Abstract
In plants, replication of RNAviruses and RNAfrom highly transcribed transgenes can trigger DNA methylation. These systems accumulate diced small RNA(smRNA) products of double-stranded RNA(dsRNA) precursors, but it is not known which RNAspecies directs methylation. The methylated PAI tryptophan biosynthetic genes in Arabidopsis allow the study of methylation signals for endogenous genes with lower expression levels. The PAI genes are arranged as a tandem inverted repeat plus two singlet genes at unlinked loci. Here we show that the predominant PAI transcript initiates at a novel unmethylated promoter that lies upstream of one of the inverted repeat PAI genes. Suppressed transcription from the upstream promoter using transgene-directed silencing reduces methylation on the singlet PAI genes, but not on the inverted repeat, consistent with an RNAmethylation signal. RNAgel blots detect normal PAI transcripts and dsRNA read-through species, but not diced smRNAs, suggesting that either precursor dsRNAs or subdetectable levels of smRNAs, below the threshold to effectively degrade PAI transcripts, serve as the PAI methylation signal. Thus, the lower expression endogenous gene system allows dissection of a RNA-directed methylation pathway distinct from RNA degradation pathways.
Keywords: Cytosine methylation, RNA-directed DNA methylation, RNA silencing, small RNAs, dicer
Genomic cytosine methylation in eukaryotes is important for control of gene expression and genome stability (for reviews, see Martienssen and Colot 2001; Bird 2002). Methylated genomic regions are typically assembled into a compacted chromatin structure that is refractory to transcription and recombination. In mammals, methylation is critical for developmental processes including genomic imprinting and X chromosome inactivation. In both mammals and plants, methylation also provides a genome defense against invasive parasitic sequences. For example, transposable element sequences in mammalian and plant genomes are often methylated (Yoder et al. 1997; Finnegan et al. 1998). In plants, loss of transposon methylation has been shown to result in activation of transposon movement with occasionally mutagenic consequences (Miura et al. 2001; Singer et al. 2001; Kato et al. 2003).
A key question is what signals direct methylation to particular genomic sequences. In both mammals and plants, tandem repeated arrays such as centromere-associated repeats or repetitive transgene insertion arrays are usually methylated (Assaad et al. 1993; Yoder et al. 1997; Finnegan et al. 1998; Garrick et al. 1998). This observation has led to the hypothesis that repeated arrays might have intrinsic structural features that make them good substrates for methylation (Assaad et al. 1993; Henikoff 1998). Additionally in plants, a growing body of evidence suggests that some types of aberrant RNAs can stimulate methylation of genomic DNA with which they share sequence identity. The original observation of RNA-directed DNA methylation came from studies with tobacco carrying replication-proficient versus replication-defective RNA viroid-encoding sequences on a genomic transgene insertion (Wassenegger et al. 1994). More recently, RNA-directed DNA methylation has been demonstrated in a number of plant systems that express high levels of RNA, including RNA virus infections and viral promoter-driven transgenes (Jones et al. 1998, 1999; Mette et al. 1999, 2000; Dalmay et al. 2000a,b; Mourrain et al. 2000; Sijen et al. 2001; Wang et al. 2001).
In both plants and animals, double-stranded RNAs (dsRNAs) have been shown to trigger an RNA-based defense response called RNA interference or RNA silencing (Matzke et al. 2001). In this process, the dsRNA is processed into small RNAs (smRNAs) of 21–25 nucleotides (nt) in length by a dicer RNAse. The smRNAs then guide degradation of any RNA species with which they share sequence complementarity. The RNA silencing process thus serves as a defense against RNA virus infections, which typically involve dsRNA replication intermediates. In plants, RNA silencing is also often associated with DNA methylation of the affected sequences, suggesting that these two silencing responses are mechanistically interrelated, with the DNA methylation perhaps providing an extra layer of protection against any viral sequences that make their way into the plant genome by reverse transcription and integration (Bender 2001). Evidence in support of a common mechanism came from genetic studies in Arabidopsis that screened for suppressors of RNA silencing of reporter transgenes (Dalmay et al. 2000b, 2001; Mourrain et al. 2000). The RNA silencing-defective mutations recovered proved to also abolish transgene DNA methylation. However, these mutations are thought to block the conversion of aberrant RNA precursors into dsRNA substrates for dicing (Dalmay et al. 2000b, 2001; Béclin et al. 2002); thus, it remains unknown which downstream RNA species— dsRNA or smRNA—mediates DNA methylation in these systems.
Duplicated tryptophan biosynthetic genes in Arabidopsis that encode the enzyme phosphoribosylanthranilate isomerase (PAI) serve as a model system to study methylation signaling for endogenous genes in plants. Arabidopsis strains vary in PAI gene arrangements and methylation status (Bender and Fink 1995; Luff et al. 1999; Melquist et al. 1999). In the Columbia (Col) strain, there are three PAI genes (PAI1, PAI2, and PAI3) at three unlinked loci in the genome, and these genes are not methylated. In the Wassilewskija (WS) strain, there is a tail-to-tail inverted repeat of two genes PAI1–PAI4 at the PAI1 locus, as well as the unlinked PAI2 and PAI3 genes; all four genes are densely methylated across their regions of shared sequence identity, which extends for several hundred base pairs upstream and downstream of the protein-coding region. In both WS and Col, only PAI1 and PAI2 encode functional PAI enzyme (Melquist et al. 1999). We previously determined that the WS PAI1–PAI4 inverted repeat locus provides the trigger for PAI gene-directed methylation because when this repeat is combined in a genome with unmethylated Col PAI genes by genetic crosses, the Col PAI genes become methylated within a few generations of inbreeding (Luff et al. 1999). Furthermore, a survey of wild isolates of Arabidopsis showed that only strains with a PAI1–PAI4 inverted repeat display PAI methylation (Melquist et al. 1999).
Given the precedent for dsRNA or diced smRNA species to stimulate DNA methylation in plants, an attractive hypothesis was that transcription through the PAI1–PAI4 locus might provide the PAI methylation signal. In fact, in WS and other PAI-methylated strains, the PAI1 gene was found to be uniquely expressed, demonstrating that this locus is indeed transcribed despite dense methylation (Melquist et al. 1999). Here we show that the WS PAI1 gene is actually transcribed from a novel upstream promoter that lies beyond the methylated region. Suppression of transcription from this promoter reduces methylation on the singlet PAI2 and PAI3 genes, consistent with an RNA-based methylation signal. These results show that even relatively low-level transcription from an endogenous promoter through an inverted repeat sequence is sufficient to provide a potent DNA methylation signal. RNA gel blot analysis reveals detectable steady-state levels of normal PAI1 transcripts and dsRNA read-through species, but not PAI smRNAs, suggesting that PAI methylation is triggered either by unprocessed precursor dsRNAs, or by low levels of smRNAs, below a threshold to effectively trigger PAI transcript degradation. Mutations in known aberrant RNA processing factors have no effect on PAI methylation, consistent with the direct production of dsRNA by read-through transcription of the PAI1–PAI4 locus. Furthermore, mutation of the DCL1 dicer gene, which has been shown to control processing of developmental micro-RNAs but not small RNAs associated with RNA silencing (Finnegan et al. 2003), has no effect on PAI methylation. Thus, if small RNAs trigger PAI DNA methylation, they are likely to be made by the same dicer(s) necessary for RNA silencing.
Results
The WS PAI1–PAI4 methylated inverted repeat is transcribed from an upstream unmethylated promoter
In the WS strain of Arabidopsis, the four PAI genes are densely methylated across the region of PAI sequence identity (Luff et al. 1999), which extends from ∼350 bp upstream of the translational start codon to downstream of the translational stop codon for each gene (Melquist et al. 1999). The flanking sequences unique to each gene are unmethylated. In particular, the PAI1–PAI4 inverted repeat is flanked by 2.9 kb of nearly perfect unmethylated direct repeat sequences (Melquist et al. 1999). The direct repeat proximal to PAI1 carries the promoter and first exon of a ribosomal protein-encoding gene S15a oriented to be transcribed in the same direction as PAI1; the direct repeat proximal to PAI4 carries the same S15a promoter/first exon sequence fused to the rest of the S15a gene (Fig. 1A). Based on eight S15a cDNA sequences available in the database, the S15a gene contains a short first exon of 39–66 bp of untranslated sequences, followed by a large intron, followed by the second exon which contains the translational start codon for the gene (Fig. 1B).
