Abstract
The C. elegans daf-2/insulin-like signaling pathway is critical for regulating development, longevity, metabolism and stress resistance. We identified the 14-3-3 protein FTT-2 to be a new regulatory component of this pathway. We found that RNAi knock down of ftt-2 specifically enhanced the daf-2-mediated dauer formation phenotype. Furthermore, ftt-2 knock down caused the nuclear accumulation of DAF-16/FOXO, the forkhead transcription factor that is the major downstream effecter of daf-2/insulin-like signaling, and enhanced the transcriptional activities of DAF-16. In contrast to ftt-2, RNAi knock down of par-5/ftt-1, the only other gene predicted to encode a 14-3-3 protein in C. elegans, did not show any notable effect on dauer formation, DAF-16 localization, or DAF-16 downstream gene transcription, underscoring the functional specification of FTT-2 and PAR-5 despite their high sequence homology. Using co-immunoprecipitation, we revealed that FTT-2 formed a complex with GFP-fused DAF-16 in C. elegans. Our results indicate that FTT-2 binds to DAF-16 in C. elegans and regulates DAF-16 by sequestering it in the cytoplasm. A similar mechanism of regulation of FOXO by 14-3-3ζ has been reported in mammalian cells, highlighting the high degree of conservation of the daf-2/insulin-like signaling pathway.
Keywords: 14-3-3, ftt-2, par-5, daf-16, FOXO, daf-2, insulin signaling
Introduction
C. elegans daf-16 encodes a forkhead transcription factor (FOXO) that regulates many biological processes, including dauer formation (Riddle and Albert, 1997), lifespan (Lin et al., 1997; Ogg et al., 1997), reproduction (Gems et al., 1998), and fat accumulation (Kimura et al., 1997). DAF-16 is the major downstream effecter of the daf-2/insulin-like signaling pathway. When the daf-2/insulin-like signaling pathway is activated, the transmembrane receptor protein tyrosine kinase DAF-2 triggers the activation of the downstream phosphatidylinositol 3- kinase AGE-1 and the protein kinase B/AKT, which leads to phosphorylation of DAF-16 and retention of DAF-16 in the cytoplasm (Brunet et al., 1999). When daf-2/insulin-like signaling is reduced or inactivated, DAF-16 becomes dephosphorylated and migrates into the nucleus to affect gene expression (Halaschek-Wiener et al., 2005; Lin et al., 2001; McElwee et al., 2003; Murphy et al., 2003). Interestingly, reduced daf-2/insulin-like signaling in C. elegans results in constitutive dauer formation (Kenyon et al., 1993; Kimura et al., 1997; Riddle et al., 1981) and dramatic lifespan extension (Kenyon et al., 1993) phenotypes that are completely dependent on daf-16 (Gottlieb and Ruvkun, 1994; Larsen et al., 1995). Therefore, nuclear translocation and eventual activation of DAF-16 likely induces gene expression changes that promote dauer formation and longevity extension (Halaschek-Wiener et al., 2005; Lee et al., 2003a; McElwee et al., 2003; Murphy et al., 2003).
The daf-2 pathway in C. elegans is entirely orthologous to the insulin/IGF-1 signaling pathways in fruit flies and mammals (Clancy et al., 2001; Holzenberger et al., 2003; Tatar et al., 2001). Three mammalian DAF-16 orthologs (FOXO1, FOXO3a and FOXO4) have been characterized to regulate apoptosis, oxidative stress response, DNA repair, and metabolism (Birkenkamp and Coffer, 2003). In mammalian cultured cells, the subcellular localization of FOXO3a is regulated by binding to the 14-3-3ζ protein. When FOXO3a is phosphorylated by protein kinase B/Akt, it is bound by 14-3-3ζ and sequestered in the cytoplasm (Brunet et al., 1999). Since the insulin/IGF-1 signaling pathway is highly conserved, it is possible that C. elegans DAF-16 is also regulated by a similar mechanism.
14-3-3 proteins are a family of highly conserved, abundant cytoplasmic proteins identified in all eukaryotic organisms examined. They are small (~30 kD), acidic proteins that usually function as hetero or homo-dimers (Jones et al., 1995). In general, 14-3-3 proteins bind to the phosphorylated form of substrate proteins. A large number of proteins are found to contain a consensus 14-3-3 recognition motif: RSXpSXP or RXXXpSXP (Yaffe et al., 1997), in which the phosphorylated serine is essential for binding (Pozuelo Rubio et al., 2004). However, 14-3-3 proteins are also capable of binding to several unphosphorylated ligands (Masters et al., 1999). By binding to their substrates, 14-3-3 proteins can induce the conformational change of the substrate proteins (Obsil et al., 2001; Yaffe, 2002), or sequester the substrate proteins in the cytoplasm (Grozinger and Schreiber, 2000), or act as a scaffold that bridges two interacting partners (Agarwal-Mawal et al., 2003). By binding to a diverse group of signaling molecules, such as Raf -1 (Fu et al., 1994; Irie et al., 1994), Cdc25 phosphatase family members (Chen et al., 2003; Forrest and Gabrielli, 2001; Peng et al., 1997) and Bad (Datta et al., 2000), 14-3-3 proteins are thought to participate in a wide variety of cellular processes, including cell cycle checkpoints, DNA repair, cell differentiation and apoptosis (Fu et al., 2000). 14-3-3 proteins typically have several isoforms in one given organism. For example, there are seven known isoforms in mammals (Ichimura et al., 1988; Martin et al., 1993) and thirteen in Arabidopsis (DeLille et al., 2001). In C. elegans, two 14-3-3 encoding genes have been identified: par-5/ftt-1 and ftt-2 (Wang and Shakes, 1996). par-5 is required for cellular asymmetry in the early C. elegans embryo. PAR-5 regulates the asymmetric cortical localization of PAR-1 and PAR-2 to the posterior and PAR-3, PAR-6 and PKC-3 to the anterior (Morton et al., 2002). Until recently, the function and protein substrates of FTT-2 were not known (Berdichevsky et al., 2006; Wang et al., 2006).
