Abstract
HIV-1 pandemic posed an unprecedent challenge to the global health and it is believed that an effective vaccine will be the final solution against HIV-1. HIV-1 envelope is the primary immunogen in developing neutralization antibody based HIV vaccine. To define the suitable Env derived immunogen, we systemically compared the immunogenicity of gp140 and gp145 in a DNA vaccination alone and a prime-boost modalities. 2 DNA vaccines and 2 recombinant Tiantan vaccinia vaccines (rTTV) were constructed for vaccination of female Balb/c mice. Elispot assay was used to read out the T cell immunity and ELISA assay was used to quantify antibody immunity. PLL (poly-L-Leucine)-ELISA assay was used in linear antibody epitope mapping. Mice primed with gp145 tended to elicit more Env-specific T cells responses than those primed with gp140, significant difference was observed in DNA immunization alone. The ultimate T cell responses in prime-boost regimen tends to be determined mainly by the priming efficacy. Linear antibody epitope mapping displayed that sera raised by gp145 priming were vigorously reactive to more peptides than that by gp140. Our data demonstrated HIV-1 Thailand B-derived gp145 may raise higher T-cell responses and broader linear peptide-specific antibody responses than gp140 does. However, it remains to be determined that how these observations are relevant to neutralization antibody activities.
Keywords: HIV-1, Vaccine, Envelope, Immunogen design
Introduction
The HIV/AIDS pandemic have globally affected millions of people, an effective vaccine remains elusive to date. Although effective antiretroviral therapy is available in the developed countries, it remains financially unaffordable for the most of HIV-1 infected individuals who largely reside in the developing countries. It is thus widely believed that an effective vaccine is the only solution to restrain the global HIV-1 epidemic.
Due to the great challenge to develop an effective vaccine able to induce neutralizing antibodies [1,2], a large effort to develop HIV-1 vaccine has switched to the induction of CD8+ cytotoxic T lymphocyte (CTL) responses against the conserved internal viral antigens, such as Gag and Pol [3,4,5]. Interestingly, the addition of the Env to Gag-Pol in vaccines dramatically increased the capacity of vaccinee to contain SHIV replication [6]. This was not resulted from the different magnitude of T-cell immune responses because both Gag-Pol-Env and Gag-Pol immunized groups mounted comparable T-cell immune responses against Gag and Pol; rather, the induction of additional T-cell immune responses against Env at least partially accounted for the improvement of viral control. Furthermore, the inclusion of Env accelerated the induction of neutralizing antibodies against the challenge virus by 2~3 weeks, which may also have significantly impacted on the viral replication in the Gag-Pol-Env immunized group [6]. In addition, since the neutralizing antibodies could protect target cells from HIV-1 infection, the vaccine effort to induce neutralizing antibodies continues. Env is the only immunogen able to induce neutralization antibodies, it is rationalized that Env is included in the vaccine design [7–14].
Previous study showed that the full-length gp160 of Env was cytotoxic for expressing cells and not suitable for the use as vaccine immunogen. Therefore, the concept to truncate gp160 to gp145 or gp140, which will remove the cytotoxic activity [15] while retaining its ability to induce appreciable antibody immune responses, has been developed. However, it remains unknown how to optimize Env immunogen in vaccine design and what is the modality of vaccination for best use of Env immunogen.
Here we employed a modality of DNA priming/recombinant Tiantan vaccinia boosting to systemically compare the immunogenicity of HIV-1 gp140 and gp145, meanwhile, we also compared modalities of homologous or heterologous priming and boosting. The recombinant Tiantan vaccinia in this study is replicative and derived from the vaccine strain which has been used for decades in billions of Chinese in the campaign to eradicate smallpox. Recently, this replicative vector was used in other vaccine development, such as EBV vaccine [16, 17] and its safety profile has been well documented both in the campaign and in the recent clinical trials [16–18].
Materials and Methods
DNA vaccine construction
Humanized gene sequences encoding gp145 of HIV-1rl42 (Thailand B) were synthesized as oligonucleotides and then assembled into the whole gene by the overlapping PCR [19]. Gp140 gene was acquired by using PCR amplication from the assembled gp145 gene by exclusion of the transmembrane domain. Both gene sequences were confirmed by sequencing double strands of sense and antisense gene DNAs and then cloned into the expression vector pDRVI-sv1.0 to generate two DNA vaccines, sv140rl and sv145rl. Plasmid vector pDRVI-sv1.0 was generated by inserting a 72-bp SV40-derived enhancer into the upstream of cytomegalovirus immediate-early promoter in pVRC2000 (kindly provided by Dr. Gary Nabel at Vaccine Research Center, NIAID, NIH).
Construction of recombinant Tiantan Vaccinia vectored vaccines
Gp140 and gp145 genes were transferred to pSC65 shuttle plasmid (with a Lac z gene as screening marker), which is designed to recombine specifically with TK gene of Tiantan vaccinia virus. Subconfluent monolayers of 143TK− cells were grown in Eagle’s media containing 10% fetus bovine serum and 1% Penicillin-Streptomycin-L-glutamine. Then, the cells were washed with Eagle’s media containing glutamine and antibiotics in the absence of fetus bovine serum. 5×106 pfu vaccinia virus per well were inoculated and incubated for 1 hour at 37°C and 0.5% CO2. Thereafter, the vaccinia infected cells were further transfected with recombinant shuttle plasmids with Lipofectamine 2000 (CAT#11668-019, Invitrogen). After 48 hour incubation, the transfection media were removed and all wells were covered with 2% melted low melting temperature (LMP) agarose mixed with equal volume 2×Eagle’s media containing 100μg/ml x-gal. The blue Lac Z positive colonies were picked up and further purified in 143TK− cells under the pressure of selection media (Eagle’s media containing 50μg/ml BuDR). The purified recombinant Tiantan vaccinias were confirmed by PCR amplification of inserted gp140 and gp145 and the generated vaccines were designated as rTTV140rl and rTTV145rl, respectively. All rTTVs were expanded in primary chicken embryo cells.
Expression of HIV immunogen by vaccines in vitro
Hela cells were plated in 6-wells plate. 8μg of each plamid of sv140rl or sv145rl, was used to transfect Hela cells. Cells were harvested 48 hours after transfection. Similarly, rTTVs infected primary chicken embryo cells were also harvested 48 hours after infection. All cells were lysed with SDS-PAGE loading buffer and cell lysates were used to determine the expression capacity with Western blotting. 1:100 diluted HIV-1 positive human serum was used as first antibody and 1:1500 diluted HRP-linked goat anti-human IgG(CAT# ZB-2304,Beijing Zhongshan Biotech) was used as second antibody in this assay. Though the Western blotting was carried out in parallel, the transfection and infection were performed separately for different vaccines, thus the cell numbers used for Western blotting may significantly vary. Therefore, this assay was designed to determine whether the immunogens can be properly expressed by vaccines, not the expression efficacy.
Animals and Vaccination
All animal experiments were reviewed and approved by the Institutional Animal Care and Use Committee (IACUC) at China CDC and were performed in accordance with relevant guidelines and regulations. Six-week-old female BALB/c mice were vaccinated as scheduled in Table 1. 100 μg of purified plasmid DNA suspended in 100μl PBS was inoculated intramuscularly into tibialis anterior three times at week 0, 2 and 4. Then at week 6, either 100μg plasmid DNA or a recombinant Tiantan vaccinia virus was injected intramuscularly into tibialis anterior for boosting. The mice were sacrificed 3 weeks after the last inoculation. Sera were collected and stored at 4°C for future quantification of antibodies, and splenocytes were freshly collected for Elispot assay.
Table 1.
