Abstract
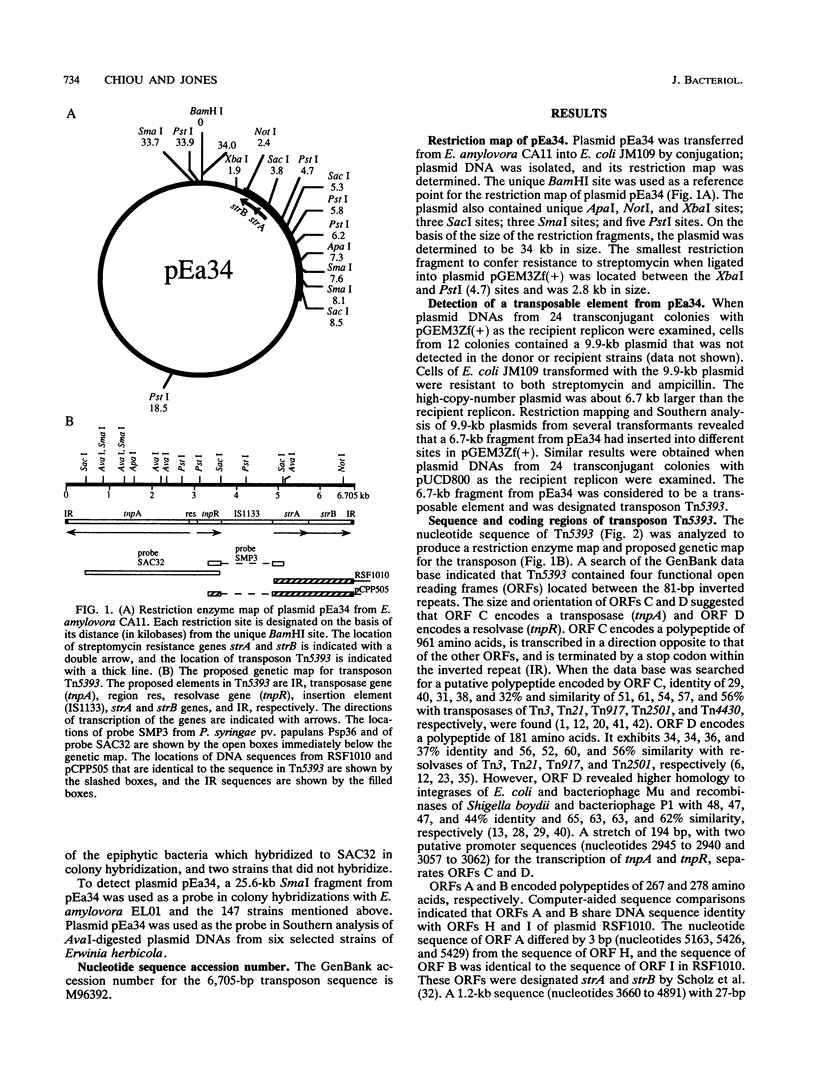
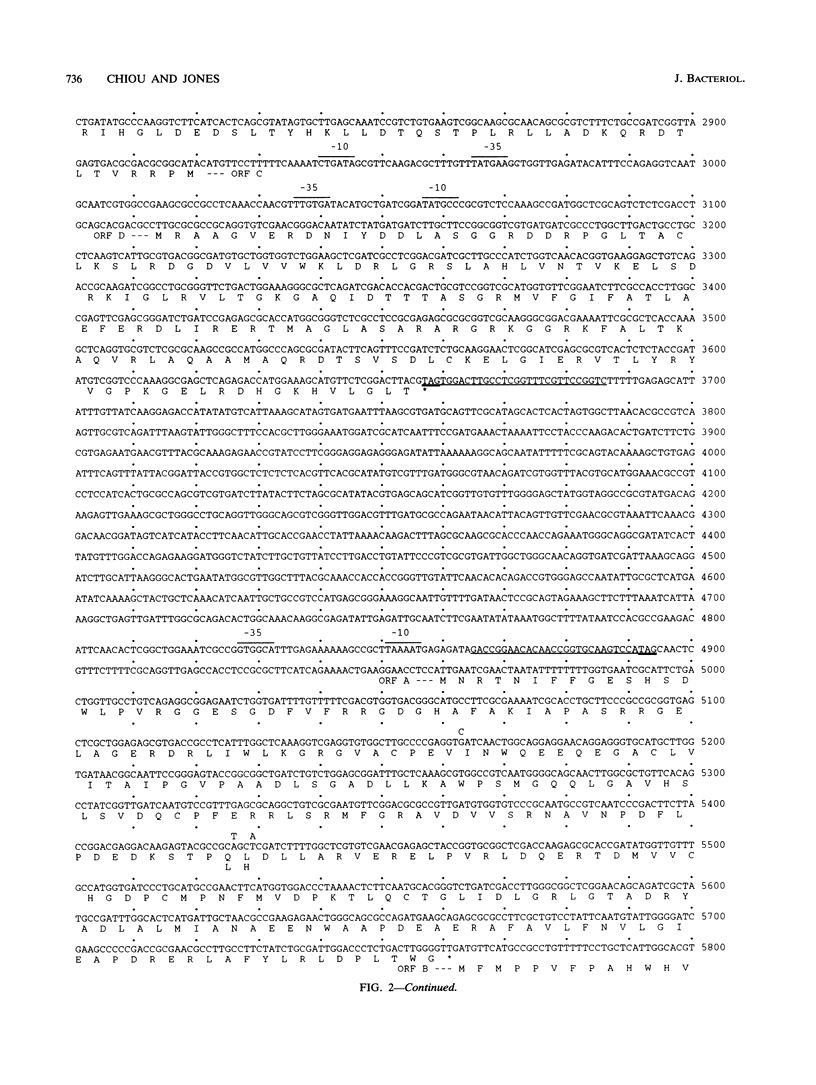
A class II Tn3-type transposable element, designated Tn5393 and located on plasmid pEa34 from streptomycin-resistant strain CA11 of Erwinia amylovora, was identified by its ability to move from pEa34 to different sites in plasmids pGEM3Zf(+) and pUCD800. Nucleotide sequence analysis reveals that Tn5393 consists of 6,705 bp with 81-bp terminal inverted repeats and generates 5-bp duplications of the target DNA following insertion. Tn5393 contains open reading frames that encode a putative transposase (tnpA) and resolvase (tnpR) of 961 and 181 amino acids, respectively. The two open reading frames are separated by a putative recombination site (res) consisting of 194 bp. Two streptomycin resistance genes, strA and strB, were identified on the basis of their DNA sequence homology to streptomycin resistance genes in plasmid RSF1010. StrA is separated from tnpR by a 1.2-kb insertion element designated IS1133. The tnpA-res-tnpR region of Tn5393 was detected in Pseudomonas syringae pv. papulans Psp36 and in many other gram-negative bacteria harboring strA and strB. Except for some strains of Erwinia herbicola, these other gram-negative bacteria lacked insertion sequence IS1133. The prevalence of strA and strB could be accounted for by transposition of Tn5393 to conjugative plasmids that are then disseminated widely among gram-negative bacteria.
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- An F. Y., Clewell D. B. Tn917 transposase. Sequence correction reveals a single open reading frame corresponding to the tnpA determinant of Tn3-family elements. Plasmid. 1991 Mar;25(2):121–124. doi: 10.1016/0147-619x(91)90023-p. [DOI] [PubMed] [Google Scholar]
- Devereux J., Haeberli P., Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984 Jan 11;12(1 Pt 1):387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diver W. P., Grinsted J., Fritzinger D. C., Brown N. L., Altenbuchner J., Rogowsky P., Schmitt R. DNA sequences of and complementation by the tnpR genes of Tn21, Tn501 and Tn1721. Mol Gen Genet. 1983;191(2):189–193. doi: 10.1007/BF00334812. [DOI] [PubMed] [Google Scholar]
- Falkenstein H., Bellemann P., Walter S., Zeller W., Geider K. Identification of Erwinia amylovora, the Fireblight Pathogen, by Colony Hybridization with DNA from Plasmid pEA29. Appl Environ Microbiol. 1988 Nov;54(11):2798–2802. doi: 10.1128/aem.54.11.2798-2802.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gay P., Le Coq D., Steinmetz M., Berkelman T., Kado C. I. Positive selection procedure for entrapment of insertion sequence elements in gram-negative bacteria. J Bacteriol. 1985 Nov;164(2):918–921. doi: 10.1128/jb.164.2.918-921.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grinsted J., de la Cruz F., Schmitt R. The Tn21 subgroup of bacterial transposable elements. Plasmid. 1990 Nov;24(3):163–189. doi: 10.1016/0147-619x(90)90001-s. [DOI] [PubMed] [Google Scholar]