Using gene-specific RNAi knock down, we show that the C. elegans 14-3-3 protein FTT-2 regulates DAF-16 activities by forming a protein complex with DAF-16 and preventing DAF-16 from entering the nucleus to regulate transcription. Our results indicate that the DAF-16 sub-cellular localization is regulated by a conserved mechanism similar to that of FOXO in mammalian cells. In contrast to ftt-2, par-5, the only other gene predicted to encode a 14-3-3-like protein in C. elegans, has no effect on dauer formation, DAF-16 localization or DAF-16 transcriptional activities, highlighting the functional specification of two highly homologous 14-3-3 members.
Materials and Methods
Strains and maintenance
The strains used in this paper were as follow: wild type N2 (from the C. elegans Genetic Center), daf-16 (mgDf47), rrf-3(pk1426), daf-2 (e1370), daf-16(mgDf47); xrls87[daf-16a::gfp::DAF-16B, rol-6(su1006)], daf-2(e1370); daf-16(mgDf47); xrls87[daf-16a::gfp::DAF-16B, rol-6(su1006)], muIs84[pAD76(sod-3::gfp)]. All strains were maintained on NGM plates seeded with Escherichia coli OP50 as the food source.
Construction of ftt-2 and par-5 specific RNAi constructs
The sequences corresponding to the 3′ end and 3′UTR of the predicted ftt-2 and par-5 transcripts were amplified from genomic DNA of N2 worms by PCR. The primers used for the par-5 RNAi construct: Forward primer: 5′-tggacatctgacgttggagctga -3′; Reverse primer: 5′-ggaatgacaatagtgacggagtga -3′. The primers used for the ftt-2 RNAi construct: Forward primer: 5′-acgctgccaccgatgacactg -3′; Reverse primer: 5′-aagggggaaaagccgtaacaaaa -3′. The ftt-2 primers and the par-5 forward primer are kind gifts from the Kemphues lab (K. Kemphues, Cornell University, Ithaca NY). The RNAi constructs were generated by inserting the ftt-2 or par-5 PCR products into the L4440 vector (a kind gift from Dr. A. Fire, Stanford). The RNAi plasmids were transformed into Escherichia coli HT115 (Timmons et al., 2001) for feeding RNAi experiments. All other feeding RNAi clones have been described previously (Lee et al., 2003b).
RNA interference
Feeding RNAi was performed as described (Lee et al., 2003b). Briefly, RNAi bacteria were grown in Luria both with 50 μg ml−1 ampicillin at 37°C for 10–16 hrs, seeded onto NGM plates containing 2mM IPTG, and induced at room temperature overnight.
Dauer assay
Dauer assay for daf-2(e1370) strain was performed as described (Lee et al., 2003b). daf-2(e1370) (Gems et al., 1998) worms at the L4 stage were put onto RNAi plates and allowed to lay egg over night. The resulting self-progeny were allowed to develop at 22°C. At ~96 hrs after egg lay, the number of dauers and adult worms on each plate were scored. Five plates were scored for each RNAi treatment. Dauer assay for N2 or rrf-3(pk1426) strain was performed at 25°C and the number of dauers in each population was scored at ~ 72 hrs after egg lay. The dauer assays were repeated three times.
DAF-16::GFP localization assay
The construction and characterization of the daf-16(mgDf47); xrls87[daf-16a::gfp::DAF-16B, rol-6(su1006)] transgenic strain have been described (Lee et al., 2001). DAF-16::GFP transgenic worms at the L4 stage were put onto the RNAi bacteria and allowed to lay egg at 16°C over night. Self-progeny were allowed to develop on the RNAi bacteria at 16°C till they were at the L3 stage. The worms were cultured at 16°C to allow for a longer time for RNAi to take effect before the worms reached the L3 stage for scoring. The GFP expression of the DAF-16::GFP worms feeding on the various RNAi bacteria was monitored using a fluorescent microscope (Leica MZFL III) and images were captured using a Hamamatsu ORCA-ER camera and OpenLab software.
Lifespan assay
Unless stated otherwise, lifespan assays were performed at 22°C. N2 or daf-2(e1370) L4 larvae worms were allowed to lay egg over night on RNAi plates and the progeny grew on RNAi plates at 16°C until the young adult stage. The young adult worms were transferred onto RNAi plates containing 0.1g/ml FUDR to prevent the growth of progeny and shifted to 22°C. The adult population was scored every day or every other day. Animals that failed to respond to a gentle prodding with a platinum wire were scored as dead. Day 0 of adult lifespan is the day that the adult worms were exposed to FUDR. Statistical analysis was performed with the Kaplan-Meier method to test the null hypothesis (SPSS v.11). The lifespan assays were repeated two times and similar results were observed in both experiments.