Figure 1.
Genome and transcript structure for the WS PAI1–PAI4 locus. (A) An overview of the genome structure at the WS PAI1–PAI4 locus is shown, with black arrows indicating the PAI genes including noncoding PAI-identical sequences; red arrows indicate the flanking 2.9-kb direct repeats, a blue arrow indicates the S15a gene, and blue dots represent the S15a promoter region. The box around the PAI sequences indicates the region of dense DNA methylation. Below this diagram, the structure of the S15aIR transgene construct is shown, with lines indicating the endogenous genomic regions that will be targeted for methylation by this construct. (B) The 5′-end transcript structures of S15a and PAI1 are shown, with blue sequences representing S15a and black sequences representing PAI1. Wide lines indicate exons and thin lines indicate introns. The positions of ATG translational start codons are indicated by asterisks. Diagonal slashes indicate that the body of the gene beyond the start codon is not drawn to scale.
In previous studies, we showed that the PAI1 gene in the inverted repeat is the sole expressed PAI gene in WS (Melquist et al. 1999). An attractive hypothesis to explain this result was that cis-acting sequences unique to PAI1—specifically the nearby unmethylated S15a promoter—might control PAI1 expression. In fact, our previous analysis of PAI cDNAs isolated by hybridization from a WS cDNA library identified one PAI1 cDNA that carried 33 bp of sequence at the S15a transcription start site spliced to more proximal PAI1 sequences, consistent with this hypothesis (Melquist et al. 1999). We performed 5′ rapid amplification of cDNA ends (5′ RACE) PCR analysis on WS RNA to determine where PAI1 transcripts initiate (Table 1). We found two general types of 5′-end species, both of which initiated in the upstream unmethylated S15a sequences and both of which were spliced from the S15a first exon donor site to more proximal PAI1 sequences. One species was spliced to a cryptic acceptor site that lies 96 bp upstream of the PAI1 translational start codon. The other species was spliced to the normal S15a acceptor site, and then spliced again from a nearby cryptic donor site to the cryptic acceptor site at the 96-bp upstream position, resulting in an extra 26 bp exon between the S15a first exon and PAI1 sequences. Therefore, in both cases, the bulk of the RNA between the S15a transcription start site, which lies ∼500 bp upstream of the PAI1 methylation boundary and ∼850 bp upstream of the PAI1 translational start codon, is spliced out of the processed PAI1 transcript. These results support the hypothesis that the unmethylated S15a promoter drives PAI1 expression in WS. These results also indicate that the dense methylation of the PAI1 sequences beginning 500 bp downstream of the S15a transcription initiation site does not impede expression of PAI1.
Table 1.
5′ RACE determination of transcript starts for WS PAI1 and Col PAI1 and PAI2
| Transcript | First upstream exona | Second upstream exona | First PAI exona |
|---|---|---|---|
| WS PAI1 | -868 to -803 | -386 to -361 | -96 to +10 |
| WS PAI1 | -868 to -803 | -386 to -361 | -96 to +10 |
| WS PAI1 | -855 to -803 | -386 to -361 | -96 to +10 |
| WS PAI1 | -851 to -803 | -386 to -361 | -96 to +10 |
| WS PAI1 | -868 to -803 | NA | -96 to +10 |
| WS PAI1 | -863 to -803 | NA | -96 to +10 |
| WS PAI1 | -860 to -803 | NA | -96 to +10 |
| WS PAI1 | -860 to -803 | NA | -96 to +10 |
| Col PAI1 | -847 to -777 | -386 to -361 | -96 to +10 |
| Col PAI1 | -845 to -777 | -386 to -361 | -96 to +10 |
| Col PAI1 | -845 to -777 | -383 to -361 | -96 to +10 |
| Col PAI1 | -842 to -777 | -386 to -361 | -96 to +10 |
| Col PAI1 | -834 to -777 | -386 to -361 | -96 to +10 |
| Col PAI1 | -834 to -777 | NA | -96 to +10 |
| Col PAI1 | NA | NA | -117 to +10 |
| Col PAI1 | NA | NA | -93 to +10 |
| Col PAI2 | NA | NA | -133 to +10 |
| Col PAI2 | NA | NA | -122 to +10 |
| Col PAI2 | NA | NA | -117 to +10 |
| Col PAI2 | NA | NA | -117 to +10 |
| Col PAI2 | NA | NA | -117 to +10 |
| Col PAI2 | NA | NA | -114 to +10 |
| Col PAI2 | NA | NA | -108 to +10 |
| Col PAI2 | NA | NA | -99 to +10 |
Numbering is relative to the first base of the PAI ATG translational start codon as +1 on genomic sequences. The WS genomic sequence (GenBank accession no. U34757) carries an extra 26 bp in the first intron region upstream of PAI1 relative to the Col genomic sequence (GenBank accession no. AF130878). The PAI sequence identity/methylation boundary ends at -355 in the WS genomic sequence.
Anunmethylated PAI1 gene can be transcribed from either the upstream promoter or a more proximal promoter
We also analyzed 5′ ends of PAI transcripts isolated from the Col strain of Arabidopsis, which carries a singlet PAI1 gene rather than the PAI1–PAI4 inverted repeat and lacks PAI gene methylation (Bender and Fink 1995; Melquist et al. 1999). The sequences upstream of Col PAI1, including the S15a promoter regions, are similar to those found in WS. Therefore, analysis of PAI transcripts in Col allows us to understand where PAI1 transcripts initiate when the PAI1-proximal upstream sequences are unmethylated.
Previous analysis of Col PAI cDNAs revealed that both PAI1 and PAI2 are expressed, with much lower expression of the divergent nonfunctional PAI3 gene (Melquist et al. 1999). A similar result was found for PAI cDNA analysis in the Landsberg erecta strain (Ler), which, like Col, carries three unmethylated PAI genes. From this analysis, one Col PAI1 cDNA initiated 101 bp upstream of the translational start codon, and one Ler PAI1 cDNA initiated at the S15a start site and was spliced directly to the cryptic acceptor site that lies 96 bp upstream of the start codon; the remaining cDNAs were truncated and uninformative. Additionally, two Col PAI1 cDNA sequences available in the database initiated at the S15a promoter and were spliced to proximal PAI sequences. These results suggest that when the PAI1 gene is unmethylated, transcripts can still initiate at the upstream S15a promoter, but that they can also initiate at a more proximal site ∼100 bp upstream of the translation start site.
To further explore this possibility, we performed 5′ RACE PCR analysis of Col PAI transcripts (Table 1). This analysis detected three types of PAI1 species: both splice forms of transcripts that initiated at the S15a start as in WS (Fig. 1B), plus transcripts that initiated at start sites more proximal to PAI1, either 117 or 93 bp upstream of the translational start codon within the region of PAI sequence identity. These results are consistent with there being two possible transcription start regions—one at S15a and one in more proximal PAI sequences—when the PAI1 gene is unmethylated.
Previous cDNA analysis in Col revealed PAI2 transcripts that initiated at 102 and 106 bp upstream of the translation start codon (Melquist et al. 1999). Col PAI 5′ RACE PCR analysis also revealed PAI2 transcripts that initiate at proximal sites ranging from 133 to 99 bp upstream of the translation start (Table 1). These patterns, together with the patterns observed for Col PAI1, indicate that proximal PAI promoter sequences are contained in the region of PAI sequence identity. Thus, in WS, where the proximal promoters are methylated and silenced, only the more distal unmethylated S15a promoter upstream of PAI1 is available as a transcription start site.
The PAI1–PAI4 inverted repeat generates both normal PAI1 and longer 3′ read-through polyadenylated transcripts
RNA gel blot analysis of PAI transcripts in total RNA prepared from either WS or Col revealed a prominent band of ∼1200 nt (Fig. 2A), the predicted size of the spliced PAI1 or PAI2 transcripts (Melquist et al. 1999). However, uniquely in WS, a heterogeneous population of higher molecular weight species was also detected, with a major band at ∼1900 nt and lower levels of longer species. Both the prominent PAI transcript band and the longer transcripts were recovered predominantly in the polyadenylated RNA fraction.
