Abstract
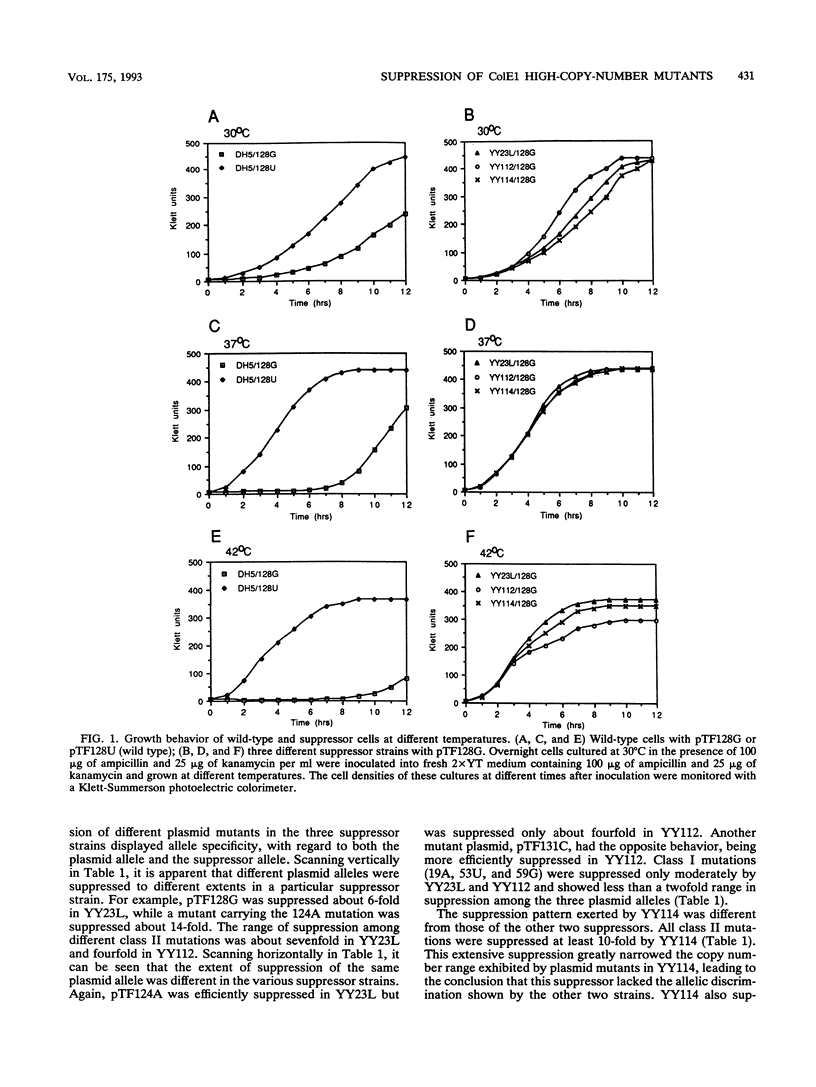
We isolated three Escherichia coli suppressor strains that reduce the copy number of a mutant ColE1 high-copy-number plasmid. These mutations lower the copy number of the mutant plasmid in vivo up to 15-fold; the wild-type plasmid copy number is reduced by two- to threefold. The suppressor strains do not affect the copy numbers of non-ColE1-type plasmids tested, suggesting that their effects are specific for ColE1-type plasmids. Two of the suppressor strains show ColE1 allele-specific suppression; i.e., certain plasmid copy number mutations are suppressed more efficiently than others, suggesting specificity in the interaction between the suppressor gene product and plasmid replication component(s). All of the mutations were genetically mapped to the chromosomal polA gene, which encodes DNA polymerase I. The suppressor mutational changes were identified by DNA sequencing and found to alter single nucleotides in the region encoding the Klenow fragment of DNA polymerase I. Two mutations map in the DNA-binding cleft of the polymerase region and are suggested to affect specific interactions of the enzyme with the replication primer RNA encoded by the plasmid. The third suppressor alters a residue in the 3'-5' exonuclease domain of the enzyme. Implications for the interaction of DNA polymerase I with the ColE1 primer RNA are discussed.