Immunoprecipitation and Western Blot
N2, daf-16::gfp, daf-2(e1370); daf-16::gfp, or Psod-3::gfp mixed staged worms were collected by washing with M9 buffer, packed by centrifugation at 3000g for 30 sec and frozen in liquid nitrogen in ~100 μl aliquot. Psod-3::gfp worms were starved for 1 day to induce the GFP expression at a comparable level before collected. daf-2(e1370); daf-16::gfp worms were shifted to 25°C for 6 hrs before collected to induce nuclear localization of DAF-16::GFP. Depending on the size of the frozen worm pellet, approximately 4X volume of lysis buffer (50 mM HEPES pH 7.5, 1 mM EDTA, 150 mM NaCl, 10% Glycerol, 0.1% Triton X-100, 1 mM sodium fluoride and protease inhibitor cocktail) was added. The worm pellets were allowed to thaw at room temperature completely and then frozen in liquid nitrogen again. The thaw-and-freeze procedures were repeated four times. The worm lysate was then sonicated using Markson Ultrasonic Processor for 10 × 15 sec bursts with 30 sec pauses at the output of 60. The debris was removed by centrifugation. The worm lysate was subsequently pre-cleared with 1/20 volume of protein A slurry (Pierce) over night at 4°C. The pre-cleared lysate was incubated with antibody (10 μl antibody per 400 μg total protein) for 1.5 hr at 4°C. 1/10 volume of BSA-blocked protein A slurry was then added and incubated for 1.5 hr at 4°C. The protein A beads were then washed with lysis buffer for six times and the bound proteins were eluted by boiling in 2X sample buffer for 5 min. The anti-FTT-2 and anti-PAR-5 antibodies used for immunoprecipitation are kind gifts from Dr. Andy Golden (NIDDK, National Institutes of Health, Bethesda, MD). The anti-FTT-2 antibody was generated against a small peptide at the extreme C-terminus of FTT-2 (CAATDDTDANETEGGN), and the anti-PAR-5 antibody has been previously reported (Morton et al., 2002). Protein samples eluted from the protein A beads were separated on a 10% SDS gel and transferred onto nitrocellulose membrane (BA85 Protran® BioScience) followed by standard Western blot procedure. The primary antibodies used: anti-ACTIN (mouse, Chemicon International), anti-GFP (goat, Rockland), anti-FTT-2 (rabbit, a gift from Dr. Andy Golden) and anti-PAR-5 (rabbit, a gift from Dr. Andy Golden). The secondary antibodies are: anti-goat (Rockland), anti-mouse (Santa Cruz) and anti-rabbit (Rockland). The immunoprecipitation experiments were repeated three times.
Real-time PCR
Synchronized young adult worms of the indicated genotypes were grown at 16°C and collected by washing with M9 buffer and frozen in liquid nitrogen. Total RNA of ~100 μl worm pellet was isolated using the RNeasy® Mini Kit (Qiagen) and quality control was done by both gel electrophoresis and UV absorbance measurement. cDNAs were synthesized with random hexamers by using SuperScript™ III First-Strand Kit (Invitrogen) according to the manufacturer’s protocol. Real-time PCR reactions were performed in a 20 μl volume using iQ™ SYBR® Green Supermix (BIO-RAD) in a 96-well plate. Duplicates for each sample were included for one single reaction. The real-time PCR primers for act-1 are: Forward primer: 5′-CCAGGAATTGCTGATCGTATGCAGAA –3′; Reverse primer: 5′-TGGAGAGGGAAGCGAGGATAGA –3′ (product length: 133 bp). Primers for sod-3 are: Forward primer: 5′-TCGCACTGCTTCAAAGCTTGTTCAA –3′; Reverse primer: 5′-CCAAATCTGCATAGTCAGATGGGAGAT –3′ (product length: 98 bp). Primers for ftt-2: Forward primer: 5′-TCGACAAGTTCCTCATTCCA -3′; Reverse primer: 5′-TAGCTTTGCTGCGACTTCTC -3′ (product length: 145 bp). Primers for par-5: Forward primer: 5′-AAGTCCCAGAAGGCTTACCA -3′; Reverse primer: 5′-TGGCTGCATCTTGTCCTTAG -3′ (product length: 57 bp). Primers for C24B9.9: Forward primer: 5′-AAAAAGCCATGTTCCCGAAT -3′; Reverse primer5′-GCTGCGAAAAGCAAGAAAAT -3′ (product length: 137 bp). Primers for F53C3.12: Forward primer5′-CGTGTACAGAGACCCCGAAT -3′; Reverse primer: 5′-TGAAGTGCCACGTATTTGGA -3′ (product length: 92 bp). act-1 was used as the internal control and the RNA level of a gene of interest was normalized to the act-1 level for comparison. PCR reaction was initiated at 95°C for 10 minutes for denaturation followed by a 40-cycles consisting of 15 sec at 95°C and 60 sec at 60°C. The real-time PCR experiments were repeated three to four times using independent RNAi worms and RNA preparations. Dauer worms grown at 22°C were not used for RT-PCR experiments because the asynchrony among the different RNAi worms caused large variations in mRNA expression.
Brood size assay
The brood size of RNAi treated worms was determined at 22°C. For each RNAi, ~10 N2 L4 larvae worms were singled onto each RNAi plate and allowed to lay egg over night and then transferred to fresh RNAi plate each subsequent days to lay egg until reproduction ended. The total number of eggs laid and hatched on each plate was counted every day. The brood size assays were repeated two times.
Developmental rate assay
The developmental rate of each RNAi worm population was determined at 22°C. For each RNAi, six gravid adults were allowed to lay egg for 1 hr. The times required for the progeny to reach adulthood were scored. The developmental rate assays were repeated two times.
Results and Discussions
RNAi inactivation of ftt-2, but not par-5, enhances dauer formation
To identify new components of the daf-2/insulin-like signaling pathway, we used feeding RNAi to knock down a select group of molecules annotated as being involved in signal transduction (Rual et al., 2004) and specifically examined their potential role in C. elegans dauer formation. In general, feeding RNAi gene knock down is not an efficient way to reveal a possible dauer phenotype, probably because feeding RNAi does not work well in the nervous systems (Tavernarakis et al., 2000; Timmons et al., 2001). Interestingly, at the semi-restrictive temperature 22°C, the daf-2(e1370) mutant worms form 100% dauer for ~72 hr and then exit dauer to develop into gravid/sterile adults. This represents a sensitized genetic background for using feeding RNAi to identify new players in daf-2-mediated dauer regulation. We found that RNAi knock down of the two 14-3-3 encoding genes, ftt-2 and par-5, greatly enhanced the dauer arrest phenotype of daf-2(e1370). At 22°C, 100% of the daf-2(e1370) worms fed with ftt-2 RNAi and 45% of the daf-2(e1370) worms fed with par-5 RNAi remained as dauer at the 96 hr time point, whereas only 4.5% of the daf-2(e1370) worms fed with the empty vector control L4440 RNAi remained as dauer at this time point.
