Abstract
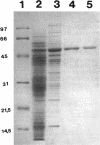
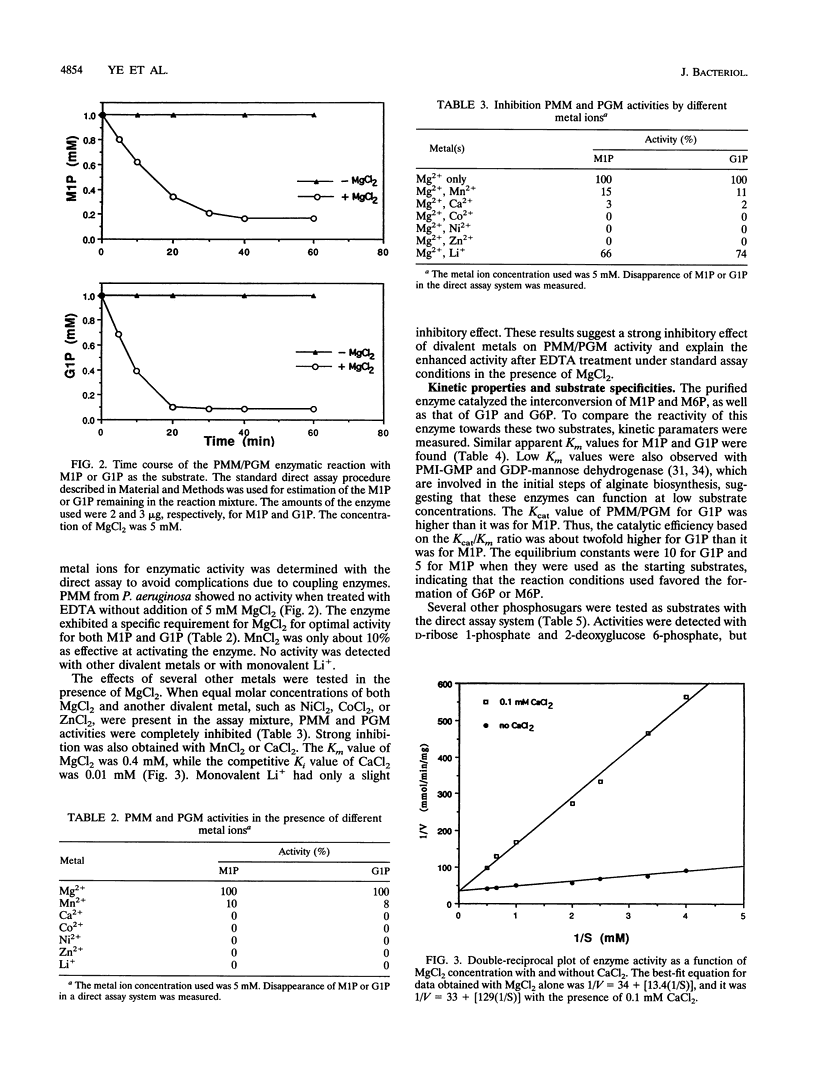
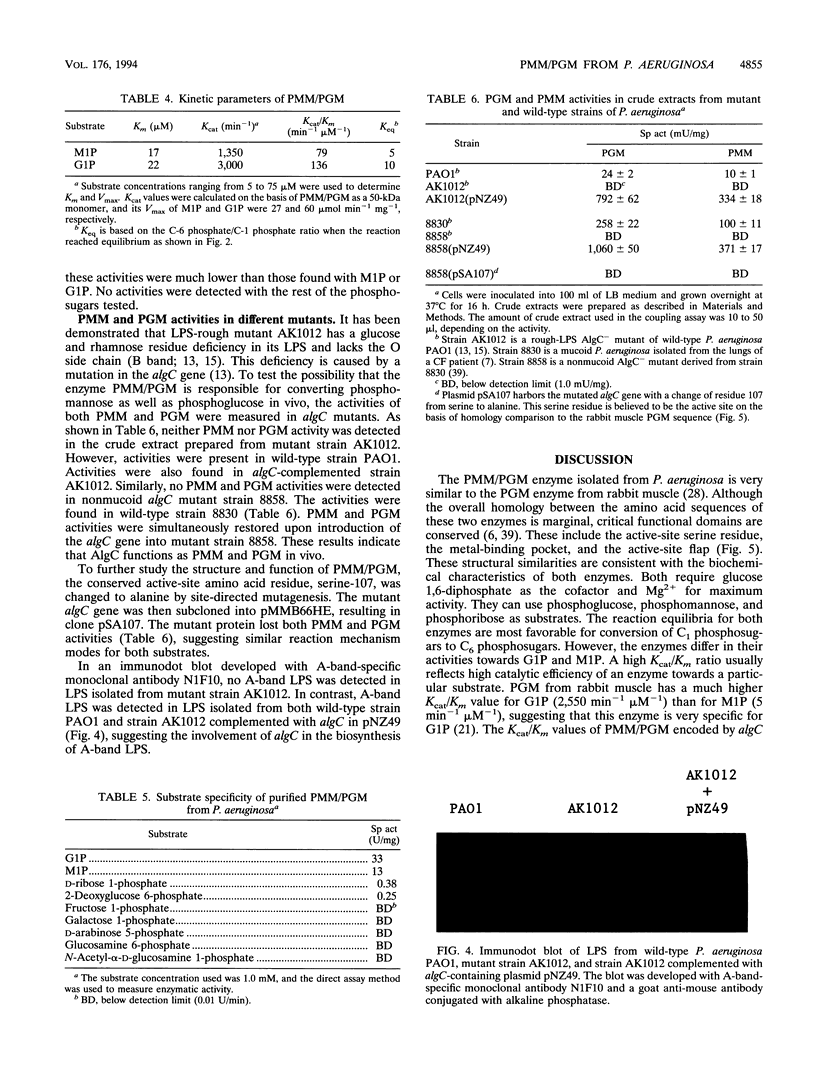
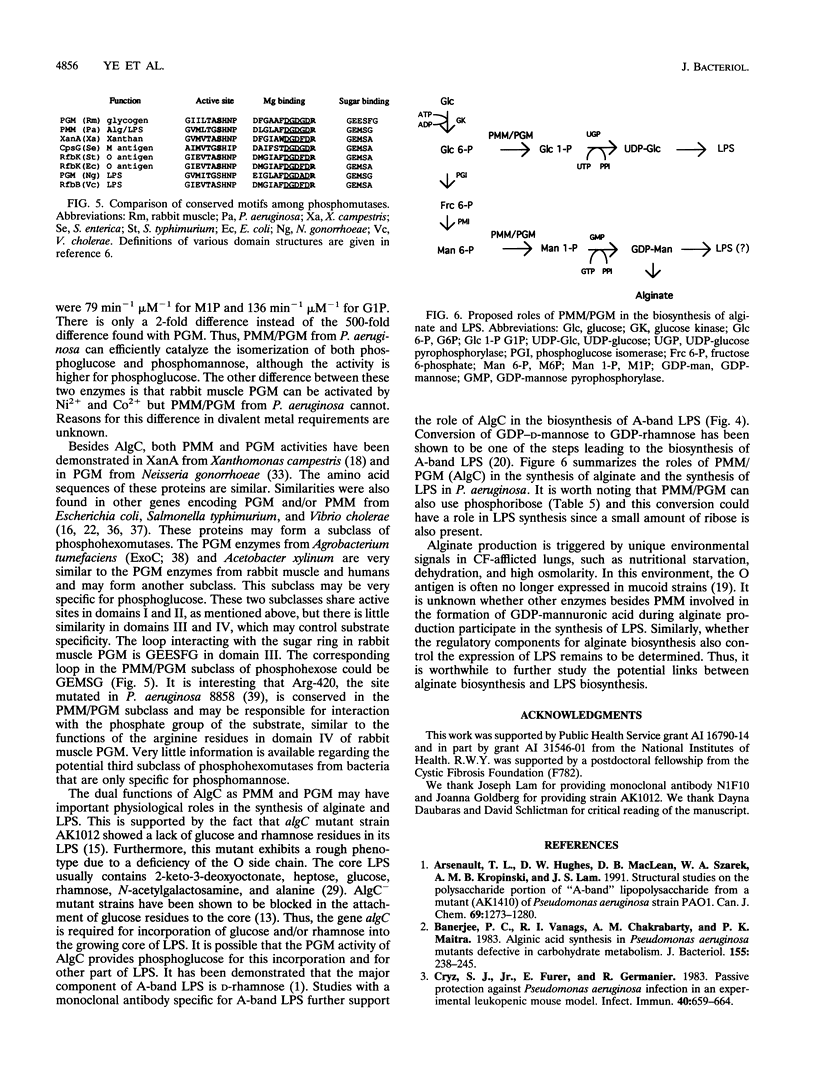
The algC gene from Pseudomonas aeruginosa has been shown to encode phosphomannomutase (PMM), an essential enzyme for biosynthesis of alginate and lipopolysaccharide (LPS). This gene was overexpressed under control of the tac promoter, and the enzyme was purified and its substrate specificity and metal ion effects were characterized. The enzyme was determined to be a monomer with a molecular mass of 50 kDa. The enzyme catalyzed the interconversion of mannose 1-phosphate (M1P) and mannose 6-phosphate, as well as that of glucose 1-phosphate (G1P) and glucose 6-phosphate. The apparent Km values for M1P and G1P were 17 and 22 microM, respectively. On the basis of Kcat/Km ratio, the catalytic efficiency for G1P was about twofold higher than that for M1P. PMM also catalyzed the conversion of ribose 1-phosphate and 2-deoxyglucose 6-phosphate to their corresponding isomers, although activities were much lower. Purified PMM/phosphoglucomutase (PGM) required Mg2+ for maximum activity; Mn2+ was the only other divalent metal that showed some activation. The presence of other divalent metals in addition to Mg2+ in the reaction inhibited the enzymatic activity. PMM and PGM activities could not be detected in nonmucoid algC mutant strain 8858 and in LPS-rough algC mutant strain AK1012, while they were present in the wild-type strains as well as in algC-complemented mutant strains. This evidence suggests that AlgC functions as PMM and PGM in vivo, converting phosphomannose and phosphoglucose in the biosynthesis of both alginate and LPS.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Banerjee P. C., Vanags R. I., Chakrabarty A. M., Maitra P. K. Alginic acid synthesis in Pseudomonas aeruginosa mutants defective in carbohydrate metabolism. J Bacteriol. 1983 Jul;155(1):238–245. doi: 10.1128/jb.155.1.238-245.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cryz S. J., Jr, Fürer E., Germanier R. Passive protection against Pseudomonas aeruginosa infection in an experimental leukopenic mouse model. Infect Immun. 1983 May;40(2):659–664. doi: 10.1128/iai.40.2.659-664.