Figure 2.
PAI steady-state transcript analysis. (A) Total RNA (Total) from Col or WS was separated into polyadenylated (polyA+) and nonpolyadenylated (polyA–) fractions, gel blotted, and probed with the PAI probe. The blot was stripped and reprobed with β-tubulin (TUB) to control for loading of polyA+ species. The ethidium bromide (EtBr) stained gel is shown to control for loading of polyA–species. Molecular weight markers in kilonucleotides are indicated in the left margin. (B) Duplicate blots of polyadenylated RNA prepared from Col, WS, or Cvi-0 electrophoresed in parallel on the same gel were hybridized with either a PAI antisense strand (AS) or a PAI sense (S) strand RNA probe. Both a short (AS-S) and a long (AS-L) exposure of the same antisense strand-probed blot are shown, in the first case to resolve the predominant lower molecular weight species, and in the second case to clearly show the less-abundant higher molecular weight species. A single exposure of the sense strand-probed blot is shown. A nonspecific band detected by the sense strand probe is marked with an asterisk in the right margin. This band serves as an internal loading control for the sense strand blot. In addition, reprobing of both blots with a TUB probe indicated approximately equal loading of all samples (data not shown).
In previous studies, we found that Arabidopsis strains carrying methylated PAI1–PAI4 inverted repeats with different amounts of sequence separating the PAI1 and PAI4 genes displayed corresponding shifts in the size range of PAI higher molecular weight species detected by RNA gel blot (Melquist et al. 1999). These strains do not differ significantly from either Col or WS in their PAI1 5′ sequences or exon/intron structures. Taken together, these observations argue in favor of the longer species detected uniquely in PAI1–PAI4-containing strains as corresponding to PAI1 3′ read-through transcripts. However, we were unable to clone WS PAI1 transcripts with unusually long 3′ ends using 3′ RACE analysis (Table 2), perhaps because these palindromic species can form secondary structures that inhibit reverse transcription and/or PCR amplification, or because longer species are out-competed by the more abundant shorter species in the transcript population.
Table 2.
3 RACE determination of transcript ends for WS PAI1 and Col PAI1 and PAI2
| Transcript | PAI fifth exona |
|---|---|
| WS PAI1 | +1257 to +1546 |
| WS PAI1 | +1257 to +1613 |
| WS PAI1 | +1257 to +1613 |
| WS PAI1 | +1257 to +1613 |
| WS PAI1 | +1257 to +1613 |
| WS PAI1 | +1257 to +1613 |
| WS PAI1 | +1257 to +1613 |
| WS PAI1 | +1257 to +1615 |
| Col PAI1 | +1257 to +1613 |
| Col PAI1 | +1257 to +1613 |
| Col PAI1 | +1257 to +1613 |
| Col PAI1 | +1257 to +1613 |
| Col PAI1 | +1257 to +1616 |
| Col PAI1 | +1257 to +1636 |
| Col PAI1 | +1257 to +1694 |
| Col PAI1 | +1257 to +1725 |
| Col PAI2 | +1257 to +1604 |
| Col PAI2 | +1257 to +1613 |
| Col PAI2 | +1257 to +1613 |
| Col PAI2 | +1257 to +1613 |
| Col PAI2 | +1257 to +1613 |
| Col PAI2 | +1257 to +1616 |
| Col PAI2 | +1257 to +1733 |
| Col PAI2 | +1257 to +1745 |
Numbering is relative to the first base of the PAI ATG translation start codon as +1 on genomic sequences. Polyadenylation occurs after the last base position given for each PAI fifth exon.
To determine whether the high molecular weight PAI RNAs correspond to 3′ read-through RNAs with palindromic PAI4 sequences, we performed RNA gel blot analysis of polyadenylated transcripts using a PAI cDNA sense strand RNA probe (Fig. 2B). In this analysis, we included RNA prepared from Cvi-0, which has a PAI1–PAI4 inverted repeat separated by 839 bp more central sequence than in WS (Melquist et al. 1999). The PAI sense strand probe detected high molecular weight species in both WS and Cvi-0, but not in Col samples, with the longest species in each case corresponding in size to the longest species detected with a PAI antisense strand probe. These results show that the high molecular weight RNAs that accumulate in strains carrying a PAI1–PAI4 inverted repeat include transcripts that read through from PAI1 into the palindromic PAI4 sequences. However, the sense probe had weak or no hybridization to shorter species in WS and Cvi-0, presumably because these species do not extend far enough into palindromic PAI4 sequences to be effectively detected. In this regard, it should be noted that the PAI cDNA sense strand probe will not hybridize to central material between PAI1 and PAI4 or to PAI intron sequences. The PAI sense strand probe cross-hybridized to an unrelated transcript of ∼800 nt in all samples tested.
The endogenous S15a promoter canbe methylated and silenced by a transcribed inverted repeat of S15a sequences
To examine the role of transcription through the WS PAI1–PAI4 inverted repeat in maintaining PAI gene methylation, we used a transgene-directed promoter silencing strategy (Mette et al. 2000; Sijen et al. 2001) to silence the S15a promoter. The basic design of this strategy is to introduce a highly transcribed inverted repeat of target promoter sequences on a transgene into the plant genome. This type of construct was shown to produce dsRNA and smRNAs corresponding to the target sequence, and to direct cytosine methylation and silencing of the target sequence. In our S15aIR construct, we drove expression of an inverted repeat of 430 bp upstream of and including the S15a transcription start site from the strong Cauliflower Mosaic Virus 35S promoter (Fig. 1A). The S15aIR construct was transformed into the WS genome, and lines with single-copy inserts were isolated.
The WS(S15aIR) transgenic strain displayed strong reduction in PAI steady-state transcript levels and undetectable levels of S15a transcripts by RNA gel blot analysis (Fig. 3A). Additionally, WS(S15aIR) plants displayed a number of typical PAI-deficient phenotypes (Bender and Fink 1995; Bartee and Bender 2001) including blue fluorescence under ultraviolet (UV) light due to accumulation of an early intermediate in the tryptophan pathway, reduced size, reduced fertility, and increased bushiness (Fig. 3B). There was no obvious consequence of reduced S15a transcripts, either because there is still enough residual expression of the gene to provide ribosomal protein function, and/or because the gene is redundant with three other predicted S15a genes elsewhere in the genome. Together, the RNA gel blot analysis and fluorescent phenotype of the WS(S15aIR) strain indicate that the S15a promoter is efficiently silenced by the transgene.
Figure 3.

Expression and phenotypic analysis of WS(S15aIR) plants. (A) Total RNA gel blot analysis of PAI1 and S15a transcript levels in parental WS and WS(S15aIR) plants, with β-tubulin (TUB) as a loading control. (B) Visible (top) and UV (bottom) photographs of representative adult WS and WS(S15aIR) plants.
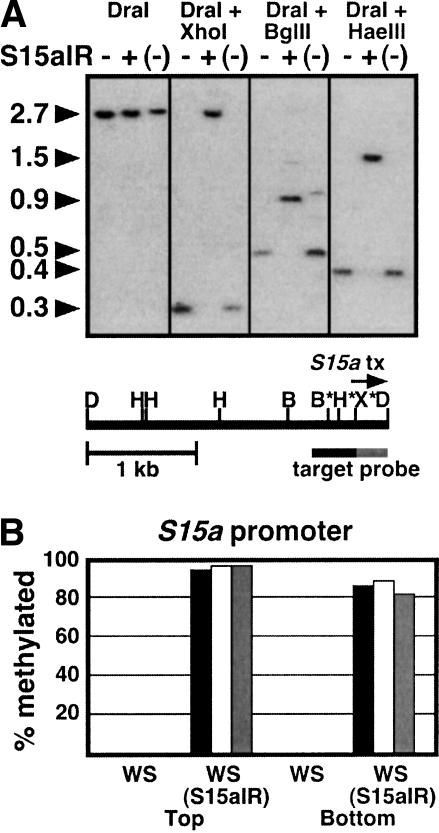
To confirm that the S15aIR transgene induces methylation on the target promoter sequence, as previously shown in transgene silencing systems, we assayed S15a methylation with two different approaches. First, we cleaved WS(S15aIR) genomic DNA with methylation-sensitive restriction endonucleases, and used DNA gel blot analysis with a probe specific for the endogenous S15a loci to detect cleavage at these target sites. This analysis showed that three different enzymes, XhoI (5′-CTCGAG-3′), BglII (5′-AGATCT-3′), and HaeIII (5′-GGCC-3′), were all inhibited from cleaving WS(S15aIR) but not WS DNA in the target S15a sequences (Fig. 4A). Reprobing of the blot with a transgene-specific probe lying just downstream of the S15aIR construct revealed that the S15a sequences on the transgene were also inhibited from cleavage (data not shown). These results indicate that both the endogenous S15a sequences and the transgene S15a sequences are methylated in the WS(S15aIR) strain.