ftt-2 and par-5 are the two putative 14-3-3 encoding genes in C. elegans. The predicted transcripts of these two genes share ~78.2% sequence identity at the nucleotide level and ~85.9% sequence identity at the amino acid level (Wang and Shakes, 1997). The ftt-2 and par-5 RNAi constructs we initially used for the dauer screen include the full-length sequence of ftt-2 and par-5 respectively. Stretches of consecutive identical sequence as long as 19 base pairs (bp) are detected between the ftt-2 and par-5 RNAi constructs and we suspected that these two RNAi constructs might cross-react and caused the knock down of both ftt-2 and par-5. In order to evaluate the specific roles of ftt-2 and par-5 in dauer formation, we generated gene-specific RNAi constructs that targeted unique fragments corresponding to the 3′ coding and UTR regions of either ftt-2 or par-5 (Morton et al., 2002). Sequence alignment indicates that the longest stretch of sequence identity between the gene-specific ftt-2 and par-5 RNAi constructs is only 6 bp. To verify the specificity of the RNAi constructs, we used reverse-transcription real-time PCR (RT-PCR) to quantify the ftt-2 and par-5 mRNA levels in wild type (N2) and daf-2(e1370) animals that were undergoing RNAi against either the ftt-2 or par-5 gene (Fig. 1A and 1B). We detected a specific 2-fold knock down of the ftt-2 and par-5 mRNA levels in worms exposed to the corresponding gene-specific RNAi. Furthermore, N2 worms fed with the par-5-specific RNAi bacteria for two generations showed severe embryonic lethality (data not shown), similar to that reported previously (Morton et al., 2002). In contrast, N2 worms treated with the ftt-2 specific RNAi for two generations exhibited reduced brood size but did not show noticeable embryonic lethality (Fig. 4). The molecular and phenotypic evidence strongly supports that the ftt-2 and par-5 RNAi constructs we generated specifically targeted the corresponding genes.
Figure 1.
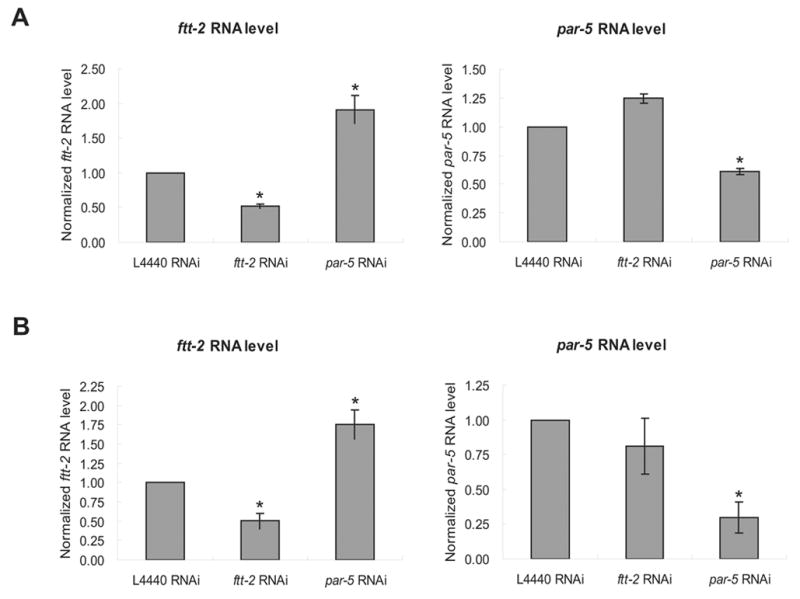
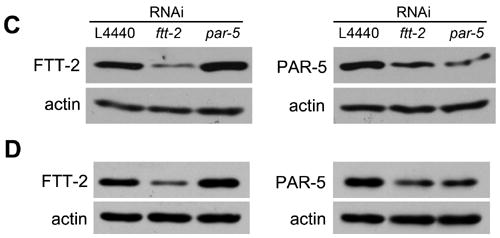
The ftt-2 and par-5 RNAi constructs are gene-specific. N2 (A) or daf-2(e1370) (B) worms were exposed to feeding RNAi bacteria starting as L1 at 16°C. Total RNA was extracted from young adult RNAi worms for real-time-PCR analysis. The y-axis indicates the relative RNA levels normalized to the RNA expression levels of the internal control act-1. The relative RNA levels for worms treated with the L4440 control RNAi is set as 1. The average of four independent experiments is shown and the error bars represent standard error of the mean (SEM). P-value of <0.01 (*) was determined by Student’s t-test. FTT-2 or PAR-5 immunoblotting was performed using young adult N2 worms exposed to the indicated RNAi bacteria at 16°C (C) or daf-2(e1370) dauer worms exposed to the indicated RNAi bacteria at 22°C (D).
Figure 4.