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cryz S. J., Jr, Pitt T. L., Fürer E., Germanier R. Role of lipopolysaccharide in virulence of Pseudomonas aeruginosa. Infect Immun. 1984 May;44(2):508–513. doi: 10.1128/iai.44.2.508-513.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cryz S. J., Jr, Sadoff J. C., Ohman D., Fürer E. Characterization of the human immune response to a Pseudomonas aeruginosa O-polysaccharide-toxin A conjugate vaccine. J Lab Clin Med. 1988 Jun;111(6):701–707. [PubMed] [Google Scholar]
- Dai J. B., Liu Y., Ray W. J., Jr, Konno M. The crystal structure of muscle phosphoglucomutase refined at 2.7-angstrom resolution. J Biol Chem. 1992 Mar 25;267(9):6322–6337. [PubMed] [Google Scholar]
- Darzins A., Chakrabarty A. M. Cloning of genes controlling alginate biosynthesis from a mucoid cystic fibrosis isolate of Pseudomonas aeruginosa. J Bacteriol. 1984 Jul;159(1):9–18. doi: 10.1128/jb.159.1.9-18.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans L. R., Linker A. Production and characterization of the slime polysaccharide of Pseudomonas aeruginosa. J Bacteriol. 1973 Nov;116(2):915–924. doi: 10.1128/jb.116.2.915-924.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franklin M. J., Chitnis C. E., Gacesa P., Sonesson A., White D. C., Ohman D. E. Pseudomonas aeruginosa AlgG is a polymer level alginate C5-mannuronan epimerase. J Bacteriol. 1994 Apr;176(7):1821–1830. doi: 10.1128/jb.176.7.1821-1830.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franklin M. J., Ohman D. E. Identification of algF in the alginate biosynthetic gene cluster of Pseudomonas aeruginosa which is required for alginate acetylation. J Bacteriol. 1993 Aug;175(16):5057–5065. doi: 10.1128/jb.175.16.5057-5065.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fürste J. P., Pansegrau W., Frank R., Blöcker H., Scholz P., Bagdasarian M., Lanka E. Molecular cloning of the plasmid RP4 primase region in a multi-host-range tacP expression vector. Gene. 1986;48(1):119–131. doi: 10.1016/0378-1119(86)90358-6. [DOI] [PubMed] [Google Scholar]