Female Balb/c mice vaccination schedule
| Vaccination and sacrifice schedule | ||||||
|---|---|---|---|---|---|---|
| Group | No. of Mice | Week 0 | Week 2 | Week 4 | Week 6 | Week 9 |
| DNA vaccination (100μg/mouse) | ||||||
| A | 5 | sv140rl | sv140rl | Sv140rl | sv140rl | sacrificed |
| B | 5 | sv145rl | sv145rl | Sv145rl | sv145rl | sacrificed |
| C | 5 | sv1.0 | sv1.0 | sv1.0 | sv1.0 | sacrificed |
| DNA priming (100μg/mouse) | Vaccinia boosting (4×106pfu/mouse) | |||||
| D | 6 | sv1.0 | sv1.0 | sv1.0 | Wild type TTV | sacrificed |
| E | 7 | sv140rl | sv140rl | Sv140rl | rTTV140rl | sacrificed |
| F | 7 | sv140rl | sv140rl | Sv140rl | rTTV145rl | sacrificed |
| G | 6 | sv145rl | sv145rl | Sv145rl | rTTV140rl | sacrificed |
| H | 7 | sv145rl | sv145rl | Sv145rl | rTTV145rl | sacrificed |
Peptide pools
A set of peptides spanning the gp145 gene of consensus B clade (kindly provided by NIH Research and Reference Reagent Program, NIAIDS, NIH) were used as stimuli, the homogenicity between Env of HIV-1rl42 and of consensus B is 90%. Peptides (15 mers with overlapping by 11 amino acids with its next peptide) were used at a final concentration of 4μg/ml for each peptide. All peptides were synthesized as free acids and were more than 80% purity. Lyophilized peptides were reconstituted to 1mg/mL in 90% RPMI 1640 with 10% DMSO for the peptide mixture. 4 peptide pools were formulated, including Env 1 (the first 48 peptides), Env 2 (the second 48 peptides), Env 3 (the third 48 peptides), Env 4 (the forth 48 peptides), and used in Elispot assay.
Elispot assay
3 weeks after the final inoculation of DNA vaccine or rTTV vaccines, mice were sacrificed and splenocytes were harvested. IFN-γ Elispot kits (Cat# CT317-T20, U-Cytech Biosciences, the Netherlands) was used to determine vaccine-elicited IFN-γ responses in Balb/c mice. 96-well plates were coated with purified anti-mouse IFN-γ monoclonal antibodies at the concentration of 5μg/ml in 100μl/well, and incubated at 4□ overnight, then washed 4 times with 200μl/well PBST (PBS containing 0.05% Tween20), and blocked with 200μl/well block solution (supplied in the kit). Mice splenocytes were isolated and red blood cells (RBC) were lysed by RBC Lysis Buffer (139.6 mmol/L NH4Cl,16.96 mmol/LTris, pH 7.2). Cells were then washed 2 times with R10 (RPMI-1640 containing 10% fetus bovine serum and 1% Penicillin-Streptomycin-L-glutamine) and re-suspended in R10 complete culture medium. After counting, splenocytes were then adjusted to the concentration of 4×106 cells/ml and plated into pre-coated 96-well Elispot plate at 50 μl/well (2×105 cells per well) with addition of 50μl peptide pools, the final concentration for each peptide is 4μg/ml (the final concentration of DMSO is 0.04%). 96-well Elispot plates were incubated for 30 hours at 37□ and 5% CO2. After incubation, the Elispot plates were developed according to the kit instruction. Finally, plates were air-dried and the resulting spots were counted with Immunospot Reader (CTL Inc., Cleveland, OH). Peptide pool specific IFN-γ Elispot responses were considered as positive only when the responses were 4-fold above negative control with no peptide stimulation. Spot forming cells from 4 Env peptide pools were compiled together as the total Elispot result for each mouse.
Quantification of HIV-1rl42 envelope-specific binding antibody titers
96-well flat-bottom plates (Costar, NY) were coated at 4°C overnight with 100μl HIV-1rl42 gp120 (more than 80% purity, expressed in E.coli and resolved in sodium bicarbonate buffer (pH9.6) at a final concentration of 4μg/ml) and then washed twice with PBS, and blocked at 37°C for 1 hour with block solution (PBS containing 5% skimmed dry milk). Mice sera were serially 2-fold diluted in block solution, and 100μl diluted sera were added to each well. After incubating the plates at 37°C for 1 hour, the plates were then washed 5 times with PBS-T (PBS containing 0.05% Tween20), and 100 μl/well of peroxidase-conjugated anti-mouse immunoglobulin G (diluted 1:2000 in block solution) was added and incubated for 1 hour at 37°C. The wells were washed 5 times with PBS-T again, and 100 μl of OPD (Cat# P9187, Sigma Aldrich) substrate was added and incubated for approximately 10 minutes at room temperature. Color development was terminated by the addition of 50μl/well of 2 N sulfuric acid. Antibody reactivity was then determined by measuring the optical density (OD) at 492 nm with an automated plate reader (Multiscan Ascent, Thermo corporation, Finland). Endpoint titers were determined by the last dilution whose OD was >2 folds than that at the corresponding dilution of mice sera immunized with sv1.0/wild type TTV.
Linear antibody epitope mapping
The method of PLL-ELISA was employed in this study [20,21]. PLL (poly-L-leucine, 30–70 kDa, Sigma Aldrich) was dissolved at the concentration of 40 μg/ml in the sodium bicarbonate buffer, 50μl/well PLL solution was added to 96-well ELISA plate and incubated at room temperature for 1 hour. The plate was washed once with PBS, incubated with 50μl per well of 1% (v/v) glutaraldehyde (Sigma Aldrich) in PBS at room temperature for 15 minutes for activation of PLL and washed twice with PBS. Then 50 μl/well of testing peptides at the concentration of 10μg/ml in PBS was added to the wells pre-coated with PLL and activated with glutaraldehyde. The plate was sealed, incubated overnight at 4°C and then washed twice with PBS. Reactive aldehyde sites were blocked by the addition of 1 M glycine, 200μl/well, for 1 hour at room temperature. The plates were washed twice again with PBS and blocked with 1M glycine,100μl/well, for 1 hour at room temperature, then incubated with 100μl/well of 0.5% nonfat dry milk:0.5% gelatin (dissolved in PBS) for 1 hour at room temperature. Due to the limited volume of mice sera from each mouse, mice sera from the same group were combined together and diluted at the ratio of 1:100 in PBS containing 5% nonfat dry milk, 50μl/well diluted sera was added into plate in duplicates and incubated at 37°C for 1 hour. The plates were then washed 4 times with PBS-T, incubated with 50μl/well diluted HRP-linked second antibody (1:2000 diluted in PBS containing 2% nonfat dry milk, CAT# ZB-2305, Beijing Zhongshan Biotech) at 37°C for 1 hour, washed 5 times again with PBS-T, and finally incubated with 50μl/well OPD(Sigma Aldrich) substrate at room temperature in dark place for about 10 minutes. The reaction was terminated with 50μl/well 2N sulfuric acid, and the optical density (OD) for each well was read with an automated plate reader (Multiscan Ascent, Thermo corporation, Finland) at 492 nm. Results of experimental settings were displayed as original OD values and >2-fold OD values of mock vector control groups against the corresponding peptide were considered as positive.
Results
Construction and in vitro expression of DNA and rTTV vaccines
To compare the immunogenicity between HIV-1rl42 gp140 and gp145, we constructed 2 DNA vaccines, sv140rl and sv145rl, and 2 rTTV vaccines, rTTV140rl and rTTV145rl. All the constructs were confirmed by PCR (data not shown). In vitro expression of these vaccines were examined by Western blot (Fig. 1). As expected, all these vaccines expressed a protein banded between 130kDa and 170kDa whereas control plasmid or rTTV did not, indicating that inserted env genes are able to express corresponding Env. Since this assay was not designed to compare the expression efficacies of different vaccines, the transfection and infection were carried out separately for different vaccines and varied cell number may have been used to prepare cell lysate for Western blotting, thus it is expected we may observe difference of expression between gp140 and gp145 in Fig. 1.