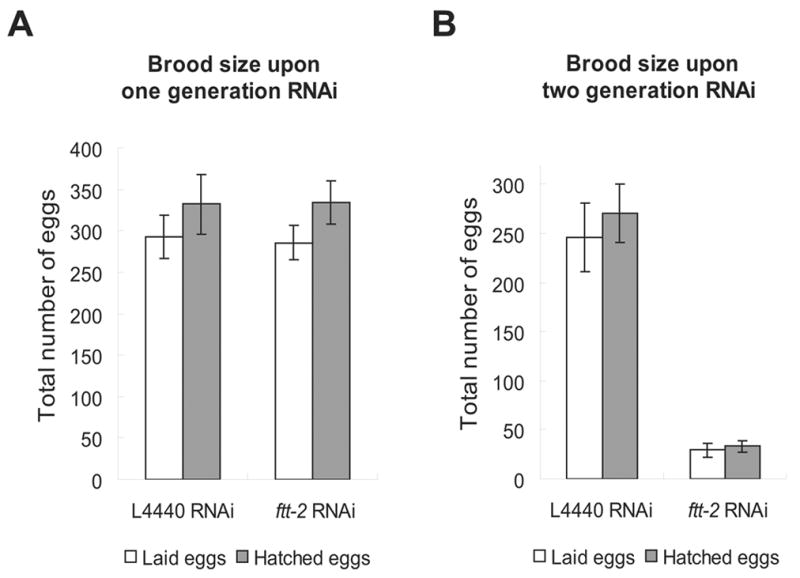
The brood size of ftt-2 RNAi worms upon one generation (A) or two generations (B) of RNAi treatment. The average brood size of ~10 worms for each RNAi treatment is shown. Error bars represent SEM.
We also used anti-FTT-2 and anti-PAR-5 immunoblotting to confirm that the FTT-2 and PAR-5 protein levels were reduced in the daf-2(e1370) dauer worms treated with the gene-specific RNAi at 22°C (Fig. 1D). We found that the anti-FTT-2 signal was specifically reduced in daf-2(e1370) worms treated with the ftt-2 RNAi, but not in worms treated with the par-5 RNAi. In contrast, we noticed that the anti-PAR-5 signal was reduced in worms treated with either the par-5 or ftt-2 RNAi. Similar results were also observed in N2 worms (Fig. 1C). Taken together the immunoblotting, the RT-PCR, and the phenotypic results, we propose that the anti-FTT-2 antibody specifically recognizes FTT-2, whereas the anti-PAR-5 antibody may cross-react with both PAR-5 and FTT-2.
We retested whether specific knock downs of either par-5 or ftt-2 were able to enhance the dauer arrest phenotype of daf-2(e1370) at 22°C. daf-2(e1370) worms were exposed to par-5 or ftt-2 RNAi starting as embryos and allowed to develop at 22°C. The empty vector L4440 was used as a negative control, and the daf-2 RNAi was included as a positive control. At 96 hr, when most of the worms treated with L4440 RNAi exited dauer and developed into sterile/gravid adults, the majority of the ftt-2 RNAi worms remained in the dauer stage (Table 1). In fact, the ftt-2 RNAi treated daf-2(e1370) worms remained arrested as dauer for the entire length of our experiments, which were usually carried out for ~240 hr, similar to that of the positive control daf-2 RNAi worms. In contrast, worms treated with par-5 RNAi behaved similarly to worms treated with L4440 RNAi (Table 1). These results indicate that despite the high sequence homology between par-5 and ftt-2, ftt-2 has the unique function of interacting with the daf-2 pathway and affecting dauer formation.
Table 1.
RNAi inactivation of ftt-2 promotes dauer formation in daf-2(e1370) at 22°C.
| Gene inactivation by RNAi | L4440 | daf-2 | daf-16 | ftt-2 | par-5 | |
|---|---|---|---|---|---|---|
| daf-2 (e1370) at 22°C | % of worms in dauer stage | 4.5% | 100% | 0% | 99.5% | 4.6% |
| Total number of worms scored | 334 | 214 | 423 | 184 | 328 | |
| N2 at 25°C | % of worms in dauer stage | 0% | 0.8% | 0% | 0% | 0% |
| Total number of worms scored | 579 | 601 | 628 | 589 | 557 |
daf-2(e1370) mutant worms or N2 worms were exposed to the various RNAi bacteria starting as embryos and allowed to develop at the indicated temperature. Total number of worms scored and the percentage of worms remained in the dauer stage on day 4 (for daf-2(e1370)) or day 3 (for N2) are shown. ftt-2 and daf-2 RNAi dramatically enhances dauer formation in daf-2(e1370) at 22°C, whereas par-5 RNAi behaves similarly to the L4440 control RNAi. However, neither ftt-2 nor daf-2 RNAi was able to significantly enhance dauer formation in N2 worms at 25°C.
RNAi knock down of ftt-2 in wild-type N2 or rrf-3(pk1426), a strain that specifically enhances somatic RNAi effects (Simmer et al., 2002), background resulted in normal development and no observable dauer arrest phenotype at either 22°C or 25°C (Table 1 and data not shown). This is not surprising as RNAi inactivation of daf-2 did not induce dauer formation in the N2 background (Table 1). Moreover, our RT-PCR and immunoblotting experiments indicate that the gene-specific ftt-2 RNAi only causes a ~2-fold decrease in FTT-2 levels (Fig. 1). It is possible that such a modest reduction in FTT-2, while sufficient to promote dauer arrest in a sensitized background, is not able to signal dauer arrest in a wild-type background. Although a ftt-2 deletion mutant is available through the knockout consortium (http://www.wormbase.org/), we are not able to analyze its dauer formation phenotype because the homozygous ftt-2 deletion mutant is not viable (JL & SSL, unpublished data; Wang et al., 2006).
RNAi inactivation of ftt-2 promotes DAF-16::GFP nuclear localization
Our results indicate that reduced ftt-2 expression specifically enhances the dauer formation phenotype associated with reduced daf-2/insulin-like signaling. Because daf-16 is the major downstream effecter of daf-2 signaling and increased DAF-16 activity is critical for promoting dauer arrest, we investigated whether RNAi knock down of ftt-2 might affect daf-16 activity. A key step of DAF-16 regulation is the translocation of DAF-16 from the cytoplasm to the nucleus. We used feeding RNAi to inactivate ftt-2 in worms carrying an integrated gfp-fused daf-16 transgene (daf-16::gfp) (Lee et al., 2001). The daf-16::gfp worms fed with ftt-2 RNAi tended to arrest at the L3 stage, but they developed to the L3 stage at a rate similar to that of control RNAi worms. We therefore monitored the DAF-16::GFP localization in L3 animals. When the daf-16::gfp worms were fed with the control L4440 RNAi, the DAF-16::GFP fusion protein was evenly distributed in the cytoplasm and nucleus of cells throughout the body of the worm (Fig. 2A). Consistent with previous reports, when daf-2 was inactivated by RNAi, the DAF-16::GFP fusion protein became intensely localized in the nucleus of cells, although some cytoplasmic staining remained detectable (Fig. 2D) (Lin et al., 2001). Interestingly, when ftt-2 was RNAi inactivated, the DAF-16::GFP fusion protein was also dramatically enriched in the nucleus of cells (Fig. 2B), with a low level of cytoplasmic DAF-16::GFP remained detectable. In contrast, par-5 RNAi knock down did not significantly affect the localization pattern of DAF-16::GFP (Fig. 2C). These results indicate that reduced ftt-2 expression specifically induces the nuclear enrichment of DAF-16.