Figure 4.
The targeted S15a promoter is methylated in WS(S15aIR) plants. (A) Southern blot analysis of S15a promoter methylation. Genomic DNA samples were prepared from WS, WS(S15aIR), and WS with the S15aIR transgene segregated away [WS(–)], shown as –, +, and (–), respectively. The restriction map of the S15a promoter region is shown in a diagram below the blot. The methylation-insensitive enzyme DraI (D) was used to release this region from flanking sequences. Cleavage within the region was then tested with the methylation-sensitive enzymes XhoI (X), BglII (B), or HaeIII (H). Sites within the transgene target sequence (black bar below the restriction map, corresponding to the black bars shown in Fig. 1A) are denoted with asterisks. The flanking probe sequence unique to the endogenous target loci is shown as a gray bar. The start point and direction of the S15a transcript (S15a tx) are indicated by an arrow above the restriction map. The molecular weight of each band is shown in kilobases. (B) Genomic bisulfite methylation sequencing data for the targeted S15a promoter region in WS and WS(S15aIR). Eight independent top strand or bottom strand clones were sequenced from each sample. The percentage of 5-methyl-cytosines of total cytosines sequenced for the 430-bp region targeted by the transgene is shown, divided into the contexts CG (black), CNG (white), and other contexts (gray).
Second, we determined specific methylation patterns across the endogenous S15a promoter target region using sodium bisulfite genomic sequencing (Frommer et al. 1992). This analysis showed that the region contained almost no methylated cytosines in untransformed WS, but that it was densely methylated in the WS(S15aIR) strain (Fig. 4B). Methylation was found in symmetrically disposed cytosine contexts (CG and CNG) as well as in other contexts. These data are consistent with the inhibition of cleavage at restriction sites carrying cytosines in different sequence contexts (Fig. 4A).
We also investigated the consequences for PAI1 expression and S15a promoter methylation when the S15aIR transgene was removed by genetic segregation. A segregating population generated by self-pollination of a plant that was hemizygous for the S15aIR transgene revealed that all of 14 progeny that segregated away from the transgene selectable marker were nonfluorescent or very weakly fluorescent. In the case of the weakly fluorescent segregants, the fluorescence did not persist in the next generation of self-pollination. These strains are termed WS(–) to indicate that they are epigenetic variants of WS. DNA prepared from a representative WS(–) segregant showed a loss of S15a promoter methylation via Southern blot assay (Fig. 4A). These observations indicate that the S15aIR construct does not leave a stable methylation imprint behind on the S15a promoter when it is removed by segregation.
Methylation of singlet PAI genes but not the PAI1–PAI4 inverted repeat is reduced in WS(S15aIR) plants
To determine the consequences for PAI gene methylation when transcription through the PAI1–PAI4 inverted repeat is suppressed by the S15aIR transgene, we used both Southern blot and bisulfite genomic sequencing assays for methylation. For Southern blot analysis, genomic DNA was cleaved with the isoschizomers HpaII and MspI (5′-CCGG-3′), which cleave once in each PAI locus. HpaII is inhibited by methylation of either cytosine (CG or CNG) in the recognition sequence, whereas MspI is only inhibited by methylation of the outer (CNG) cytosine. This analysis showed that cleavage in the PAI2 and PAI3 genes for both enzymes was increased in WS(S15aIR) versus WS or WS(–), but that there was no significant effect on PAI1–PAI4 cleavage patterns (Fig. 5A). Bisulfite genomic sequencing of the proximal promoter regions for the PAI1 and PAI2 genes in WS(S15aIR) also showed that PAI2 methylation was reduced, with a partial reduction in CG methylation and a stronger reduction in non-CG methylation (Fig. 5B). This PAI2 methylation profile resembles that previously determined in the “Hyb4” strain, a hybrid from a cross of WS and Col that retains the WS PAI2 and PAI3 genes but which has replaced the WS PAI1–PAI4 locus with the Col singlet PAI1 gene (Fig. 5B; Luff et al. 1999). Bisulfite genomic sequencing of the PAI1 proximal promoter region in the WS(S15aIR) strain showed a similar pattern to that previously determined for wild-type WS, except for a partial reduction in CG methylation. These results indicate that transcription through the inverted repeat is required for maintaining dense methylation on singlet PAI genes but not on the inverted repeat itself.
Figure 5.
Methylation is reduced on singlet PAI genes but not on the PAI1–PAI4 inverted repeat. (A) Genomic DNA samples prepared from WS, WS(S15aIR), and WS with the S15aIR transgene segregated away [WS(–)]—shown as –, +, and (–), respectively—were digested with the methylation-sensitive isoschizomers HpaII and MspI and probed with a PAI internal cDNA fragment. Methylated bands are denoted with asterisks. (P1–P4) PAI1–PAI4; (P2) PAI2; (P3) PAI3. The bands corresponding to the singlet genes are shown with black arrows and the bands corresponding to the inverted repeat are shown with open arrows. (B) Genomic bisulfite methylation sequencing data for PAI1 and PAI2 proximal promoter regions in WS(S15aIR). Eight independent top-strand clones were sequenced for PAI1 or PAI2. The percentage of 5-methyl-cytosines of total cytosines sequenced within the region of PAI sequence identity (344 bp for PAI1 or 338 bp for PAI2) is shown, divided into the contexts CG (black), CNG (white), and other contexts (gray). For comparison, previously determined wild-type WS PAI1 and PAI2 data and PAI2 data from Hyb4—a Col/WS hybrid with Col PAI1 and WS PAI2—are shown (Luff et al. 1999).
RNA silencing mutations and a dicer mutation do not affect PAI methylation
Aberrant RNAs in plants can trigger both RNA silencing of homologous transcripts and cytosine methylation of homologous genomic DNA (see introduction). Genetic screens in Arabidopsis have identified several loci required for RNA silencing and maintenance of DNA methylation on reporter transgenes that express aberrant RNAs, including SDE1, SDE2, SDE3, and SDE4 (Dalmay et al. 2000b, 2001). The SDE1/SGS2 locus encodes a predicted RNA-dependent RNA polymerase (Dalmay et al. 2000b; Mourrain et al. 2000), and the SDE3 locus encodes a predicted RNA helicase (Dalmay et al. 2001). These gene products are thought to be processing factors that convert precursor aberrant RNA species into dsRNA during RNA silencing and RNA-directed DNA methylation (Dalmay et al. 2000b, 2001; Béclin et al. 2002).
To assess the potential role of these RNA metabolism loci in PAI gene methylation, we monitored PAI methylation levels in mutant backgrounds using HpaII/MspI Southern blot analysis. The four sde mutants were isolated in the C24 strain background, which has similar PAI gene arrangements and methylation to WS (Melquist et al. 1999). Therefore, we assayed the sde mutant isolates directly relative to the untransformed C24 control and to the parental transgenic strain “GxA” used in the sde genetic screen (Fig. 6A). This analysis showed no obvious alteration in PAI methylation patterns in any of the four sde mutants. In contrast, all four isolates were previously found to reduce methylation on the reporter transgenes used in the sde genetic screen (Dalmay et al. 2000b, 2001). Our result argues that PAI1–PAI4 directly produces a methylation-inducing RNA, such as a dsRNA, without the need for processing by SDE factors.