Figure 1.
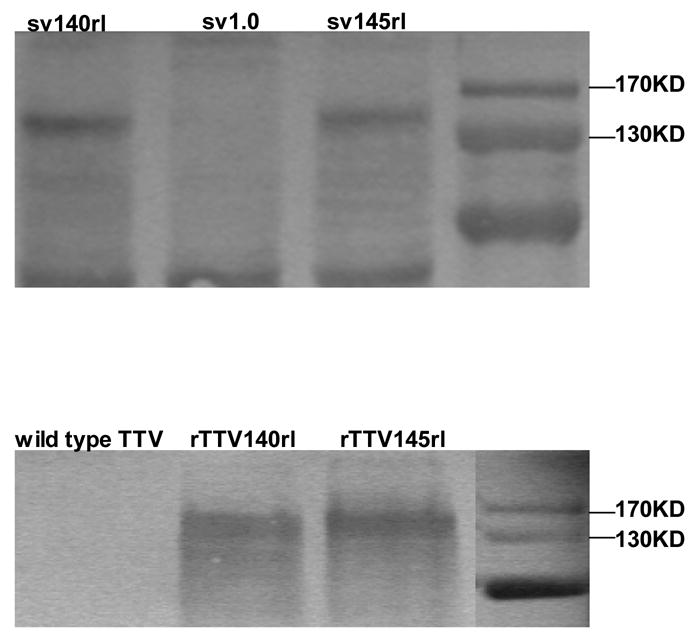
In vitro expressions of DNA vaccines encoding HIV-1rl42 gp140 and gp145 in Hela cells (upper panel) and of rTTVs in primary chicken embryo cells (bottom panel) were assayed by Western Blotting of cell lysate. Blank vector pSV1.0 and wild type Tiantan vaccinia virus was used as mock controls. Transfection and infection were carried out separately for different vaccines, cell numbers used for preparation of cell lysate were varied.
Env-specific T cell immune responses
To determine the immunogenicity, we next immunized mice with these vaccines as scheduled in Table 1. HIV-1rl42 Env-specific T cell responses were quantified with IFN-γ based ELISPOT assay after stimulation of splenocytes with HIV-1 consensus B gp145 peptide pools. All the results were subtracted by the background which is resulted from the stimulation with no peptide (<10 SFCs per million cells, supplementary figure 1). For each mouse, Env-specific T cell responses against all 4 peptide pools were compiled together as the total T-cell responses for each mouse and graphed into groups, and the significance of difference among different groups were calculated by using a statistical method of one way analysis of variance (Sigmastat3.1). Results demonstrated that all 4 groups boosted with rTTV (1554±678 SFCs/106 splenocytes for the group immunized with sv140rl plus rTTV140rl, 1678±579 SFCs/106 splenocytes for sv140rl plus rTTV145rl vaccination, 2053±786 SFCs/106 splenocytes for sv145rl plus rTTV140rl vaccination and 2068±635 SFCs/106 splenocytes for sv145rl plus rTTV145rl vaccination) based vaccine mounted higher Env-specific T-cell immune responses than that inoculated with DNA vaccine alone (433±112 SFCs/106 splenocytes for sv140rl vaccination and 1324±603 SFCs/106 splenocytes for sv145rl vaccination) (Fig. 2). Furthermore, no matter DNA vaccine immunization alone or DNA priming/rTTV boosting, priming with gp145 consistently tended to elicit more Env-specific T cells than that with gp140. However, statistical significances were only observed between the groups immunized with sv140rl DNA vaccine alone and all other 5 groups. The highest Env-specific T cell responses were reached in the group primed with sv145rl and boosted with rTTV145rl (Fig. 2).
Figure 2.
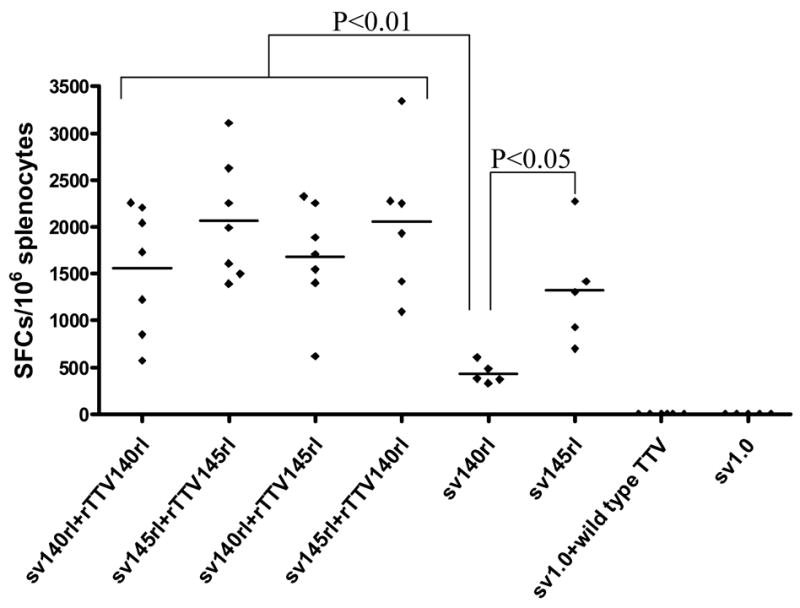
Env-specific T cell immune responses among different groups. Mice immunized with different regimens of vaccines (as scheduled in table 1) were sacrificed and splenocytes were harvested. Four peptide pools derived from consensus B clade Env were used as stimuli in Elispot assay, T-cell responses for each mouse were compiled from 4 peptide pool stimulations and then grouped in the figure. Positive Elispot control (stimulated with PMA and Inomycin) and negative control (stimulated with no peptides) were also included. Peptide pool specific IFN-γ Elispot responses were considered as positive only when the responses were 4-fold above negative control with no peptide stimulation. Statistical significances were only observed between gp140 and other 5 groups. Control groups immunized with mock vectors raise no T cell immune responses.
Env-specific binding antibody immune responses
We next examined binding antibody immune responses. HIV-1rl42 gp140- and gp145-specific binding antibodies were quantified by ELISA with bacterial expressed gp120 as the coating antigen. It was observed that the titers of Env-specific binding antibodies elicited by DNA vaccine sv140rl (ranged from 3200 to 12800 with a median of 6400) were higher than DNA vaccine sv145rl (ranged from 1200 to 6400 with a median of 3200). All the groups boosted with rTTVs (titers ranged from 600 to 38400 with a median of 19200 for group sv140rl plus rTTV140rl, 2400–38400 with a median of 9600 for group sv140rl plus rTTV 145rl, 400–19200 with a median of 8000 for group sv145rl plus rTTV140rl and 800–38400 with a median of 19200 for group sv145rl plus rTTV145rl) showed higher binding antibody titers than the groups inoculated with DNA vaccine alone. However, overall there were no statistical significant differences (using the method of one way analysis of variance) among the titers of 6 groups (Fig. 3).
Figure 3.