Figure 2.
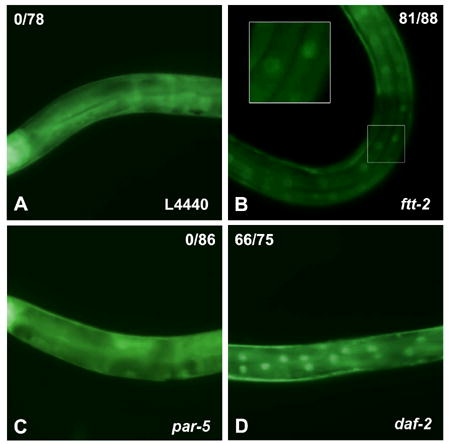
ftt-2 RNAi, but not par-5 RNAi, causes DAF-16::GFP accumulation in the nucleus. DAF-16::GFP worms were exposed to the various RNAi bacteria starting as embryos at 16°C. DAF-16::GFP expression of the RNAi worms at the L3 stage are shown. None of the 78 worms exposed to the control L4440 RNAi (panel A) nor the 86 worms exposed to par-5 RNAi (panel C) showed DAF-16::GFP nuclear localization. In contrast, 81 out of 88 worms exposed to ftt-2 RNAi (panel B) and 66 out of 75 exposed to daf-2 RNAi (panel D) exhibited prominent DAF-16::GFP nuclear localization. A small number of daf-2 RNAi or ftt-2 RNAi worms continued to exhibit cytoplasmic DAF-16::GFP, probably due to variable RNAi efficiency. The images (A–D) were captured using identical magnification and exposure time.
Enhanced DAF-16 transcriptional activities upon ftt-2 RNAi
Because nuclear translocation of DAF-16 may allow DAF-16 to access its transcriptional targets, we next investigated whether ftt-2 RNAi promoted the transcriptional activities of DAF-16. To accomplish this, we exposed N2 or daf-16(mgDf47) null mutant worms to ftt-2 RNAi and monitored the mRNA levels of sod-3, a well-characterized DAF-16 direct target gene (Honda and Honda, 1999; Wook Oh et al., 2006). Consistent with previous reports (Honda and Honda, 1999), we detected ~5-fold up-regulation of sod-3 levels in N2 worms treated with daf-2 RNAi (Fig. 3A). Importantly, when N2 worms were fed ftt-2 RNAi, the expression levels of sod-3 were upregulated about 2-fold (Fig. 3A), suggesting that the transcriptional activities of DAF-16 were elevated. The increased sod-3 expression in ftt-2 RNAi treated N2 worms was not due to a change in daf-16 levels, as the daf-16 mRNA levels were not significantly different in worms treated with the different RNAi (data not shown). Consistent with the distinction between ftt-2 and par-5 on dauer promotion and DAF-16::GFP translocation, reduction of par-5 did not affect DAF-16 transcriptional activities. Similar RNAi experiments performed in the daf-16(mgDf47) null mutant indicate that the induction of sod-3 expression observed in the ftt-2 or daf-2 RNAi worms was mediated by daf-16. In daf-16(mgDf47) null mutant worms treated with control L4440 RNAi, very low basal expression of sod-3 was observed (Fig. 3A). Furthermore, in daf-16(mgDf47) null mutant worms, neither ftt-2 nor daf-2 RNAi had any significant effect on the low basal level of sod-3 expression (Fig. 3A).
Figure 3.
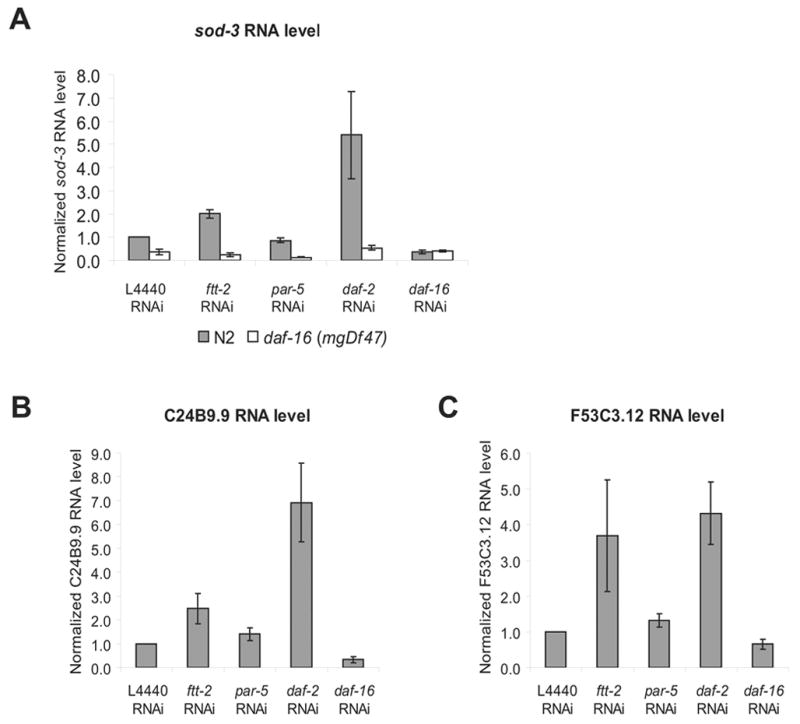
RNAi inactivation of ftt-2 results in the up-regulation of daf-16-dependent mRNA expression of sod-3 (A), C24B9.9 (B) and F53C3.12 (C). N2 worms or daf-16 (mgDf47) null mutant worms were exposed to the indicated RNAi starting as L1 at 16°C. Total RNA was extracted from young adult RNAi worms for real-time-PCR analysis. The y-axis indicates the RNA levels normalized to the RNA expression levels of the internal control act-1. The relative RNA levels for N2 worms treated with the L4440 control RNAi is set as 1. The average of four independent experiments is shown and the error bars represent SEM.