- Guerry P., van Embden J., Falkow S. Molecular nature of two nonconjugative plasmids carrying drug resistance genes. J Bacteriol. 1974 Feb;117(2):619–630. doi: 10.1128/jb.117.2.619-630.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hattori M., Sakaki Y. Dideoxy sequencing method using denatured plasmid templates. Anal Biochem. 1986 Feb 1;152(2):232–238. doi: 10.1016/0003-2697(86)90403-3. [DOI] [PubMed] [Google Scholar]
- Heffron F., McCarthy B. J., Ohtsubo H., Ohtsubo E. DNA sequence analysis of the transposon Tn3: three genes and three sites involved in transposition of Tn3. Cell. 1979 Dec;18(4):1153–1163. doi: 10.1016/0092-8674(79)90228-9. [DOI] [PubMed] [Google Scholar]
- Hiestand-Nauer R., Iida S. Sequence of the site-specific recombinase gene cin and of its substrates serving in the inversion of the C segment of bacteriophage P1. EMBO J. 1983;2(10):1733–1740. doi: 10.1002/j.1460-2075.1983.tb01650.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KING E. O., WARD M. K., RANEY D. E. Two simple media for the demonstration of pyocyanin and fluorescin. J Lab Clin Med. 1954 Aug;44(2):301–307. [PubMed] [Google Scholar]
- Kado C. I., Liu S. T. Rapid procedure for detection and isolation of large and small plasmids. J Bacteriol. 1981 Mar;145(3):1365–1373. doi: 10.1128/jb.145.3.1365-1373.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleckner N. Transposable elements in prokaryotes. Annu Rev Genet. 1981;15:341–404. doi: 10.1146/annurev.ge.15.120181.002013. [DOI] [PubMed] [Google Scholar]
- Lax A. J., Walker C. A. Plasmids related to RSF1010 from Bordetella bronchiseptica. Plasmid. 1986 May;15(3):210–216. doi: 10.1016/0147-619x(86)90039-9. [DOI] [PubMed] [Google Scholar]
- Mahillon J., Lereclus D. Structural and functional analysis of Tn4430: identification of an integrase-like protein involved in the co-integrate-resolution process. EMBO J. 1988 May;7(5):1515–1526. doi: 10.1002/j.1460-2075.1988.tb02971.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michiels T., Cornelis G. Detection and characterization of Tn2501, a transposon included within the lactose transposon Tn951. J Bacteriol. 1984 Jun;158(3):866–871. doi: 10.1128/jb.158.3.866-871.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michiels T., Cornelis G., Ellis K., Grinsted J. Tn2501, a component of the lactose transposon Tn951, is an example of a new category of class II transposable elements. J Bacteriol. 1987 Feb;169(2):624–631. doi: 10.1128/jb.169.2.624-631.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura A., Morita M., Nishimura Y., Sugino Y. A rapid and highly efficient method for preparation of competent Escherichia coli cells. Nucleic Acids Res. 1990 Oct 25;18(20):6169–6169. doi: 10.1093/nar/18.20.6169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norelli J. L., Burr T. J., Lo Cicero A. M., Gilbert M. T., Katz B. H. Homologous Streptomycin Resistance Gene Present among Diverse Gram-Negative Bacteria in New York State Apple Orchards. Appl Environ Microbiol. 