sp. strain CF600 involve conversion of 4-hydroxy-2-ketovalerate to pyruvate and acetyl coenzyme A (acetyl-CoA) by the enzymes 4-hydroxy-2-ketovalerate aldolase and aldehyde dehydrogenase (acylating) [acetaldehyde:NAD+ oxidoreductase (CoA acetylating), EC 1.2.1.10]. A procedure for purifying these two enzyme activities to homogeneity is reported here. The two activities were found to copurify through five different chromatography steps and ammonium sulfate fractionation, resulting in a preparation that contained approximately equal proportions of two polypeptides with molecular masses of 35 and 40 kDa. Amino-terminal sequencing revealed that the first six amino acids of each polypeptide were those deduced from the previously determined nucleotide sequences of the corresponding dmp operon-encoded genes. The isolated complex had a native molecular mass of 148 kDa, which is consistent with the presence of two of each polypeptide per complex. In addition to generating acetyl-CoA from acetaldehyde, CoA, and NAD+, the dehydrogenase was shown to acylate propionaldehyde, which would be generated by action of the meta-cleavage pathway enzymes on the substrates 3,4-dimethylcatechol and 4-methylcatechol. 4-Hydroxy-2-ketovalerate aldolase activity was stimulated by the addition of Mn2+ and, surprisingly, NADH to assay mixtures. The possible significance of the close physical association between these two polypeptides in ensuring efficient metabolism of the short-chain aldehyde generated by this pathway is discussed.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Assinder S. J., Williams P. A. The TOL plasmids: determinants of the catabolism of toluene and the xylenes. Adv Microb Physiol. 1990;31:1–69. doi: 10.1016/s0065-2911(08)60119-8. [DOI] [PubMed] [Google Scholar]
- BURTON R. M., STADTMAN E. R. The oxidation of acetaldehyde to acetyl coenzyme A. J Biol Chem. 1953 Jun;202(2):873–890. [PubMed] [Google Scholar]
- Brown R. E., Jarvis K. L., Hyland K. J. Protein measurement using bicinchoninic acid: elimination of interfering substances. Anal Biochem. 1989 Jul;180(1):136–139. doi: 10.1016/0003-2697(89)90101-2. [DOI] [PubMed] [Google Scholar]
- Burlingame R., Chapman P. J. Catabolism of phenylpropionic acid and its 3-hydroxy derivative by Escherichia coli. J Bacteriol. 1983 Jul;155(1):113–121. doi: 10.1128/jb.155.1.113-121.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collinsworth W. L., Chapman P. J., Dagley S. Stereospecific enzymes in the degradation of aromatic compounds by pseudomonas putida. J Bacteriol. 1973 Feb;113(2):922–931. doi: 10.1128/jb.113.2.922-931.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corkey B. E., Brandt M., Williams R. J., Williamson J. R. Assay of short-chain acyl coenzyme A intermediates in tissue extracts by high-pressure liquid chromatography. Anal Biochem. 1981 Nov 15;118(1):30–41. doi: 10.1016/0003-2697(81)90152-4. [DOI] [PubMed] [Google Scholar]
- DAGLEY S., GIBSON D. T. THE BACTERIAL DEGRADATION OF CATECHOL. Biochem J. 1965 May;95:466–474. doi: 10.1042/bj0950466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson D. T., Wang K. C., Sih C. J., Whitlock H., Jr Mechanisms of steroid oxidation by microorganisms. IX. On the mechanism of ring A cleavage in the degradation of 9,10-seco steroids by microorganisms. J Biol Chem. 1966 Feb 10;241(3):551–559. [PubMed] [Google Scholar]
- Harayama S., Rekik M., Ngai K. L., Ornston L. N. Physically associated enzymes produce and metabolize 2-hydroxy-2,4-dienoate, a chemically unstable intermediate formed in catechol metabolism via meta cleavage in Pseudomonas putida. J Bacteriol. 1989 Nov;171(11):6251–6258. doi: 10.1128/jb.171.11.6251-6258.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Matsudaira P. Sequence from picomole quantities of proteins electroblotted onto polyvinylidene difluoride membranes. J Biol Chem. 1987 Jul 25;262(21):10035–10038. [PubMed] [Google Scholar]
- Powlowski J., Shingler V. In vitro analysis of polypeptide requirements of multicomponent phenol hydroxylase from Pseudomonas sp. strain CF600. J Bacteriol. 1990 Dec;172(12):6834–6840. doi: 10.1128/jb.172.12.6834-6840.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudolph F. B., Purich D. L., Fromm H. J. Coenzyme A-linked aldehyde dehydrogenase from Escherichia coli. I. Partial purification, properties, and kinetic studies of the enzyme. J Biol Chem. 1968 Nov 10;243(21):5539–5545. [PubMed] [Google Scholar]
- Sala-Trepat J. M., Evans W. C. The meta cleavage of catechol by Azotobacter species. 4-Oxalocrotonate pathway. Eur J Biochem. 1971 Jun 11;20(3):400–413. doi: 10.1111/j.1432-1033.1971.tb01406.x. [DOI] [PubMed] [Google Scholar]
- Shingler V., Powlowski J., Marklund U. Nucleotide sequence and functional analysis of the complete phenol/3,4-dimethylphenol catabolic pathway of Pseudomonas sp. strain CF600. J Bacteriol. 1992 Feb;174(3):711–724. doi: 10.1128/jb.174.3.711-724.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wierenga R. K., Terpstra P., Hol W. G. Prediction of the occurrence of the ADP-binding beta alpha beta-fold in proteins, using an amino acid sequence fingerprint. J Mol Biol. 1986 Jan 5;187(1):101–107. doi: 10.1016/0022-2836(86)90409-2. [DOI] [PubMed] [Google Scholar]