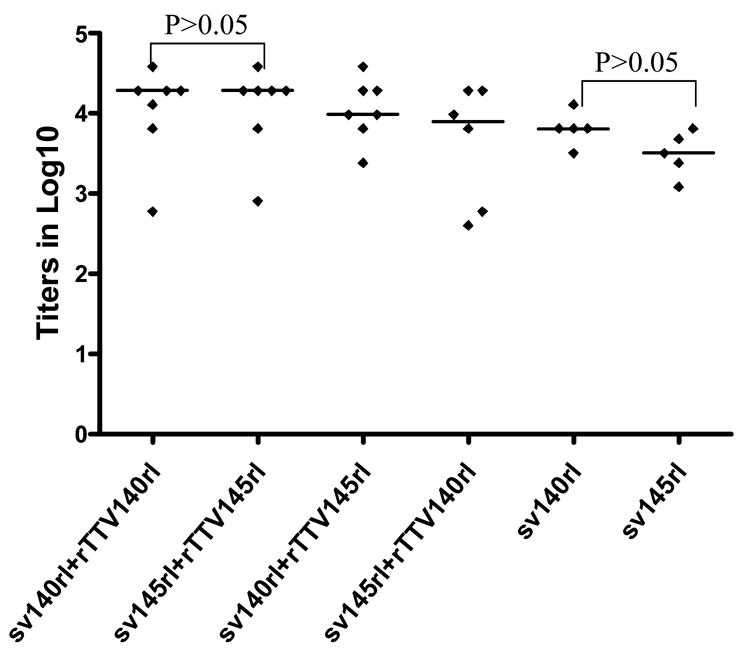
Gp120-specific binding antibodies among different groups. Mice immunized with different regimens of vaccines (as scheduled in table 1) were sacrificed and sera were collected. Gp120 expressed in E.coli was used in ELISA assay and endpoint titers were determined by the last dilution whose OD was >2 folds than that at the corresponding dilution of mice sera immunized with mock control group (sv1.0/wild type TTV). Control groups immunized with mock vectors raise no humoral responses. No significant differences were observed among all 6 groups.
Linear antibody epitope mapping
To further characterize the antibody responses, we performed assay to precisely map the linear antibody epitopes. OD values were used as the measurement of antibody responses against all linear peptides. Data showed that no significant humoral responses were detected in sera from vector control groups (sv1.0 alone or sv1.0 priming/wild type Tiantan vaccinia virus boosting) and the mean OD value of control groups was 0.2 (supplementary figure 2). Interestingly, we observed distinct responding patterns among different groups. Linear peptide epitopes with vigorous humoral responses were identified and clustered in several “hot regions” across the full amino acid sequence of HIV-1rl42 envelope, including p22~p27 of C1, p76/p77 of V3, p86/p87 and p91~94 of C3, p115~p119 and p121/p122 of C5 and p145~p147 between the 2 HR (heptad repeat) regions (Fig. 4). The intensities of responses against those regions varied among different groups, but, generally, responses in groups boosted with rTTVs mounted higher than that inoculated with DNA vaccine alone. Interestingly, sera raised by gp145 priming but not by gp140 were vigorously reactive to peptide p77, p86/p87, p91/p92/p93/p94 and p115/p116 whereas p76, p121/p122 were more reactive to sera raised by gp140. However, DNA inoculation alone may display different immunogenicity from that in the modality of priming and boosting. For example, gp145 DNA vaccine alone was incapable of whereas gp145 priming plus gp145 boosting are capable of raising vigorous responses to p77, p91/92 and to p117/p119, which implicated that rTTV may present different predominant epitopes from what presented by DNA vaccine. In addition, the order of gp140 and gp145 in the sequential inoculation may exert significant influences on the magnitude of peptide-specific response. Although gp140 plus gp145 and gp145 plus gp140 stimulated similar level of responses to peptide p22-p27, p76, p87, p118 and p145/p146, it was gp145 plus gp140 but not gp140plus gp145 stimulated significantly responses to peptide p77 and p86.
Figure 4.
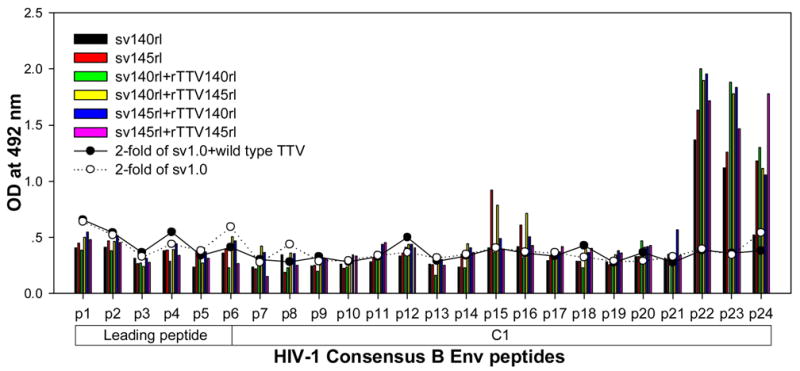
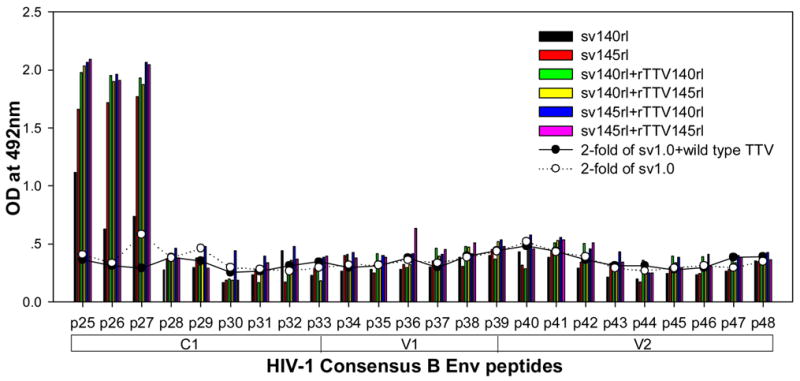
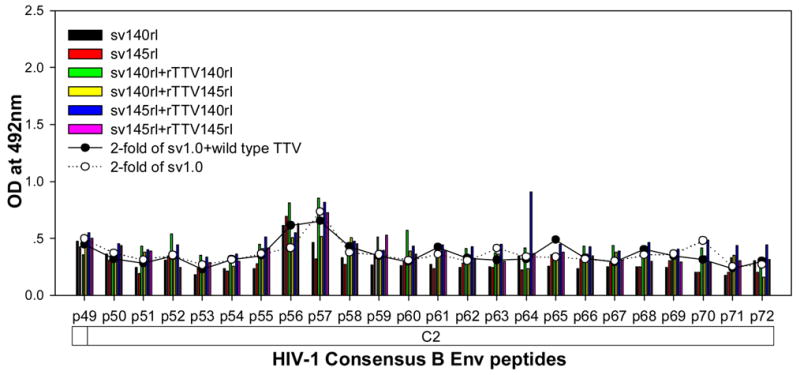
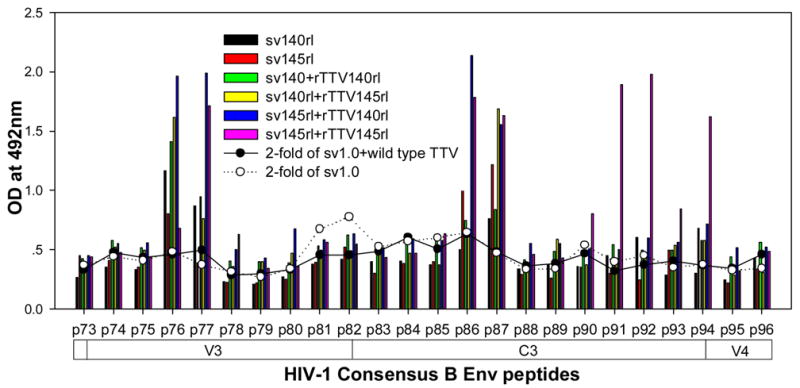
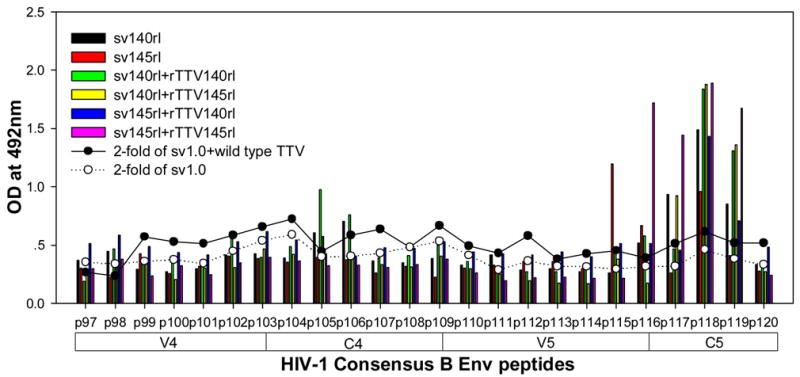
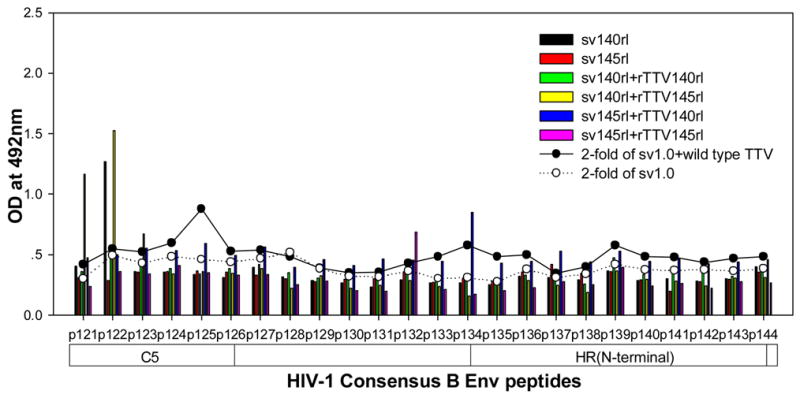
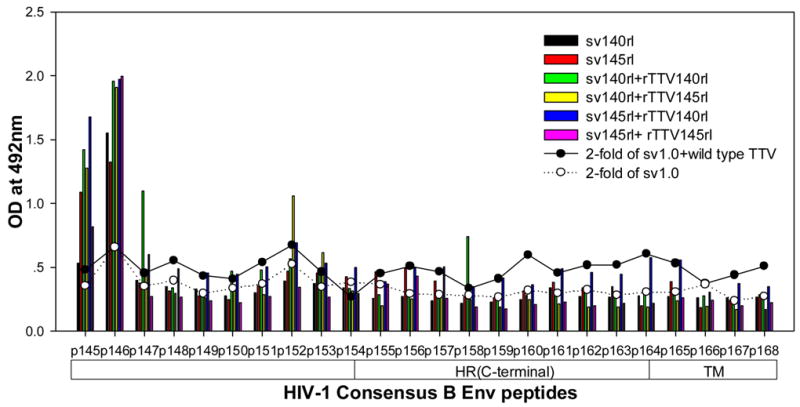
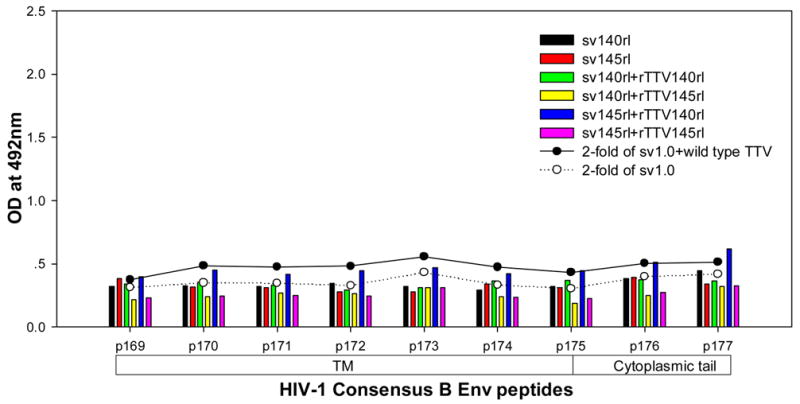
Results of linear antibody epitope mapping. For each group, the sera from mice in the same group were combined together in equal volume and mixed. The mixed serum was then assayed in duplicated wells. The averages of duplicated original OD values from vaccination groups were graphed into figures. The straight-scatter lines in the figures indicate the threshold value, which is 2-fold of the OD values from mock vector controls against each corresponding peptide(2-fold of sv1.0 is the cutoff value for groups immunized with DNA only; 2-fold of sv1.0+wild type TTV is the cutoff value for groups immunized DNA plus rTTV). Though vaccination stimulated no significant responses to the majority of peptides, several highly reactive epitopes were identified across the full amino acid sequences of HIV-1rl42 gp145, including p22~p27 of C1, P76/p77 of V3, p86/p87 of C3, p116~p119 of C5 and p145/146 between the 2 HR (heptad repeat) regions.