1991 Feb;57(2):486–491. doi: 10.1128/aem.57.2.486-491.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plasterk R. H., van de Putte P. The invertible P-DNA segment in the chromosome of Escherichia coli. EMBO J. 1985 Jan;4(1):237–242. doi: 10.1002/j.1460-2075.1985.tb02341.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plasterk R. H., van de Putte P. The invertible P-DNA segment in the chromosome of Escherichia coli. EMBO J. 1985 Jan;4(1):237–242. doi: 10.1002/j.1460-2075.1985.tb02341.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rotger R., Rubio F., Nombela C. A multi-resistance plasmid isolated from commensal Neisseria species is closely related to the enterobacterial plasmid RSF1010. J Gen Microbiol. 1986 Sep;132(9):2491–2496. doi: 10.1099/00221287-132-9-2491. [DOI] [PubMed] [Google Scholar]
- Rotger R., Rubio F., Nombela C. A multi-resistance plasmid isolated from commensal Neisseria species is closely related to the enterobacterial plasmid RSF1010. J Gen Microbiol. 1986 Sep;132(9):2491–2496. doi: 10.1099/00221287-132-9-2491. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scholz P., Haring V., Wittmann-Liebold B., Ashman K., Bagdasarian M., Scherzinger E. Complete nucleotide sequence and gene organization of the broad-host-range plasmid RSF1010. Gene. 1989 Feb 20;75(2):271–288. doi: 10.1016/0378-1119(89)90273-4. [DOI] [PubMed] [Google Scholar]
- Scholz P., Haring V., Wittmann-Liebold B., Ashman K., Bagdasarian M., Scherzinger E. Complete nucleotide sequence and gene organization of the broad-host-range plasmid RSF1010. Gene. 1989 Feb 20;75(2):271–288. doi: 10.1016/0378-1119(89)90273-4. [DOI] [PubMed] [Google Scholar]
- Shaw J. H., Clewell D. B. Complete nucleotide sequence of macrolide-lincosamide-streptogramin B-resistance transposon Tn917 in Streptococcus faecalis. J Bacteriol. 1985 Nov;164(2):782–796. doi: 10.1128/jb.164.2.782-796.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw J. H., Clewell D. B. Complete nucleotide sequence of macrolide-lincosamide-streptogramin B-resistance transposon Tn917 in Streptococcus faecalis. J Bacteriol. 1985 Nov;164(2):782–796. doi: 10.1128/jb.164.2.782-796.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinberger E. M., Cheng G. Y., Beer S. V. Characterization of a 56-kb plasmid of Erwinia amylovora Ea322: its noninvolvement in pathogenicity. Plasmid. 1990 Jul;24(1):12–24. doi: 10.1016/0147-619x(90)90021-4. [DOI] [PubMed] [Google Scholar]
- Steinberger E. M., Cheng G. Y., Beer S. V. Characterization of a 56-kb plasmid of Erwinia amylovora Ea322: its noninvolvement in pathogenicity. Plasmid. 1990 Jul;24(1):12–24. doi: 10.1016/0147-619x(90)90021-4. [DOI] [PubMed] [Google Scholar]