Figure 6.
PAI methylation is not reduced in sde or dcl1 mutant plants. (A) Genomic DNA from wild-type C24, parental GxA (a transgenic line of C24), or the indicated sde mutants in the GxA background was digested with HpaII (H) or MspI (M) and probed with the PAI internal cDNA probe. Methylated bands are denoted with asterisks. (P1–P4) PAI1–PAI4; (P2) PAI2; (P3) PAI3. Note that in C24 there are three MspI sites in PAI1–PAI4 affected by methylation (Melquist et al. 1999). (B) Representative sibling DCL1/dcl1 and dcl1/dcl1 plants in the WS pai1 reporter background are shown photographed under visible (top) and UV (bottom) light. (C) Genomic DNA from the WS pai1 parental strain or a representative line of pai1 dcl1 was digested with HpaII or MspI and probed with the PAI internal cDNA probe. PAI species are denoted as in A.
We also tested whether dicer function might be involved in PAI methylation via processing PAI1–PAI4 dsRNA read-through transcripts into smRNAs. The dicer-like1 (dcl1) allele that we used for this analysis, dcl1-9, is a kanamycin resistance insertional disruption that confers developmental pleiotropy and sterility when homozygous (Jacobsen et al. 1999; Schauer et al. 2002). This mutation has been previously shown to block processing of endogenous micro-RNAs in Arabidopsis (Park et al. 2002; Reinhart et al. 2002; Kasschau et al. 2003), but not small RNAs associated with RNA silencing triggered by a high-expression inverted repeat transgene construct (Finnegan et al. 2003). Because dcl1 was isolated in a strain background with three unmethylated PAI genes, we crossed this allele into the WS pai1 reporter background to monitor effects on PAI gene silencing and DNA methylation. The WS pai1 reporter strain carries a crippling missense mutation in the PAI1 gene, and displays strong blue fluorescence and other PAI-deficient phenotypes because the only other functional PAI gene, PAI2, is densely methylated and silenced (Bartee and Bender 2001; Bartee et al. 2001; Malagnac et al. 2002). Mutations that reduce PAI2 methylation and silencing confer reduced fluorescence in this background, which is easily monitored by inspection under UV light.
The effects of dcl1 on PAI silencing and methylation were examined by segregating dcl1/dcl1 homozygous progeny from dcl1/DCL1 heterozygous parent plants in the WS pai1 fluorescent background. The dcl1 homozygous seedlings were strongly fluorescent, similar to their DCL1 siblings (Fig. 6B). In HpaII/MspI Southern blot analysis, pai1 dcl1 genomic DNA showed a similar cleavage pattern to pai1 DCL1 parental strain genomic DNA for the pai1–PAI4 and PAI2 loci inherited from the methylated WS background (Fig. 6C). Note that one allele of the PAI3 locus was inherited from the PAI-unmethylated dcl1 parent in the cross (Materials and Methods), accounting for the presence of a PAI3 demethylated species. These results indicate that the dcl1 mutation has no significant effect on PAI silencing or methylation.
PAI smRNAs are not detected in WS
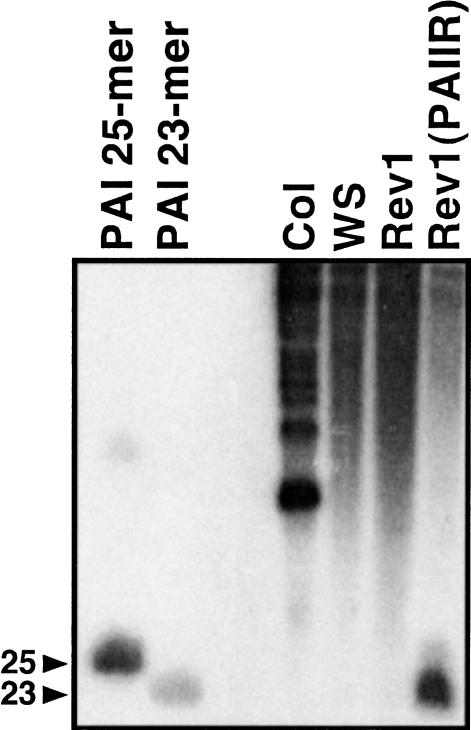
As another means of testing whether diced smRNAs might be involved in PAI methylation, we assayed directly for PAI smRNAs in WS. As a negative control for this experiment, we used RNA prepared from the Col strain, which lacks a PAI1–PAI4 inverted repeat and which lacks PAI methylation. We also used RNA prepared from the Rev1strain, which is a derivative of WS in which the PAI1–PAI4 inverted repeat locus is deleted and the residual methylation on the PAI2 and PAI3 genes has been lost (Bender and Fink 1995). As a positive control, we used RNA prepared from a transgenic strain, Rev1(PAIIR), carrying an inverted repeat of PAI cDNA sequences driven by the 35S promoter. The untransformed Rev1strain is nonfluorescent, but introduction of the PAIIR transgene in single copy induces a PAI-deficient fluorescent phenotype, presumably via RNA silencing. The PAI cDNA sequences on the transgene and the corresponding PAI endogenous sequences are methylated in the Rev1(PAIIR) strain (data not shown).
Analysis of PAI smRNAs in these strains revealed that there were no detectable 21–25 nt RNAs in the Col, WS, or Rev1backgrounds (Fig. 7). In contrast, PAI smRNAs were readily detectable in Rev(PAIIR) plants, with a prominent lower band and a fainter higher band, similar to the pattern previously observed for a GFP transgene undergoing RNA silencing (Dalmay et al. 2000b).
Figure 7.
PAI small RNAs are not detected in WS. Small RNA fractions were prepared from Col, WS, Rev1, and Rev1(PAIIR), and hybridized with a PAI cDNA antisense probe. Positions of 23 and 25 nt DNA PAI sense strand oligomers are denoted with arrows. Hybridization with a PAI cDNA sense probe gave similar results (data not shown).
Discussion
Cytosine methylation and accompanying changes in chromatin structure play an important role in regulation of eukaryotic gene expression and genome stability. A key issue is how methylation is targeted to particular genomic regions. We characterized the nature of methylation signals for the methylated endogenous PAI genes in the WS strain of Arabidopsis using genetic and biochemical methods. Previous experiments showed that the PAI1–PAI4 inverted repeat arrangement of the PAI genes in WS provides a trigger for PAI methylation (Bender and Fink 1995; Luff et al. 1999; Melquist et al. 1999). We found that this inverted repeat is transcribed from an upstream unmethylated promoter. To test the hypothesis that an RNA species produced from PAI1–PAI4 triggers PAI methylation, we suppressed transcription from the upstream promoter using a transgene that directs methylation and silencing of the promoter sequences. Suppression of transcription through PAI1–PAI4 resulted in reduced methylation on singlet PAI genes, suggesting that a PAI RNA species emanating from PAI1–PAI4 targets methylation to these genes. An alternative possibility is that the reduced methylation results as an indirect consequence of chromatin structure changes at PAI1–PAI4, but given the body of evidence for RNA-based DNA methylation signals in plants, we think this alternative model is unlikely.
In plant genomes, methylated regions have a basal level of methylation found primarily in CG dinucleotide contexts. For example, the centromere-associated repeats in Arabidopsis carry mainly CG methylation (Vongs et al. 1993). This CG patterning is thought to be the most easily maintained after each round of DNA replication. However, a subset of methylated regions in the Arabidopsis genome, such as the PAI genes, have an additional layer of methylation found in non-CG cytosine contexts. Previously, it was suggested that this non-CG methylation is a hallmark of RNA-directed DNA methylation, based on methylation patterning triggered by an RNA viroid (Pélissier et al. 1999). In accord with this view, the S15aIR transgene triggers dense CG and non-CG methylation on its target promoter (Fig. 4). Therefore, the high proportion of non-CG methylation on the WS PAI genes (Luff et al. 1999) suggests an RNA trigger for methylation. The residual CG methylation found on the singlet PAI genes when transcription is suppressed in the WS(S15aIR) strain (Fig. 5) likely reflects a basal level of maintenance methylation that persists when the signal from the inverted repeat is impaired.