We examined the mRNA levels of additional DAF-16 downstream genes (Murphy et al., 2003). As expected, the DAF-16 activated genes C24B9.9 & F53C3.12 exhibited robust expression changes when worms were treated with daf-2 RNAi vs. daf-16 RNAi (Fig. 3B & 3C). For these two genes, increased expression was also detected in ftt-2 RNAi worms, consistent with an elevation of DAF-16 activities in ftt-2 knock down worms. The results we described thus far suggest the model that RNAi inactivation of ftt-2 induces the nuclear translocation of DAF-16 and promotes the transcription of some DAF-16 downstream genes.
RNAi inactivation of ftt-2 leads to shortened lifespan
Because reduced daf-2 signaling and increased daf-16 activities are often associated with enhanced dauer formation and extended adult lifespan, we investigated whether RNAi inactivation of ftt-2 in worms also affected adult lifespan. We found that the ftt-2 RNAi knock down worms had a much shorter adult lifespan compared to the control RNAi worms (Table 2). In contrast, the par-5 RNAi knock down worms exhibited normal lifespan (Table 2). Interestingly, ftt-2 RNAi did not appear to affect the extended lifespan phenotype of daf-2(e1370) worms (Table 2). Moreover, we determined that the ftt-2 RNAi treated worms developed into adults at a normal rate (control RNAi: 46.5 +/− 2.5 hr, N= 717; ftt-2 RNAi: 44.2 +/− 2.2 hr, N=186) and exhibited no obvious developmental defects. Taken together, the results indicate that ftt-2 RNAi leads to a reduced lifespan in wild-type background, but ftt-2 RNAi does not appear to shorten lifespan by causing non-specific sickness in worms (Berdichevsky et al., 2006; Wang et al., 2006). The lifespan shortening effect of ftt-2 RNAi knock down was somewhat unexpected based on our observation that ftt-2 RNAi promoted dauer formation of daf-2(e1370) worms. On the other hand, the lifespan and dauer phenotypes associated with the daf-2/insulin-like signaling pathway have been shown to be mediated by distinct effecter complexes. For instance, the eak mutations robustly enhance the dauer formation phenotype associated with the akt-1 mutant (the serine/threonine kinase downstream of daf-2) but have little effect on the longevity of the akt-1 mutant worms (Hu et al., 2006); the smk-1 gene specifically interferes with the ability of daf-16 to affect lifespan but has little effect on daf-16-mediated dauer regulation (Wolff et al., 2006). Furthermore, because 14-3-3 proteins are known to bind to a large number of substrates, FTT-2 is most certainly involved in numerous diverse functions. RNAi inactivation of ftt-2 will likely disrupt multiple biological pathways that together may result in lifespan shortening. Two recent papers implicated 14-3-3 proteins in mediating the longevity promoting effect of SIR-2.1 by bridging the interactions of SIR-2.1 and DAF-16 in the nucleus upon stress signals (Berdichevsky et al., 2006; Wang et al., 2006). An interesting possibility is that, in the ftt-2 knock down worms, although more cytoplasmic DAF-16 are released into the nucleus to turn on some DAF-16 downstream genes, such a beneficial effect may be countered by the dissociation of the nuclear 14-3-3/SIR-2.1/DAF-16 protein complexes and result in lifespan shortening.
Table 2.
RNAi inactivation of ftt-2 shortens the lifespan of N2 worms but has no effect on the lifespan of daf-2(e1370) worms.
| Mean (days) | Median (days) | # counted | P-value | ||
|---|---|---|---|---|---|
| N2 | L4440 RNAi | 15.200 | 15.000 | 36 | N/A |
| ftt-2 RNAi | 11.665 | 11.000 | 46 | 0.000a | |
| par-5 RNAi | 16.440 | 19.000 | 39 | 0.380a | |
| daf-2 RNAi | 39.925 | 40.000 | 40 | 0.000a | |
| daf-16 RNAi | 10.216 | 10.000 | 38 | 0.000a | |
| daf-2(e1370) | L4440 RNAi | 48.604 | 50.000 | 53 | N/A |
| ftt-2 RNAi | 46.846 | 52.000 | 52 | 0.737b | |
| par-5 RNAi | 50.533 | 52.000 | 60 | 0.764b | |
| daf-2 RNAi | 45.644 | 50.000 | 45 | 0.980b | |
| daf-16 RNAi | 12.705 | 11.000 | 42 | 0.000b |
The lifespan of N2 or daf-2(e1370) worms treated with the indicated RNAi was determined at 22°C and the results of one representative experiment are shown.
Compared to N2 worms fed with L4440 RNAi bacteria;
Compared to daf-2(e1370) worms fed with L4440 RNAi bacteria.