- Timmerman K. P., Tu C. P. Complete sequence of IS3. Nucleic Acids Res. 1985 Mar 25;13(6):2127–2139. doi: 10.1093/nar/13.6.2127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timmerman K. P., Tu C. P. Complete sequence of IS3. Nucleic Acids Res. 1985 Mar 25;13(6):2127–2139. doi: 10.1093/nar/13.6.2127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tominaga A., Ikemizu S., Enomoto M. Site-specific recombinase genes in three Shigella subgroups and nucleotide sequences of a pinB gene and an invertible B segment from Shigella boydii. J Bacteriol. 1991 Jul;173(13):4079–4087. doi: 10.1128/jb.173.13.4079-4087.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tominaga A., Ikemizu S., Enomoto M. Site-specific recombinase genes in three Shigella subgroups and nucleotide sequences of a pinB gene and an invertible B segment from Shigella boydii. J Bacteriol. 1991 Jul;173(13):4079–4087. doi: 10.1128/jb.173.13.4079-4087.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner A. K., Grinsted J. DNA sequence of the transposase gene of the new category of class II transposon, Tn2501. Nucleic Acids Res. 1987 Dec 10;15(23):10049–10049. doi: 10.1093/nar/15.23.10049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner A. K., Grinsted J. DNA sequence of the transposase gene of the new category of class II transposon, Tn2501. Nucleic Acids Res. 1987 Dec 10;15(23):10049–10049. doi: 10.1093/nar/15.23.10049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward E., Grinsted J. The nucleotide sequence of the tnpA gene of Tn21. Nucleic Acids Res. 1987 Feb 25;15(4):1799–1806. doi: 10.1093/nar/15.4.1799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward E., Grinsted J. The nucleotide sequence of the tnpA gene of Tn21. Nucleic Acids Res. 1987 Feb 25;15(4):1799–1806. doi: 10.1093/nar/15.4.1799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willson P. J., Deneer H. G., Potter A., Albritton W. Characterization of a streptomycin-sulfonamide resistance plasmid from Actinobacillus pleuropneumoniae. Antimicrob Agents Chemother. 1989 Feb;33(2):235–238. doi: 10.1128/aac.33.2.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willson P. J., Deneer H. G., Potter A., Albritton W. Characterization of a streptomycin-sulfonamide resistance plasmid from Actinobacillus pleuropneumoniae. Antimicrob Agents Chemother. 1989 Feb;33(2):235–238. doi: 10.1128/aac.33.2.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanisch-Perron C., Vieira J., Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33(1):103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- Yanisch-Perron C., Vieira J., Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33(1):103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- Zheng Z. X., Chandler M., Hipskind R., Clerget M., Caro L. Dissection of the r-determinant of the plasmid R100.1: the sequence at the extremities of Tn21. Nucleic Acids Res. 1981 Dec 11;9(23):6265–6278. doi: 10.1093/nar/9.23.6265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Z. X., Chandler M., Hipskind R., Clerget M., Caro L. Dissection of the r-determinant of the plasmid R100.1: the sequence at the extremities of Tn21. Nucleic Acids Res. 1981 Dec 11;9(23):6265–6278. doi: 10.1093/nar/9.23.6265. [DOI] [PMC free article] [PubMed] [Google Scholar]