Discussion
Envelope glycoprotein is the primary target for HIV vaccine design. Although the diversity of Env protein poses a grand challenge for generating broad reactive neutralizing antibody responses and T-lymphocyte responses [22,23], Env remains critical in the induction of suppressive immunity in current AIDS vaccine candidates tested in non-human primates [24,25], and is included in the majority of HIV vaccines, especially in polyvalent vaccines and gp120 based protein vaccines [14].
However, clinical studies demonstrated that antibodies elicited by soluble gp120 have a narrow neutralization specificities, are unable to neutralize primary isolates [26–30] and thereby qualitatively different from those induced by HIV-1 infection [31, 32]. A critical deficiency of the gp120 vaccines is the absence of epitopes in the relative conservative protein of gp41 [33, 34], Therefore, it is rationalized that gp160 might be a better immunogen than gp120. However, previous study had shown that gp160 is cytotoxic and truncation from its C-terminal to less than 145 kDa could remove its cytotoxicity [15]. Thus, gp140 was thought to be a better vaccine immunogen, as it contains the entire ectodomain of HIV-1 Env and can be secreted from expressing cells. Several vaccine strategies attempted to elicit cross-reactive antibodies by incorporating an oligomeric/trimeric form of gp140 [35–40]. Immunogenicity comparison between gp120 and gp140 showed that sera raised by gp140 exhibited improved cross-reactivity to heterologous Env proteins and greater neutralizing activities compared to what to be raised by gp120 [41].
It remains unknown whether the immunogenicity of gp140 could be further enhanced by including more amino acids at its C-terminal. This study was designed to comprehensively compare the immunogenicity between a gp140 and a gp145 vaccine which added 30 aa at the C-terminal of gp140. Two DNA vaccines and two recombinant Tiantan vaccinia viruses were constructed for this study and the comparisons were performed with a DNA vaccination alone or a Prime-Boost regimen. A number of previous studies have focused on the ability of Env to elicit cross-reactive neutralizing antibodies [15,41–49], however, several primate experiments reminded us to pay attention to specific binding antibodies, which might also play important roles in vaccine induced protection [20, 21], and clinical data also identified HIV-specific antibodies but not T cell responses associated with protection [50]. In this study, we first compared the titers of gp120-specific binding antibodies between different groups. Although no significant difference were reached, the mean titer of mice immunized with gp140-encoding DNA vaccine was higher than that in mice vaccinated with gp145-encoding DNA vaccine, indicating that gp140 may be more efficient in elicting gp120-specific binding antibody responses than gp145 when used as DNA vaccine. However, since the gp120 protein used in the ELISA was expressed in bacteria and did not subject to be properly folded, this assay may underestimate the differences among different groups, which may explain why our results are different from what was observed in previous reports [47, 49].