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bachmann B. J. Linkage map of Escherichia coli K-12, edition 8. Microbiol Rev. 1990 Jun;54(2):130–197. doi: 10.1128/mr.54.2.130-197.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coll M., Frederick C. A., Wang A. H., Rich A. A bifurcated hydrogen-bonded conformation in the d(A.T) base pairs of the DNA dodecamer d(CGCAAATTTGCG) and its complex with distamycin. Proc Natl Acad Sci U S A. 1987 Dec;84(23):8385–8389. doi: 10.1073/pnas.84.23.8385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper P. K., Hanawalt P. C. Role of DNA polymerase I and the rec system in excision-repair in Escherichia coli. Proc Natl Acad Sci U S A. 1972 May;69(5):1156–1160. doi: 10.1073/pnas.69.5.1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davison J., Heusterspreute M., Chevalier N., Ha-Thi V., Brunel F. Vectors with restriction site banks. V. pJRD215, a wide-host-range cosmid vector with multiple cloning sites. Gene. 1987;51(2-3):275–280. doi: 10.1016/0378-1119(87)90316-7. [DOI] [PubMed] [Google Scholar]
- De Lucia P., Cairns J. Isolation of an E. coli strain with a mutation affecting DNA polymerase. Nature. 1969 Dec 20;224(5225):1164–1166. doi: 10.1038/2241164a0. [DOI] [PubMed] [Google Scholar]
- Delarue M., Poch O., Tordo N., Moras D., Argos P. An attempt to unify the structure of polymerases. Protein Eng. 1990 May;3(6):461–467. doi: 10.1093/protein/3.6.461. [DOI] [PubMed] [Google Scholar]
- Dooley T. P., Tamm J., Polisky B. Isolation and characterization of mutants affecting functional domains of ColE1 RNAI. J Mol Biol. 1985 Nov 5;186(1):87–96. doi: 10.1016/0022-2836(85)90259-1. [DOI] [PubMed] [Google Scholar]
- Fitzwater T., Yang Y. L., Zhang X. Y., Polisky B. Mutations affecting RNA-DNA hybrid formation of the ColE1 replication primer RNA. Restoration of RNA I sensitivity to a copy-number mutant by second-site mutations. J Mol Biol. 1992 Aug 20;226(4):997–1008. doi: 10.1016/0022-2836(92)91048-t. [DOI] [PubMed] [Google Scholar]
- Fitzwater T., Zhang X. Y., Elble R., Polisky B. Conditional high copy number ColE1 mutants: resistance to RNA1 inhibition in vivo and in vitro. EMBO J. 1988 Oct;7(10):3289–3297. doi: 10.1002/j.1460-2075.1988.tb03196.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horii T., Ogawa T., Ogawa H. Organization of the recA gene of Escherichia coli. Proc Natl Acad Sci U S A. 1980 Jan;77(1):313–317. doi: 10.1073/pnas.77.1.313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh T., Tomizawa J. Formation of an RNA primer for initiation of replication of ColE1 DNA by ribonuclease H. Proc Natl Acad Sci U S A. 1980 May;77(5):2450–2454. doi: 10.1073/pnas.77.5.2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh T., Tomizawa J. Initiation of replication of plasmid ColE1 DNA by RNA polymerase, ribonuclease H, and DNA polymerase I. Cold Spring Harb Symp Quant Biol. 1979;43(Pt 1):409–417. doi: 10.1101/sqb.1979.043.01.047. [DOI] [PubMed] [Google Scholar]
- Joyce C. M., Fujii D. M., Laks H. S., Hughes C. M., Grindley N. D. Genetic mapping and DNA sequence analysis of mutations in the polA gene of Escherichia coli. J Mol Biol. 1985 Nov 20;186(2):283–293. doi: 10.1016/0022-2836(85)90105-6. [DOI] [PubMed] [Google Scholar]
- Joyce C. M., Grindley N. D. Method for determining whether a gene of Escherichia coli is essential: application to the polA gene. J Bacteriol. 1984 May;158(2):636–643. doi: 10.1128/jb.158.2.636-643.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joyce C. M., Kelley W. S., Grindley N. D. Nucleotide sequence of the Escherichia coli polA gene and primary structure of DNA polymerase I. J Biol Chem. 1982 Feb 25;257(4):1958–1964. [PubMed] [Google Scholar]