It has been proposed that either precursor dsRNAs or diced smRNAs processed from dsRNA could constitute the RNA-directed DNA methylation signal, because smRNAs typically accumulate in plant transgene and virus RNA silencing/DNA methylation systems. The endogenous PAI gene methylation system differs from previously characterized systems in that smRNA species associated with RNA silencing are not detectable by RNA gel blot analysis (Fig. 7), and full-length PAI transcripts, including 3′ read-through species, are not efficiently degraded (Fig. 2). In addition, PAI1–PAI4-triggered methylation accumulates slowly over several generations of inbreeding (Luff et al. 1999; Malagnac et al. 2002) relative to methylation triggered by 35S promoter-driven transgenes or RNA viruses that produce high levels of RNAs (Jones et al. 1998, 1999; Mette et al. 1999, 2000; Dalmay et al. 2000a,b; Mourrain et al. 2000; Wang et al. 2001). Taken together, these differences can be accounted for by the relatively low level of transcription through the endogenous PAI1–PAI4 locus producing insufficient aberrant RNAs for effective RNA silencing or rapid DNA methylation. However, the PAI aberrant RNAs are effective for progressive DNA methylation of target loci.
Our results are consistent with either of two general models for RNA-directed DNA methylation: PAI methylation could be signaled directly by dsRNA produced by read-through transcription from PAI1 into PAI4, or it could be signaled by low levels of smRNAs. In the second case, a dicer activity other than DCL1would be involved in RNA processing, because a dcl1 mutation has no effect on PAI methylation (Fig. 6C). Recently, work from several groups has led to the view that plants contain two distinct dicer activities: the DCL1activity needed to process micro-RNAs, and a different activity needed to process inhibitory smRNAs associated with RNA silencing (siRNAs; Hamilton et al. 2002; Finnegan et al. 2003; Tang et al. 2003). Thus, if PAI methylation is triggered by smRNAs, they are most likely produced by the siRNA dicer(s). In this regard, there are three other putative dicer-encoding genes in the Arabidopsis genome (Schauer et al. 2002; Finnegan et al. 2003). Mutations in these genes have not been recovered from forward genetic screens for RNA silencing mutants, suggesting that such dicer mutants may be redundant or lethal. It should be noted that the single dicer-encoding gene in Schizosaccharomyces pombe is required for centromeric heterochromatin formation, arguing that this fungal system uses smRNAs rather than unprocessed precursors to communicate with homologous genomic DNA (Volpe et al. 2002). Regardless of whether the PAI methylation signal is unprocessed dsRNA or low levels of smRNAs, our results show that efficient DNA methylation can be separated from efficient RNA silencing on the basis of the amount of aberrant RNA produced from the affected locus.
Although we can detect PAI1–PAI4 read-through transcripts in the polyadenylated fraction of RNAs (Fig. 2B), we do not know whether the methylation trigger species, which may be subdetectable by RNA gel blot, is processed by splicing or polyadenylation. Dense methylation is found on intron as well as exon sequences of the PAI genes (Luff et al. 1999), suggesting that unspliced RNA is present in the trigger population. Capturing, cloning, and sequencing rare PAI smRNAs produced in WS might shed light on which regions of PAI1 and PAI4 are present in precursor dsRNA species.
Interestingly, PAI1–PAI4 methylation itself is not affected by reduced transcription (Fig. 5). This result is in agreement with a similar observation from a transgenic promoter silencing system (Mette et al. 1999), and with our previous observation that a promoterless pai1–pai4 inverted repeat transgene can trigger its own methylation but not the methylation of endogenous singlet PAI genes (Luff et al. 1999). A possible explanation is that the unusual inverted repeat structure of the locus might provide an intrinsic RNA-independent methylation signal. Alternatively, methylation-inducing RNA produced from the inverted repeat could act much more efficiently in cis on the inverted repeat itself than in trans on singlet PAI genes, so that the inverted repeat could maintain methylation even with extremely low levels of RNA.
We recently found that mutation of the histone methyltransferase SUVH4 confers a similar PAI methylation profile to that observed in the WS(S15aIR) strain: reduced non-CG methylation on singlet PAI genes but unaffected methylation on PAI1–PAI4 (Malagnac et al. 2002). This finding suggests that SUVH4-mediated histone modifications could act to transmit an RNA signal into efficiently maintained dense DNA methylation patterning. In this model, the methylation on PAI1–PAI4 would be SUVH4 independent either because it is RNA independent, or because there is a sufficiently high accumulation of cis-acting RNA to bypass the need for SUVH4.
We have shown that a signal for dense methylation of identical sequences at unlinked positions in a plant genome can be generated by relatively low levels of transcription through an inverted repeat gene arrangement, given sufficient generations for the DNA methylation to accumulate. This signal comprises either unprocessed dsRNAs or diced smRNAs, which presumably interact with homologous DNA sequences and recruit cytosine methyltransferases. By extension, other sequence arrangements that can produce even low levels of dsRNA, such as dispersed transposable elements that are transcribed through on sense or antisense strands from flanking promoters, could potentially provide a DNA methylation signal. For DNA-based transposons that move via a cut-and-paste mechanism, methylation of transposon ends could prevent movement by blocking access of transposase proteins. Thus, even levels of dsRNA too low to be effectively processed by the RNA silencing machinery into an RNA degradation signal could still target a defense against invasive repeated sequences.
Materials and methods
RNA extraction and analysis
Total RNA was isolated from 4-week-old plants as previously described (Nagy et al. 1988). Polyadenylated RNA was separated from nonpolyadenylated RNA using the Dynabeads mRNA extraction kit (Dynal). RNA gel blots were performed as previously described (Melquist et al. 1999). The PAI probe was an internal 0.7-kb PstI Col PAI1 (At1g07780) cDNA fragment (Bender and Fink 1995). The S15a (At1g07770) probe was an internal 400-bp cDNA fragment. A β-tubulin (At5g44340) cDNA probe was used to control for loading differences. Ambion Millenium RNA markers were used to determine transcript length.
For RACE analysis, 1μg of total RNA was used as input for first-strand synthesis with the SMART RACE cDNA amplification kit (Clontech). A PAI-specific reverse primer was used to amplify the 5′ ends of transcripts: P135 (5′-GATTCCCCCAGC TAAGAGCCACCC-3′). This primer is a perfect match to all the PAI genes in Col and WS. A PAI-specific forward primer was used to amplify the 3′ ends of transcripts: P134 (5′-TGAGCTGC AGcGTTTCCAACACAGAG-3′). The P134 primer has complete identity with WS PAI1 and PAI4; one mismatch with WS PAI2, Col PAI1, and Col PAI2 (lowercase); and an additional mismatch with WS and Col PAI3 (underlined). Modified amplification conditions from manufacturer's protocols were one cycle of 94°C for 5 min; five cycles of 94°C for 30 sec, 70°C for 1min, and 72°C for 3 min; five cycles of 94°C for 30 sec, 68°C for 1min, and 72°C for 3 min; 32 cycles of 94°C for 30 sec, 60°C for 1min, and 72°C for 3 min; and one cycle of 72°C for 10 min. PCR products were run on 0.8% agarose gels. DNA was isolated from generous areas surrounding prominent bands and cloned using the pGEM T-EASY vector system (Promega). For each species, eight clones were sequenced to obtain an overview of the population (Tables 1, 2). The 5′ and 3′ RACE data agree with previous PAI cDNA sequencing results (Melquist et al. 1999).