In contrast to the responses of binding antibodies, the results of linear antibody epitope mapping showed distinct response patterns among the 6 groups (Fig. 4). Groups immunized with DNA+rTTV showed broader and more vigorous reactivity than groups immunized with DNA only, and it seemed that gp145 could raise antibodies recognizing more linear antibody epitopes and thereby mounted broader antibody responses than gp140. The difference was not due to gp145 has more peptide epitopes at the C-terminal than gp140 because those epitopes primed by gp145 were located within gp140. A more reasonable explanation might be that the different cell membrane anchoring ability of gp140 and gp145 account for this differences, which has been reported before [15]. Consensus B Env derived peptides were used in our assay, which is 90% homogeneous to the Env of HIV-1rl42. The epitopes with specific antibody binding activity in our experiments were mainly located at the high homolog regions: 13 peptides(p23~27/p90~93/p117~119/p121) have 100% homolog in amino acid sequence with HIV-1rl42; 6 peptides(p22/p86~87/p94/p116/p122) have one amino acid different from HIV-1rl42 and 3 peptides(p76~p77/p115) have more than two amino acids different from HIV-1rl42(information detailed in supplementary Fig. 3). Therefore, we may underestimate the breadth of humoral responses for HIV-1rl42 gp140 and gp145 vaccines at those regions with low homolog. However, this underestimation was not likely to be important for an effective vaccine because the non-cross-reactive epitopes will not be epitopes providing any significant protection for vaccinee.
Two observations should be noticed in our studies. Firstly, DNA vaccination alone may be unable to fully evaluate the immunogenicity of vaccines because we observed that DNA inoculation alone was unable to stimulate vigorous humoral responses to certain epitopes whereas the modality of priming and boosting was capable, thus a strong boost is necessary to overcome this deficiency. Secondly, we tested the modality of heterologous priming and boosting, we observed that the order of gp140 and gp145 in the sequential inoculation could exert significant influences on the magnitude and specificities of humoral responses. In our experiments, we observed that priming with gp145 and boosting with gp140 raised broader epitope-specific humoral immune responses than that by gp140 priming and gp145 boosting. Therefore, the modality of priming and boosting with heterologous immunogens and the order of inoculations may be highly relevant to the development of HIV vaccine [51]. However, it remains to be addressed the exact underlying mechanism for these observations and how these differences could influence neutralizing activities. In vitro neutralizing assay before and after the absorption of sera by peptides will be able to answer the latter question. Interestingly, based on the crystal structure of gp120 [52], linear peptide epitopes with vigorous humoral responses could be located at both the inner domain (p22~p27/p115~p119/p121/p122) and the outer domain(p76/p77/p86/p87/p91~94) of gp120, which indicates that not only the “outer” epitopes, but also the “inner” epitopes can elicit specific binding antibody response. Besides, the peptide 76 and 77 are located on the tip of V3 loop which partially overlaps with the neutralization epitope identified by the antibody 447-52D [53].
As known, Env-specific T-cell responses also play important role in suppressing HIV replication. One recent study indicated that T cell responses elicted by envelope contributed to vaccine induced protection from challenging viruses carrying heterologous envelope in rhesus monkeys [24]. Clinical data of our lab also showed that HIV-specific CD8 T cell responses targeting envelope are quite high in individuals with chronic HIV-I B’ infection (unpublished data). These results imply that Env-elicited T cell responses may play a role in fighting against HIV-1. In present study, HIV-specific T cell responses induced by gp140 and gp145 were also compared. Results showed that HIV-specific T cell responses of mice immunized with gp145 raised higher T cell responses than that with gp140, significant difference were only observed in the immunization with DNA vaccine alone. This observation is different from previous report in which gp140 elicited a stronger cellular responses than gp145 did [15]. The discrepancy may be explained by the fact that a functional cytotoxicity assay was employed in the previous report, which mainly define the cytotoxic T cells, whereas INF-gamma based Elispot assay was used in our study, which quantified IFN-gamma producing T cells. As known, IFN-gamma production and cytotoxic activity could be two independent events for CD8+ T cells [54, 55]. Besides, our data also showed animals primed by sv145rl always had stronger specific T cell responses than those primed by sv140rl (Fig. 2), no matter boosted by rTTV140rl or rTTV145rl, which indicates that the ultimate T cell response of prime-boost regimen may mainly depend on priming stage.
Two limitations should be noticed in this study. First, it remains to be determined whether our data are relevant to the neutralizing activities; Second, during antibody epitope mapping, OD values were used to display the magnitude of antibody responses and may underestimate the differences of antibody responses among different groups. Further studies to address these issues are required.
Supplementary Material
Supplementary Fig. 1 Examples of raw data from Elispot assay. Negative control stimulated <10 spots in any mice and positive control stimulated more than countable spots. Interestingly, peptide pool IV stimulated no more responses than negative control in mice 4 and 7.
Supplementary Fig. 2 Epitope mapping results from mock control groups. Results showed that no significant humoral responses were detected in sera from mock control groups. The mean OD value of mock control was 0.2.
Supplementary Fig. 3 Homolog comparison between HIV-1 consensus B Env peptides and HIV-1rl42 gp140/145. Epitopes identified in our experiments were located not only at high homolog region, such as p23~p27, but also at low homolog regions, for example p76/p77/p115.
* Epitopes identified in this experiment.
Acknowledgments
We thank Danli Duan, Ning Zhang for their suggestions and technical support in rTTVs construction and purification. We thank Xianggang Huang, Li Ren, Chao Qiu and Hong Peng for their kind help in animal experiments.
Sponsorship: This research was supported by project 30571709, National Natural Science Foundation of China and by China Comprehensive Integrated Programs for Research on AIDS (CIPRA, U19AI51915), the National Institute of Allergy and Infectious Diseases, National Institute of Health, USA
Footnotes
Yanmin Wan and Lan Wu equally contributed to this work.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Pitisuttithum P, Berman PW, Phonrat B, Suntharasamai P, Raktham S, et al. PhaseI/II study of a candidate vaccine designed against the B and E subtypes of HIV-1. J AIDS. 2004;37:1160–65. doi: 10.1097/01.qai.0000136091.72955.4b. [DOI] [PubMed] [Google Scholar]
- 2.Desrosiers RC. Prospects for an AIDS vaccine. Nat Med. 2004;10:221–23. doi: 10.1038/nm0304-221. [DOI] [PubMed] [Google Scholar]
- 3.Gotch FM, Imami N, Hardy G. Candidate vaccines for immunotherapy in HIV. HIV Med. 2001;2:260–265. doi: 10.1046/j.1468-1293.2001.00077.x. [DOI] [PubMed] [Google Scholar]
- 4.Ross RW, Wright ME, Tavel JA. Ongoing trials of immunebased therapies for HIV infection in adults. Exp Opin Biol Ther. 2001;1:413–424. doi: 10.1517/14712598.1.3.413. [DOI] [PubMed] [Google Scholar]