- Kelley W. S., Joyce C. M. Genetic characterization of early amber mutations in the Escherichia coli polA gene and purification of the amber peptides. J Mol Biol. 1983 Mar 15;164(4):529–560. doi: 10.1016/0022-2836(83)90049-9. [DOI] [PubMed] [Google Scholar]
- Kingsbury D. T., Helinski D. R. Temperature-sensitive mutants for the replication of plasmids in Escherichia coli: requirement for deoxyribonucleic acid polymerase I in the replication of the plasmid ColE 1 . J Bacteriol. 1973 Jun;114(3):1116–1124. doi: 10.1128/jb.114.3.1116-1124.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacatena R. M., Cesareni G. Base pairing of RNA I with its complementary sequence in the primer precursor inhibits ColE1 replication. Nature. 1981 Dec 17;294(5842):623–626. doi: 10.1038/294623a0. [DOI] [PubMed] [Google Scholar]
- Lerner C. G., Inouye M. Low copy number plasmids for regulated low-level expression of cloned genes in Escherichia coli with blue/white insert screening capability. Nucleic Acids Res. 1990 Aug 11;18(15):4631–4631. doi: 10.1093/nar/18.15.4631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J. D., Parkinson J. S. Genetics and sequence analysis of the pcnB locus, an Escherichia coli gene involved in plasmid copy number control. J Bacteriol. 1989 Mar;171(3):1254–1261. doi: 10.1128/jb.171.3.1254-1261.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopilato J., Bortner S., Beckwith J. Mutations in a new chromosomal gene of Escherichia coli K-12, pcnB, reduce plasmid copy number of pBR322 and its derivatives. Mol Gen Genet. 1986 Nov;205(2):285–290. doi: 10.1007/BF00430440. [DOI] [PubMed] [Google Scholar]
- March J. B., Colloms M. D., Hart-Davis D., Oliver I. R., Masters M. Cloning and characterization of an Escherichia coli gene, pcnB, affecting plasmid copy number. Mol Microbiol. 1989 Jul;3(7):903–910. doi: 10.1111/j.1365-2958.1989.tb00239.x. [DOI] [PubMed] [Google Scholar]
- Masters M., March J. B., Oliver I. R., Collins J. F. A possible role for the pcnB gene product of Escherichia coli in modulating RNA: RNA interactions. Mol Gen Genet. 1990 Jan;220(2):341–344. doi: 10.1007/BF00260507. [DOI] [PubMed] [Google Scholar]
- Masukata H., Tomizawa J. A mechanism of formation of a persistent hybrid between elongating RNA and template DNA. Cell. 1990 Jul 27;62(2):331–338. doi: 10.1016/0092-8674(90)90370-t. [DOI] [PubMed] [Google Scholar]
- Masukata H., Tomizawa J. Control of primer formation for ColE1 plasmid replication: conformational change of the primer transcript. Cell. 1986 Jan 17;44(1):125–136. doi: 10.1016/0092-8674(86)90491-5. [DOI] [PubMed] [Google Scholar]
- Masukata H., Tomizawa J. Effects of point mutations on formation and structure of the RNA primer for ColE1 DNA replication. Cell. 1984 Feb;36(2):513–522. doi: 10.1016/0092-8674(84)90244-7. [DOI] [PubMed] [Google Scholar]
- Matson S. W., Capaldo-Kimball F. N., Bambara R. A. On the processive mechanism of Escherichia coli DNA Polymerase I. The polA5 mutation. J Biol Chem. 1978 Nov 10;253(21):7851–7856. [PubMed] [Google Scholar]
- Naito S., Kitani T., Ogawa T., Okazaki T., Uchida H. Escherichia coli mutants suppressing replication-defective mutations of the ColE1 plasmid. Proc Natl Acad Sci U S A. 1984 Jan;81(2):550–554. doi: 10.1073/pnas.81.2.550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ollis D. L., Brick P., Hamlin R., Xuong N. G., Steitz T. A. Structure of large fragment of Escherichia coli DNA polymerase I complexed with dTMP. 1985 Feb 28-Mar 6Nature. 313(6005):762–766. doi: 10.1038/313762a0. [DOI] [PubMed] [Google Scholar]