Small RNAs were isolated according to previously described methods (Hamilton and Baulcombe 1999; Dalmay et al. 2000a; Mette et al. 2000). For total RNA preparation, 1g of frozen ground tissue was resuspended in 5 mL extraction buffer (50 mM Tris-HCl at pH 8.5, 10 mM EDTA at pH 8.0, 100 mM NaCl, 2% SDS) and incubated at 55°C for 5 min. Samples were placed on ice for 2 min, and then extracted twice with phenol:chloroform. After the second extraction, samples were ethanol precipitated, and the pellets were resuspended in 3.87 mL of water and incubated for 15 min at 65°C. Enrichment of the small RNA fraction was performed as described (Dalmay et al. 2000a). Fifty micrograms of RNA was run at 400 volts for 4 h on a 15% polyacrylamide, 7 M Urea, 0.5× TBE gel and transferred to membrane as described (Hamilton and Baulcombe 1999; Dalmay et al. 2000a). Blots were hybridized with PAI1 0.7-kb PstI internal cDNA antisense or sense strand RNA probes in 50% formamide, 7% SDS, 50 mM sodium phosphate buffer (pH 7.2), 0.3 M NaCl, 5× Denhardt's solution, and 100 μg/mL salmon sperm DNA overnight at 40°C and washed three times with 2× SSC, 0.2% SDS at 50°C.
Plant strains
To make the WS(S15aIR) line, WS was transformed with the S15aIR construct by an Agrobacterium-mediated in planta method (Clough and Bent 1998). The construct was made by inserting a PCR-amplified 430-bp segment of the S15a promoter into the pFGC1008 vector (http://www.chromdb.org/fgc1008.html) in the AscI–SwaI and in the BamHI–SpeI polylinker sites to create an inverted repeat of this segment flanking a central GUS gene-derived spacer sequence. Primers used were S15a5 (5′-GCACTAGTGGCGCGCCGGAAGTTGATTGAAGTAG-3′) and S15a6 (5′-GCGGATCCATTTAAATTGTACCTTAAGAT TGGGAG-3′). Three independent single-copy lines were identified by Southern blot analysis with end-specific probes. Data for a single representative line are shown. The Rev1(PAIIR) line was made by similar methods, using a 735-bp fragment of a Col PAI1 cDNA inserted as an inverted repeat in the pFGC1008 vector. Primers used were PAIIRF (5′-GGACTAGTGGCGCGC CACTGATCTCCATGT-3′) and PAIIRR (5′-CGGGATCCATT TAAATACCGCTACTAACAT-3′). Note that the PAI segment in this construct is coextensive with the PAI cDNA probe used for RNA blot analysis (Fig. 7).
To introduce the dcl1-9 insertion allele from a strain background with unmethylated Ler alleles at all three PAI loci into the WS pai1 reporter strain background (Bartee and Bender 2001), the two strains were crossed and F2 progeny were selected for kanamycin resistance diagnostic of the dcl1 insertion (either heterozygous or homozygous) and screened for blue fluorescence diagnostic of homozygous methylated WS pai1-PAI4 and WS PAI2 loci. Eight fluorescent DCL1/dcl1 F2 individuals were identified based on normal floral morphology, and their PAI genotypes at all three PAI loci were determined with PCR-based markers (Luff et al. 1999). F3 progeny segregating for the dcl1 mutation were analyzed from each of the eight lines, and in every case the dcl1 homozygous mutants showed similar fluorescence intensity to their DCL1 siblings throughout all stages of growth. For two representative lines, which were both heterozygous Ler/WS at the PAI3 locus, eight F3 adult plants displaying the characteristic dcl1 morphological defects were pooled to prepare genomic DNA for Southern blot analysis of methylation.
DNA analysis
Southern blots were performed as previously described (Melquist et al. 1999) except that the low molecular weight fragments shown in Figure 4 were resolved on a 1.5% agarose gel. Bisulfite sequencing was performed as previously described (Luff et al. 1999) except that genomic DNA was cleaved with EcoRV prior to treatment, and only 5 μg of DNA was used. PAI PCR products were cloned into pBluescript KS II+ (Stratagene) and S15a promoter PCR products were cloned into pGEM T-EASY (Promega) for sequencing. For each region, eight independent clones were sequenced and the compiled data are shown in Figures 4 and 5. The PAI1 top strand was amplified with primers PITF (5′-GCTCTAGATGYAGAATTYTGTGYATTTG-3′) and PITR (5′-CGGGATCCTRTTRACATCTTAATTTCAC-3′). The PAI2 top strand was amplified with primers P2TF (5′-GCTCTA GATTAATGTTTYGAAGATGATAAG-3′) and PITR. These primers are the same as those used for previous PAI proximal promoter region methylation analysis (Luff et al. 1999; Bartee and Bender 2001; Bartee et al. 2001; Malagnac et al. 2002). They amplify segments extending from upstream sequences unique to each gene through the PAI-identical regions just upstream of the translation start codon. The S15a top strand was amplified with primers S15aTF2 (5′-GCTCTAGAGGGAAAAGGAAGTT GATTGAAG-3′) and S15aTR (5′-CGGGATCCAATTARCARC TATCTCACACTTC-3′). The S15a bottom strand was amplified with primers S15aBF2 (5′-CTTCCCCRCCTTRRRTTTCTT CTCC-3′) and S15aBR2 (5′-GATAAGYAGATYAGATTYTG-3′). These primers immediately flank the region targeted for methylation by the S15aIR construct.
Acknowledgments
We thank the Arabidopsis Biological Resource Center for providing the dcl1-9 mutant strain (CS3828), D. Baulcombe for providing the GxA and sde mutant strains, and M.F. Mette and M. Matzke for advice on small RNA isolation and detection. This work was supported by National Institutes of Health grant GM61148 to J.B. and by National Institutes of Environmental Health Sciences Training Grant ES 07141 to S.M.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Article published online ahead of print. Article and publication date are at http://www.genesdev.org/cgi/doi/10.1101/gad.1081603.
References
- Assaad, F.F., Tucker, K.L., and Signer, E.R. 1993. Epigenetic repeat-induced gene silencing (RIGS) in Arabidopsis. Plant Mol. Biol. 22: 1067–1085. [DOI] [PubMed] [Google Scholar]
- Bartee, L. and Bender, J. 2001. Two Arabidopsis methylationdeficiency mutations confer only partial effects on a methylated endogenous gene family. Nucleic Acids Res. 29: 2127–2134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartee, L., Malagnac, F., and Bender, J. 2001. Arabidopsis cmt3 chromomethylase mutations block non-CG methylation and silencing of an endogenous gene. Genes & Dev. 15: 1753–1758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Béclin, C., Boutet, S., Waterhouse, P., and Vaucheret, H. 2002. A branched pathway for transgene-induced RNA silencing in plants. Curr. Biol. 12: 684–688. [DOI] [PubMed] [Google Scholar]
- Bender, J. 2001. A vicious cycle: RNA silencing and DNA methylation in plants. Cell 106: 129–132. [DOI] [PubMed] [Google Scholar]
- Bender, J. and Fink, G.R. 1995. Epigenetic control of an endogenous gene family is revealed by a novel blue fluorescent mutant of Arabidopsis. Cell 83: 725–734. [DOI] [PubMed] [Google Scholar]
- Bird, A. 2002. DNA methylation patterns and epigenetic memory. Genes & Dev. 16: 6–21. [DOI] [PubMed] [Google Scholar]
- Clough, S.J. and Bent, A.F. 1998. Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743. [DOI] [PubMed] [Google Scholar]
- Dalmay, T., Hamilton, A., Mueller, E., and Baulcombe, D.C. 2000a. Potato Virus X amplicons in Arabidopsis mediate genetic and epigenetic gene silencing. Plant Cell 12: 369–379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalmay, T., Hamilton, A., Rudd, S., Angell, S., and Baulcombe, D.C. 2000b. An RNA-dependent RNA polymerase gene in Arabidopsis is required for posttranscriptional gene silencing mediated by a transgene but not by a virus. Cell 101: 543–553. [DOI] [PubMed] [Google Scholar]