- 5.Girard M, Habel A, Chanel C. New prospects for the development of a vaccine against human immunodeficiency virus type 1: an overview. C R Acad Sci. 1999;III 322:959–966. doi: 10.1016/s0764-4469(00)87193-0. [DOI] [PubMed] [Google Scholar]
- 6.Amara RR, Smith JM, Staprans SI, Montefiori DC, Montefiori DC, Altman JD, et al. Critical role for Env as well as Gag-Pol in control of a simian-human immunodeficiency virus 89.6P challenge by a DNA prime/recombinant modified vaccinia virus Ankara vaccine. J Virol. 2002;76(12):6138–46. doi: 10.1128/JVI.76.12.6138-6146.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Binley JM, Sanders RW, Clas B, Schuelke N, Master A, Guo Y, et al. A recombinant human immunodeficiency virus type 1 envelope glycoprotein complex stabilized by an intermolecular disulfide bond between the gp120 and gp41 subunits is an antigenic mimic of the trimeric virion-associated structure. J Virol. 2000;74(2):627–643. doi: 10.1128/jvi.74.2.627-643.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kwong PD, Wyatt R, Sattentau QJ, Sodroski J, Hendrickson WA. Oligomeric modeling electrostatic analysis of the gp120 envelope glycoprotein of human immunodeficiency virus. J Virol. 2000;74(4):1961–1972. doi: 10.1128/jvi.74.4.1961-1972.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Moore JP, Binley J. HIV Envelope’s letters boxed into shape. Nature. 1998;393 (6686):630–631. doi: 10.1038/31359. [DOI] [PubMed] [Google Scholar]
- 10.Moore JP, Sodroski J. Antibody cross-competition analysis of the human immunodeficiency virus type 1 gp120 exterior envelope glycoprotein. J Virol. 1996;70(3):1863–1872. doi: 10.1128/jvi.70.3.1863-1872.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Parren PW, Moore JP, Burton DR, Sattentau QJ. The neutralizing antibody response to HIV-1: viral evasion and escape from humoral immunity. AIDS. 1999;13 (Suppl A):S137–S162. [PubMed] [Google Scholar]
- 12.Reitter JN, Means RE, Desrosiers RC. A role for carbohydrates in immune evasion in AIDS. Nat Med. 1998;4 (6):679–684. doi: 10.1038/nm0698-679. [DOI] [PubMed] [Google Scholar]
- 13.Wyatt R, Kwong PD, Desjardins E, Sweet RW, Robinson J, Hendrickson WA, et al. The antigenic structure of the HIV gp120 envelope glycoprotein. Nature. 1998;393(6686):705–711. doi: 10.1038/31514. [DOI] [PubMed] [Google Scholar]
- 14.Andrew J McMichael. HIV vaccine. Annu Rev Immunol. 2006;24:227–55. doi: 10.1146/annurev.immunol.24.021605.090605. [DOI] [PubMed] [Google Scholar]
- 15.Chakrabarti BK, Kong WP, Wu BY, Yang ZY, Friborg J, Ling X, et al. Modifications of the Human Immunodeficiency Virus Envelope Glycoprotein Enhance Immunogenicity for Genetic Immunization. J Virol. 2002;76(11):5357–5368. doi: 10.1128/JVI.76.11.5357-5368.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gu Shuyan, Huang Tianmin, et al. A preliminary study on the immunogenicity in rabbits and in human volunteers of a recombinant vaccinia virus expressing Epstein-Barr virus membrane antigen. Chin Med Sci J. 1991;6(4):241–243. [PubMed] [Google Scholar]
- 17.Gu S, Tian-Min H, Li R, Yi-Hong M, Chi-Ming C, Hua L, Wolf H. Immunogenicty in human volunteers of a recombinant vaccinia virus expressing Epstein-Barr virus membrane antigen. Vaccine. 1992:349–355. [Google Scholar]
- 18.Zhao K, et al. Archive of TianTan Vaccinia Research. 1975. [Google Scholar]
- 19.Ren L, Qiu C, Huang X, Pan X, Xu J, Shao Y. Construction and immunogenecity analysis of HIV-1 B’ gag-env DNA vaccine. Chinese Journal of Microbiology and Immunology. 2006;26(7):654–658. [Google Scholar]
- 20.Cole KS, Paliotti MJ, Murphey-Corb M, Montelaro RC. Maturation of envelope-specific antibody responses to linear determinants in monkeys inoculated with attenuated SIV. J Med Primatol. 2000;29:220–230. doi: 10.1034/j.1600-0684.2000.290315.x. [DOI] [PubMed] [Google Scholar]
- 21.Cole KS, Rowles JL, Jagerski BA, Murphey-Corb M, Unangst T, Clements JE, et al. Evolution of Envelope-Specific Antibody Responses in Monkeys Experimentally Infected or Immunized with Simian Immunodeficiency Virus and Its Association with the Development of Protective Immunity. J Virol. 1997;71(7):5069–5079. doi: 10.1128/jvi.71.7.5069-5079.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Letvin NL, Barouch DH, Montefiori DC. Prospects for vaccine protection against HIV-1 infection and AIDS. Annu Rev Immunol. 2002;20:73–99. doi: 10.1146/annurev.immunol.20.081501.094854. [DOI] [PubMed] [Google Scholar]
- 23.Mascola JR. Defining the protective antibody response for HIV-1. Curr Mol Med. 2003;3:209–216. doi: 10.2174/1566524033479799. [DOI] [PubMed] [Google Scholar]
- 24.Amara RR, Smith JM, Staprans SI, et al. Critical role for Env as well as Gag-Pol in control of a simian-human immunodeficiency virus 89.6P challenge by a DNA prime/recombinant modified vaccinia virus Ankara vaccine. J Virol. 2002;76(12):6138–46. doi: 10.1128/JVI.76.12.6138-6146.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Letvin NL, Huang Y, Chakrabarti BK, Xu L, Seaman MS, Beaudry K, et al. Heterologous Envelope Immunogens Contribute to AIDS Vaccine Protection in Rhesus Monkeys. J Virol. 2004;78(14):7490–7497. doi: 10.1128/JVI.78.14.7490-7497.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Belshe RB, Gorse GJ, Mulligan MJ, Evans TG, Keefer MC, Excler JL, et al. Induction of immune responses to HIV-1 by canarypox virus (ALVAC) HIV-1 and gp120 SF-2 recombinant vaccines in uninfected volunteers. NIAID AIDS Vaccine Evaluation Group. AIDS. 1998;12:2407–2415. doi: 10.1097/00002030-199818000-00009. [DOI] [PubMed] [Google Scholar]
- 27.Belshe RB, Graham BS, Keefer MC, Gorse GJ, Wright P, Dolin R, et al. Neutralizing antibodies to HIV-1 in seronegative volunteers immunized with recombinant gp120 from the MN strain of HIV-1. NIAID AIDS Vaccine Clinical Trials Network. JAMA. 1994;272:475–480. doi: 10.1001/jama.272.6.475. [DOI] [PubMed] [Google Scholar]
- 28.Graham BS, McElrath MJ, Connor RI, Schwartz DH, Gorse GJ, Keefer MC, et al. Analysis of intercurrent human immunodeficiency virus type 1 infections in phase I and II trials of candidate AIDS vaccines. AIDS Vaccine Evaluation Group and the Correlates of HIV Immune Protection Group. J Infect Dis. 1998;177:310–319. doi: 10.1086/514209. [DOI] [PubMed] [Google Scholar]
- 29.Mascola JR, Snyder SW, Weinslow OS, Belay SM, Belshe RB, Scwartz DH, et al. Immunization with envelope subunit vaccine products elicits neutralizing antibodies against laboratory-adapted but not primary isolates of human immunodeficiency virus type 1. J Infect Dis. 1996;173:340–348. doi: 10.1093/infdis/173.2.340. [DOI] [PubMed] [Google Scholar]
- 30.Matthews TJ. Dilemma of neutralization resistance of HIV-1 field isolates and vaccine development. AIDS Res Hum Retroviruses. 1994;10:631–632. doi: 10.1089/aid.1994.10.631. [DOI] [PubMed] [Google Scholar]
- 31.Beddows S, Lister S, Cheingsong R, Bruck C, Weber J. Comparison of the antibody repertoire generated in healthy volunteers following immunization with a monomeric recombinant gp120 construct derived from a CCR5/CXCR4-using human immunodeficiency virus type 1 isolate with sera from naturally infected individuals. J Virol. 1999;73:1740–1745. doi: 10.1128/jvi.73.2.1740-1745.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.VanCott TC, Bethke FR, Burke DS, Redfield RR, Birx DL. Lack of induction of antibodies specific for conserved, discontinuous epitopes of HIV-1 envelope glycoprotein by candidate AIDS vaccines. J Immunol. 1995;155:4100–4110. [PubMed] [Google Scholar]