- Goldberg J. B., Hatano K., Pier G. B. Synthesis of lipopolysaccharide O side chains by Pseudomonas aeruginosa PAO1 requires the enzyme phosphomannomutase. J Bacteriol. 1993 Mar;175(6):1605–1611. doi: 10.1128/jb.175.6.1605-1611.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatano K., Goldberg J. B., Pier G. B. Pseudomonas aeruginosa lipopolysaccharide: evidence that the O side chains and common antigens are on the same molecule. J Bacteriol. 1993 Aug;175(16):5117–5128. doi: 10.1128/jb.175.16.5117-5128.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarrell K. F., Kropinski A. M. Pseudomonas aeruginosa bacteriophage phi PLS27-lipopolysaccharide interactions. J Virol. 1981 Nov;40(2):411–420. doi: 10.1128/jvi.40.2.411-420.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang X. M., Neal B., Santiago F., Lee S. J., Romana L. K., Reeves P. R. Structure and sequence of the rfb (O antigen) gene cluster of Salmonella serovar typhimurium (strain LT2). Mol Microbiol. 1991 Mar;5(3):695–713. doi: 10.1111/j.1365-2958.1991.tb00741.x. [DOI] [PubMed] [Google Scholar]
- Knirel YuA, Vinogradov E. V., Kocharova N. A., Paramonov N. A., Kochetkov N. K., Dmitriev B. A., Stanislavsky E. S., Lányi B. The structure of O-specific polysaccharides and serological classification of Pseudomonas aeruginosa (a review). Acta Microbiol Hung. 1988;35(1):3–24. [PubMed] [Google Scholar]
- Köplin R., Arnold W., Hötte B., Simon R., Wang G., Pühler A. Genetics of xanthan production in Xanthomonas campestris: the xanA and xanB genes are involved in UDP-glucose and GDP-mannose biosynthesis. J Bacteriol. 1992 Jan;174(1):191–199. doi: 10.1128/jb.174.1.191-199.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam M. Y., McGroarty E. J., Kropinski A. M., MacDonald L. A., Pedersen S. S., Høiby N., Lam J. S. Occurrence of a common lipopolysaccharide antigen in standard and clinical strains of Pseudomonas aeruginosa. J Clin Microbiol. 1989 May;27(5):962–967. doi: 10.1128/jcm.27.5.962-967.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lightfoot J., Lam J. S. Chromosomal mapping, expression and synthesis of lipopolysaccharide in Pseudomonas aeruginosa: a role for guanosine diphospho (GDP)-D-mannose. Mol Microbiol. 1993 May;8(4):771–782. doi: 10.1111/j.1365-2958.1993.tb01620.x. [DOI] [PubMed] [Google Scholar]
- Lowry O. H., Passonneau J. V. Phosphoglucomutase kinetics with the phosphates of fructose, glucose, mannose, ribose, and galactose. J Biol Chem. 1969 Feb 10;244(3):910–916. [PubMed] [Google Scholar]