- Dalmay, T., Horsefield, R., Braunstein, T.H., and Baulcombe, D.C. 2001. SDE3 encodes an RNA helicase required for post-transcriptional gene silencing in Arabidopsis. EMBO J. 20: 2069–2077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finnegan, E.J., Genger, R.K., Peacock, W.J., and Dennis, E.S. 1998. DNA methylation in plants. Annu. Rev. Plant Physiol. Plant Mol. Biol. 49: 223–247. [DOI] [PubMed] [Google Scholar]
- Finnegan, E.J., Margis, R., and Waterhouse, P.M. 2003. Posttranscriptional gene silencing is not compromised in the Arabidopsis CARPEL FACTORY (DICER-LIKE1) mutant, a homolog of Dicer-1from Drosophila. Curr. Biol. 13: 236–240. [DOI] [PubMed] [Google Scholar]
- Frommer, M., McDonald, L.E., Millar, D.S., Collis, C.M., Watt, F., Grigg, G.W., Molloy, P.L., and Paul, C.L. 1992. A genomic sequencing protocol that yields a positive display of 5-methylcytosine residues in individual DNA strands. Proc. Natl. Acad. Sci. 89: 1827–1831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrick, D., Fiering, S., Martin, D.I.K., and Whitelaw, E. 1998. Repeat-induced gene silencing in mammals. Nat. Genet. 18: 56–59. [DOI] [PubMed] [Google Scholar]
- Hamilton, A.J. and Baulcombe, D.C. 1999. A species of small antisense RNA in posttranscriptional gene silencing in plants. Science 286: 950–952. [DOI] [PubMed] [Google Scholar]
- Hamilton, A., Voinnet, O., Chappell, L., and Baulcombe, D. 2002. Two classes of short interfering RNA in RNA silencing. EMBO J. 21: 4671–4679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henikoff, S. 1998. Conspiracy of silence among repeated transgenes. Bioessays 20: 532–535. [DOI] [PubMed] [Google Scholar]
- Jacobsen, S.E., Running, M.P., and Meyerowitz, E.M. 1999. Disruption of an RNA helicase/RNAse III gene in Arabidopsis causes unregulated cell division in floral meristems. Development 126: 5231–5243. [DOI] [PubMed] [Google Scholar]
- Jones, A.L., Thomas, C.L., and Maule, A.J. 1998. De novo methylation and co-suppression induced by a cytoplasmically replicating plant RNA virus. EMBO J. 17: 6385–6393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones, L., Hamilton, A.J., Voinnet, O., Thomas, C.L., Maule, A.J., and Baulcombe, D.C. 1999. RNA–DNA interactions and DNA methylation in post-transcriptional gene silencing. Plant Cell 11: 2291–2301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasschau, K.D., Xie, Z., Allen, E., Llave, C., Chapman, E.J., Krizan, K.A., and Carrington, J.C. 2003. P1/HC-Pro, a viral suppressor of RNA silencing, interferes with Arabidopsis development and miRNA function. Dev. Cell 4: 205–217. [DOI] [PubMed] [Google Scholar]
- Kato, M., Miura, A., Bender, J., Jacobsen, S.E., and Kakutani, T. 2003. Role of CG and non-CG methylation in immobilization of transposons in Arabidopsis. Curr. Biol. 13: 421–426. [DOI] [PubMed] [Google Scholar]
- Luff, B., Pawlowski, L., and Bender, J. 1999. An inverted repeat triggers cytosine methylation of identical sequences in Arabidopsis. Mol. Cell 3: 505–511. [DOI] [PubMed] [Google Scholar]
- Malagnac, F., Bartee, L., and Bender, J. 2002. An Arabidopsis SET domain protein required for maintenance but not establishment of DNA methylation. EMBO J. 21: 6842–6852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martienssen, R.A. and Colot, V. 2001. DNA methylation and epigenetic inheritance in plants and filamentous fungi. Science 293: 1070–1074. [DOI] [PubMed] [Google Scholar]
- Matzke, M., Matzke, A.J.M., and Kooter, J.M. 2001. RNA: Guiding gene silencing. Science 293: 1080–1083. [DOI] [PubMed] [Google Scholar]
- Melquist, S., Luff, B., and Bender, J. 1999. Arabidopsis PAI gene arrangements, cytosine methylation and expression. Genetics 153: 401–413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mette, M.F., van der Winden, J., Matzke, M.A., and Matzke, A.J.M. 1999. Production of aberrant promoter transcripts contributes to methylation and silencing of unlinked homologous promoters intrans. EMBO J. 18: 241–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mette, M.F., Aufsatz, W., van der Winden, J., Matzke, M.A., and Matzke, A.J.M. 2000. Transcriptional silencing and promoter methylation triggered by double-stranded RNA. EMBO J. 19: 5194–5201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miura, A., Yonebayashi, S., Watanabe, K., Toyama, T., Shimada, H., and Kakutani, T. 2001. Mobilization of transposons by a mutation abolishing full DNA methylation in Arabidopsis. Nature 411: 212–214. [DOI] [PubMed] [Google Scholar]
- Mourrain, P., Béclin, C., Elmayan, T., Feuerbach, F., Godon, C., Morel, J.-B., Jouette, D., Lacombe, A.-M., Nikic, S., Picault, N., et al. 2000. Arabidopsis SGS2 and SGS3 genes are required for posttranscriptional gene silencing and natural virus resistance. Cell 101: 533–542. [DOI] [PubMed] [Google Scholar]
- Nagy, F., Kay, S.A., and Chua, N.-H. 1988. Analysis of gene expression in transgenic plants. In Plant molecular biology manual (eds. S.B. Gelvin and R.A. Schilperoort), pp. B4/11–B4/12. Kluwer Academic Publishers, Dordrecht, Netherlands.
- Park, W., Li, J., Song, R., Messing, J., and Chen, X. 2002. CARPEL FACTORY, a dicer homolog, and HEN1, a novel protein, act in microRNA metabolism in Arabidopsis thaliana. Curr. Biol. 12: 1484–1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pélissier, T., Thalmeir, S., Kempe, D., Sänger, H.-L., and Wassenegger, M. 1999. Heavy de novo methylation at symmetrical and non-symmetrical sites is a hallmark of RNA-directed DNA methylation. Nucleic Acids Res. 27: 1625–1634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhart, B.J., Weinstein, E.G., Rhoades, M.W., Bartel, B., and Bartel, D.P. 2002. MicroRNAs in plants. Genes & Dev. 16: 1616–1626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schauer, S.E., Jacobsen, S.E., Meinke, D.W., and Ray, A. 2002. DICER-LIKE1: Blind men and elephants in Arabidopsis development. Trends Plant Sci. 7: 487–491. [DOI] [PubMed] [Google Scholar]
- Sijen, T., Vijn, I., Rebocho, A., van Blokland, R., Roelofs, D., Mol, J.N.M., and Kooter, J.M. 2001. Transcriptional and posttranscriptional gene silencing are mechanistically related. Curr. Biol. 11: 436–440. [DOI] [PubMed] [Google Scholar]
- Singer, T., Yordan, C., and Martienssen, R.A. 2001. Robertson's Mutator transposons in A. thaliana are regulated by the chromatin-remodeling gene Decrease inDNA Methylation (DDM1). Genes & Dev. 15: 591–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang, G., Reinhart, B.J., Bartel, D.P., and Zamore, P.D. 2003. A biochemical framework for RNA silencing in plants. Genes & Dev. 17: 49–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volpe, T.A., Kidner, C., Hall, I.M., Teng, G., Grewal, S.I.S., and Martienssen, R.A. 2002. Regulation of heterochromatic silencing and histone H3 lysine-9 methylation by RNAi. Science 297: 1833–1837. [DOI] [PubMed] [Google Scholar]
- Vongs, A., Kakutani, T., Martienssen, R.A., and Richards, E.J. 1993. Arabidopsis thaliana DNA methylation mutants. Science 260: 1926–1928. [DOI] [PubMed] [Google Scholar]
- Wang, M.-B., Wesley, S.V., Finnegan, E.J., Smith, N.A., and Waterhouse, P.M. 2001. Replicating satellite RNA induces sequence-specific DNA methylation and truncated transcripts in plants. RNA 7: 16–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wassenegger, M., Heimes, S., Riedel, L., and Sänger, H.L. 1994. RNA-directed de novo methylation of genomic sequences in plants. Cell 76: 567–576. [DOI] [PubMed] [Google Scholar]
- Yoder, J.A., Walsh, C.P., and Bestor, T.H. 1997. Cytosine methylation and the ecology of intragenomic parasites. Trends Genet. 13: 335–340. [DOI] [PubMed] [Google Scholar]