- Polesky A. H., Dahlberg M. E., Benkovic S. J., Grindley N. D., Joyce C. M. Side chains involved in catalysis of the polymerase reaction of DNA polymerase I from Escherichia coli. J Biol Chem. 1992 Apr 25;267(12):8417–8428. [PubMed] [Google Scholar]
- Polesky A. H., Steitz T. A., Grindley N. D., Joyce C. M. Identification of residues critical for the polymerase activity of the Klenow fragment of DNA polymerase I from Escherichia coli. J Biol Chem. 1990 Aug 25;265(24):14579–14591. [PubMed] [Google Scholar]
- Polisky B., Zhang X. Y., Fitzwater T. Mutations affecting primer RNA interaction with the replication repressor RNA I in plasmid CoIE1: potential RNA folding pathway mutants. EMBO J. 1990 Jan;9(1):295–304. doi: 10.1002/j.1460-2075.1990.tb08108.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhodes D., Klug A. Sequence-dependent helical periodicity of DNA. Nature. 1981 Jul 23;292(5821):378–380. doi: 10.1038/292378a0. [DOI] [PubMed] [Google Scholar]
- Selzer G., Tomizawa J. I. Specific cleavage of the p15A primer precursor by ribonuclease H at the origin of DNA replication. Proc Natl Acad Sci U S A. 1982 Dec;79(23):7082–7086. doi: 10.1073/pnas.79.23.7082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singer M., Baker T. A., Schnitzler G., Deischel S. M., Goel M., Dove W., Jaacks K. J., Grossman A. D., Erickson J. W., Gross C. A. A collection of strains containing genetically linked alternating antibiotic resistance elements for genetic mapping of Escherichia coli. Microbiol Rev. 1989 Mar;53(1):1–24. doi: 10.1128/mr.53.1.1-24.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamm J., Polisky B. Characterization of the ColE1 primer-RNA1 complex: analysis of a domain of ColE1 RNA1 necessary for its interaction with primer RNA. Proc Natl Acad Sci U S A. 1985 Apr;82(8):2257–2261. doi: 10.1073/pnas.82.8.2257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomizawa J. I., Itoh T. The importance of RNA secondary structure in CoIE1 primer formation. Cell. 1982 Dec;31(3 Pt 2):575–583. doi: 10.1016/0092-8674(82)90313-0. [DOI] [PubMed] [Google Scholar]
- Tomizawa J. Control of ColE1 plasmid replication: initial interaction of RNA I and the primer transcript is reversible. Cell. 1985 Mar;40(3):527–535. doi: 10.1016/0092-8674(85)90201-6. [DOI] [PubMed] [Google Scholar]
- Tomizawa J., Itoh T. Plasmid ColE1 incompatibility determined by interaction of RNA I with primer transcript. Proc Natl Acad Sci U S A. 1981 Oct;78(10):6096–6100. doi: 10.1073/pnas.78.10.6096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomizawa J., Itoh T., Selzer G., Som T. Inhibition of ColE1 RNA primer formation by a plasmid-specified small RNA. Proc Natl Acad Sci U S A. 1981 Mar;78(3):1421–1425. doi: 10.1073/pnas.78.3.1421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong E. M., Muesing M. A., Polisky B. Temperature-sensitive copy number mutants of CoIE1 are located in an untranslated region of the plasmid genome. Proc Natl Acad Sci U S A. 1982 Jun;79(11):3570–3574. doi: 10.1073/pnas.79.11.3570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woolfson D. N., Williams D. H. The influence of proline residues on alpha-helical structure. FEBS Lett. 1990 Dec 17;277(1-2):185–188. doi: 10.1016/0014-5793(90)80839-b. [DOI] [PubMed] [Google Scholar]