- Marolda C. L., Valvano M. A. Identification, expression, and DNA sequence of the GDP-mannose biosynthesis genes encoded by the O7 rfb gene cluster of strain VW187 (Escherichia coli O7:K1). J Bacteriol. 1993 Jan;175(1):148–158. doi: 10.1128/jb.175.1.148-158.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- May T. B., Shinabarger D., Boyd A., Chakrabarty A. M. Identification of amino acid residues involved in the activity of phosphomannose isomerase-guanosine 5'-diphospho-D-mannose pyrophosphorylase. A bifunctional enzyme in the alginate biosynthetic pathway of Pseudomonas aeruginosa. J Biol Chem. 1994 Feb 18;269(7):4872–4877. [PubMed] [Google Scholar]
- May T. B., Shinabarger D., Maharaj R., Kato J., Chu L., DeVault J. D., Roychoudhury S., Zielinski N. A., Berry A., Rothmel R. K. Alginate synthesis by Pseudomonas aeruginosa: a key pathogenic factor in chronic pulmonary infections of cystic fibrosis patients. Clin Microbiol Rev. 1991 Apr;4(2):191–206. doi: 10.1128/cmr.4.2.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pindar D. F., Bucke C. The biosynthesis of alginic acid by Azotobacter vinelandii. Biochem J. 1975 Dec;152(3):617–622. doi: 10.1042/bj1520617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowe P. S., Meadow P. M. Structure of the Core oligosaccharide from the lipopolysaccharide of Pseudomonas aeruginosa PAC1R and its defective mutants. Eur J Biochem. 1983 May 2;132(2):329–337. doi: 10.1111/j.1432-1033.1983.tb07366.x. [DOI] [PubMed] [Google Scholar]
- Roychoudhury S., Chakrabarty K., Ho Y. K., Chakrabarty A. M. Characterization of guanosine diphospho-D-mannose dehydrogenase from Pseudomonas aeruginosa. Structural analysis by limited proteolysis. J Biol Chem. 1992 Jan 15;267(2):990–996. [PubMed] [Google Scholar]
- Roychoudhury S., May T. B., Gill J. F., Singh S. K., Feingold D. S., Chakrabarty A. M. Purification and characterization of guanosine diphospho-D-mannose dehydrogenase. A key enzyme in the biosynthesis of alginate by Pseudomonas aeruginosa. J Biol Chem. 1989 Jun 5;264(16):9380–9385. [PubMed] [Google Scholar]