- 33.Evans LA, Thomson-Honnebier G, Steimer K, Paoletti E, Perkus ME, Hollander H, Levy JA. Antibody-dependent cellular cytotoxicity is directed against both the gp120 and gp41 envelope proteins of HIV. AIDS. 1989;3:273–276. doi: 10.1097/00002030-198905000-00004. [DOI] [PubMed] [Google Scholar]
- 34.Gnann JWJ, Nelson JA, Oldstone MBA. Fine mapping of an immunodominant domain in the transmembrane glycoprotein of human immunodeficiency virus. J Virol. 1987;61:2639–2641. doi: 10.1128/jvi.61.8.2639-2641.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Beddows S, Schulke N, Kirschner M, et al. Evaluating the immunogenicity of a disulfide-stabilized, cleaved, trimeric form of the envelope glycoprotein complex of human immunodeficiency virus type 1. J Virol. 2005;79(14):8812–27. doi: 10.1128/JVI.79.14.8812-8827.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bower JF, Yang X, Sodroski J, Ross TM. Elicitation of neutralizing antibodies with DNA vaccines expressing soluble stabilized human immunodeficiency virus type 1 envelope glycoprotein trimers conjugated to C3d. J Virol. 2004;78(9):4710–9. doi: 10.1128/JVI.78.9.4710-4719.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sanders RW, Vesanen M, Schuelke N, et al. Stabilization of the soluble, cleaved, trimeric form of the envelope glycoprotein complex of human immunodeficiency virus type 1. J Virol. 2002;76(17):8875–89. doi: 10.1128/JVI.76.17.8875-8889.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yang X, Farzan M, Wyatt R, Sodroski J. Characterization of stable, soluble trimer containing complete ectodomains of HIV-1 envelope glycoprotein. J Virol. 2000;74(12):5716–25. doi: 10.1128/jvi.74.12.5716-5725.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yang X, Lee J, Mahony EM, Kwong PD, Wyatt R, Sodroski J. Highly stable trimers formed by human immunodeficiency virus type 1 envelope glycoproteins fused with the trimeric motif of T4 bacteriophage fibritin. J Virol. 2002;76(9):4634–42. doi: 10.1128/JVI.76.9.4634-4642.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bower JF, Li YX, Wyatt R, Ross TM. HIV-1 Envgp140 trimers elicit neutralizing antibodies without efficient induction of conformational antibodies. Vaccine. 2006;24:5442–5451. doi: 10.1016/j.vaccine.2006.03.063. [DOI] [PubMed] [Google Scholar]
- 41.Earl P, Sugiura W, Montefiori DC, Broder CC, Lee SA, Wild C, et al. Immunogenicity and Protective Efficacy of Oligomeric Human Immunodeficiency Virus Type 1 gp140. J Virol. 2001;75 (2):645–653. doi: 10.1128/JVI.75.2.645-653.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Trkola A, Purtscher M, Muster T, Ballaun C, Buchacher A, Sullivan N, et al. Human monoclonal antibody 2G12 defines a distinctive neutralization epitope on the gp120 glycoprotein of human immunodeficiency virus type 1. J Virol. 1996;70:1100–8. doi: 10.1128/jvi.70.2.1100-1108.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wyatt R, Moore J, Accola M, Desjardin E, Robinson J, Sodroski J. Involvement of the V1/V2 variable loop structure in the exposure of human immunodeficiency virus type 1 gp120 epitopes induced by receptor binding. J Virol. 1995;69:5723–5733. doi: 10.1128/jvi.69.9.5723-5733.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wyatt R, Sullivan N, Thali M, Repke H, Ho D, Robinson J. Functional and immunologic characterization of human immunodeficiency virus type 1 envelope glycoproteins containing deletions of the major variable regions. J Virol. 1993;67:4557–4565. doi: 10.1128/jvi.67.8.4557-4565.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cao J, Sullivan N, Desjardin E, Parolin C, Robinson J, Wyatt R. Replication and neutralization of human immunodeficiency virus type 1 lacking the V1 and V2 variable loops of the gp120 envelope glycoprotein. J Virol. 1997;71:9808–9812. doi: 10.1128/jvi.71.12.9808-9812.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kim YB, Han DP, Cao C, Cho MW. Immunogenicity and Ability of Variable Loop-Deleted Human Immunodeficiency Virus Type 1 Envelope Glycoproteins to Elicit Neutralizing Antibodies. Virology. 2002;305:124–137. doi: 10.1006/viro.2002.1727. [DOI] [PubMed] [Google Scholar]
- 47.Yang Z, Chakrabarti BK, Xu L, Welcher B, Kong WP, Leung K, et al. Selective Modification of Variable Loops Alters Tropism and Enhances Immunogenicity of Human Immunodeficiency Virus Type 1 Envelope. J Virol. 2004;78(8):4029–4036. doi: 10.1128/JVI.78.8.4029-4036.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Stamatatos L, Cheng-Mayer C. An envelope modification that renders a primary, neutralization resistant, clade B HIV-1 isolate highly susceptible to neutralization by sera from other clades. J Virol. 1998;72:7840–7845. doi: 10.1128/jvi.72.10.7840-7845.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kiszka I, Kmieciak D, Gzy J, Naito T, Bolesta E, Sieron A, et al. Effect of the V3 loop deletion of envelope glycoprotein on cellular responses and protection against challenge with recombinant vaccinia virus expressing gp160 of primary human immunodeficiency virus type 1 isolates. J Virol. 2002;76(9):4222–4232. doi: 10.1128/JVI.76.9.4222-4232.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nguyen M, Pean P, Lopalco L, Nouhin J, Phoung V, Ly N, et al. HIV-specific antibodies but not t-cell responses are associated with protection in seronegative partners of HIV-1-infected individuals in Cambodia. J Acquir Immune Defic Syndr. 2006;42(4):412–9. doi: 10.1097/01.qai.0000222289.97825.35. [DOI] [PubMed] [Google Scholar]
- 51.Jianqing Xu, Li Ren, Xianggang Huang, Chao Qiu, Yong Liu, Ying Liu, et al. Sequential priming and boosting with heterologous HIV immunogens predominantly stimulated T cell immunity against conserved epitopes. AIDS. 2006;20:2293–2303. doi: 10.1097/QAD.0b013e328010ad0c. [DOI] [PubMed] [Google Scholar]
- 52.Kwong PD, Wyatt R, Robinson J, Sweet RW, Sodroski J, Hendrickson WA. Structure of an HIVgp120 envelope glycoprotein in complex with the CD4 receptor and a neutralizing human antibody. Nature. 1998;393:684–659. doi: 10.1038/31405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Conley AJ, Gorny MK, Kessler JA, II, Boots LJ, Ossorio-Castro M, Koenig S, et al. Neutralization of primary human immunodeficiency virus type 1 isolates by the broadly reactive anti-V3 monoclonal antibody, 447-52D. J Virol. 1994;68:6994–7000. doi: 10.1128/jvi.68.11.6994-7000.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bartholdy C, Christensen JP, Wodarz D, Thomsen AR. Persistent virus infection despite chronic cytotoxic T-lymphocyte activation in gamma interferon-deficient mice infected with lymphocytic choriomeningitis virus. J Virol. 2000;74:10304–11. doi: 10.1128/jvi.74.22.10304-10311.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Belz GT, Xie W, Doherty PC. Diversity of epitope and cytokine profiles for primary and secondary influenza a virus-specific CD8+ T cell responses. J Immunol. 2001;166:4627–33. doi: 10.4049/jimmunol.166.7.4627. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Fig. 1 Examples of raw data from Elispot assay. Negative control stimulated <10 spots in any mice and positive control stimulated more than countable spots. Interestingly, peptide pool IV stimulated no more responses than negative control in mice 4 and 7.
Supplementary Fig. 2 Epitope mapping results from mock control groups. Results showed that no significant humoral responses were detected in sera from mock control groups. The mean OD value of mock control was 0.2.
Supplementary Fig. 3 Homolog comparison between HIV-1 consensus B Env peptides and HIV-1rl42 gp140/145. Epitopes identified in our experiments were located not only at high homolog region, such as p23~p27, but also at low homolog regions, for example p76/p77/p115.
* Epitopes identified in this experiment.