- Sandlin R. C., Stein D. C. Role of phosphoglucomutase in lipooligosaccharide biosynthesis in Neisseria gonorrhoeae. J Bacteriol. 1994 May;176(10):2930–2937. doi: 10.1128/jb.176.10.2930-2937.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinabarger D., Berry A., May T. B., Rothmel R., Fialho A., Chakrabarty A. M. Purification and characterization of phosphomannose isomerase-guanosine diphospho-D-mannose pyrophosphorylase. A bifunctional enzyme in the alginate biosynthetic pathway of Pseudomonas aeruginosa. J Biol Chem. 1991 Feb 5;266(4):2080–2088. [PubMed] [Google Scholar]
- Shinabarger D., May T. B., Boyd A., Ghosh M., Chakrabarty A. M. Nucleotide sequence and expression of the Pseudomonas aeruginosa algF gene controlling acetylation of alginate. Mol Microbiol. 1993 Sep;9(5):1027–1035. doi: 10.1111/j.1365-2958.1993.tb01232.x. [DOI] [PubMed] [Google Scholar]
- Stevenson G., Lee S. J., Romana L. K., Reeves P. R. The cps gene cluster of Salmonella strain LT2 includes a second mannose pathway: sequence of two genes and relationship to genes in the rfb gene cluster. Mol Gen Genet. 1991 Jun;227(2):173–180. doi: 10.1007/BF00259668. [DOI] [PubMed] [Google Scholar]
- Stroeher U. H., Karageorgos L. E., Morona R., Manning P. A. Serotype conversion in Vibrio cholerae O1. Proc Natl Acad Sci U S A. 1992 Apr 1;89(7):2566–2570. doi: 10.1073/pnas.89.7.2566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sá-Correia I., Darzins A., Wang S. K., Berry A., Chakrabarty A. M. Alginate biosynthetic enzymes in mucoid and nonmucoid Pseudomonas aeruginosa: overproduction of phosphomannose isomerase, phosphomannomutase, and GDP-mannose pyrophosphorylase by overexpression of the phosphomannose isomerase (pmi) gene. J Bacteriol. 1987 Jul;169(7):3224–3231. doi: 10.1128/jb.169.7.3224-3231.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uttaro A. D., Cangelosi G. A., Geremia R. A., Nester E. W., Ugalde R. A. Biochemical characterization of avirulent exoC mutants of Agrobacterium tumefaciens. J Bacteriol. 1990 Mar;172(3):1640–1646. doi: 10.1128/jb.172.3.1640-1646.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zielinski N. A., Chakrabarty A. M., Berry A. Characterization and regulation of the Pseudomonas aeruginosa algC gene encoding phosphomannomutase. J Biol Chem. 1991 May 25;266(15):9754–9763. [PubMed] [Google Scholar]