FTT-2 forms a complex with DAF-16 in vivo
14-3-3 proteins usually function by binding to phosphorylated ligands (Fu et al., 2000). Recent studies in cultured mammalian cells indicate that the 14-3-3ζ isoform binds to phosphorylated FOXO3a, one of the DAF-16 mammalian homologs, and sequesters FOXO3a in the cytoplasm (Brunet et al., 1999). Furthermore, in vitro pull-down assays show that the mammalian 14-3-3ζ isoform is able to bind to phosphorylated DAF-16 (Cahill et al., 2001). Therefore, it is possible that FTT-2 forms a complex with DAF-16 in C. elegans. We used co-immunoprecipitation (co-IP) experiments to test this possibility. Because we do not have an antibody that could detect endogenous DAF-16 robustly, we used worm extracts from the daf-16::gfp strains and an anti-FTT-2 antibody for immunoprecipitation. The co-IP experiments showed that when FTT-2 was specifically immunoprecipitated, DAF-16::GFP was also recovered (Fig. 5A). In the same experiment, a GFP protein driven by the sod-3 promoter (Psod-3::gfp) was not co-immunoprecipitated with FTT-2, indicating that FTT-2 and DAF-16::GFP forms a specific complex in worms. Furthermore, co-IP experiments carried out using worm extracts from animals treated with ftt-2 RNAi showed that a much reduced level of FTT-2 was immunoprecipitated and consequently no detectable DAF-16::GFP was co-immunoprecipitated (Fig. 5B). We also carried out co-IP experiments using daf-2(e1370); daf-16::gfp worms that were cultured at the non-permissive temperature for six hours. Under this condition, the majority of DAF-16::GFP was observed in the nuclei of cells, presumably due to the inactivation of DAF-2 at 25°C. We found that reduced levels of DAF-16::GFP was co-immunoprecipitated with FTT-2 in daf-2(e1370); daf-16::gfp worm extracts compared to that of daf-16::gfp, even though similar levels of FTT-2 was immunoprecipitated in both extracts (Fig. 5A). These results are consistent with the model that when daf-2/insulin-like signaling is inactivated, DAF-16 becomes dephosphorylated and dissociates from 14-3-3 binding.
Figure 5.
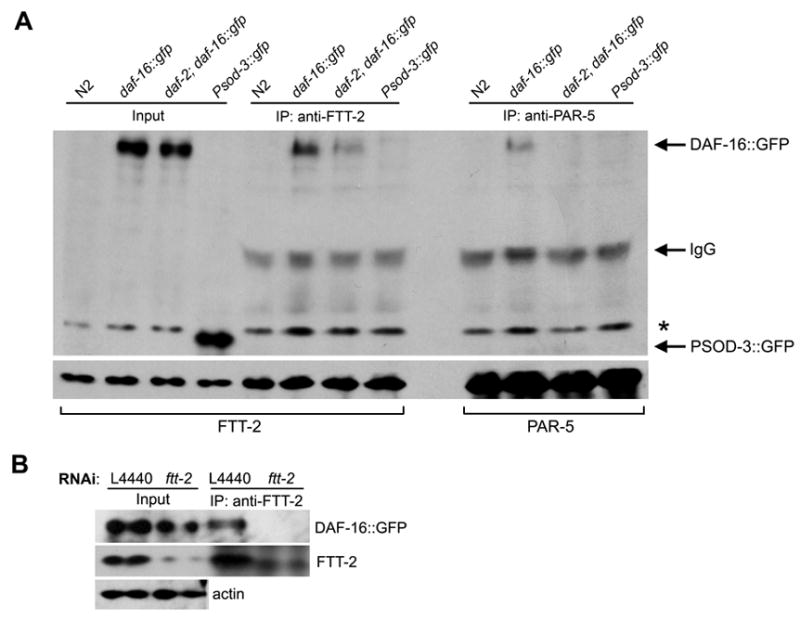
FTT-2 forms a complex with DAF-16::GFP in C. elegans. (A) Worm extracts from mixed staged daf-16::gfp worms (daf-16(mgDf47); xrls87[daf-16a::gfp::DAF-16B, rol-6(su1006)]) or daf-2(e1370); daf-16::gfp worms that had been incubated at 25°C for 6 hrs were immunoprecipitated with an anti-FTT-2 or an anit-PAR-5 antibody and the precipitated proteins were immunoblotted with an anti-GFP antibody (upper panel) and an anti-FTT-2 or an anti-PAR-5 antibody (lower panel). Extracts from N2 and Psod-3::gfp worms were used as negative controls. *: Non-specific band. (B) Worm extracts were prepared from daf-16::gfp L3 larvae treated with ftt-2 RNAi or control L4440 RNAi and immunoprecipitated with an anti-FTT-2 antibody. Upper panel: immunoblotting with an anti-GFP antibody; middle panel: immunoblotting with an anti-FTT-2 antibody; lower panel: immunoblotting with an anti-actin antibody. Actin was included as a loading control for the input extracts.
Our co-IP results together with the previously published in vitro pull-down evidence (Cahill et al., 2001) suggest that FTT-2 likely directly binds to phosphorylated DAF-16 in C. elegans. Because 14-3-3 proteins are abundantly distributed in the cytoplasm, the binding of FTT-2 with DAF-16 may sequester DAF-16 in the cytoplasm. We hypothesize that when ftt-2 is depleted by RNAi, DAF-16 is free to move into the nucleus and execute its transcriptional role.
In our co-IP experiments, a low level of DAF-16::GFP also co-immunoprecipitated with PAR-5 when anti-PAR-5 was used for immunoprecipitation. The FTT-2 and PAR-5 antibodies we used were generated against two distinct FTT-2 and PAR-5 peptides (kind gifts from Dr. Golden). However, as described earlier, in immunoblotting experiments, we found that whereas the anti-FTT-2 antibody appears to be highly specific, the anti-PAR-5 antibody may cross-react with the FTT-2 protein (Fig. 1). Therefore, the weak binding we detected between PAR-5 and DAF-16::GFP may be due to cross-reactivity of the anti-PAR-5 antibody to FTT-2. Alternatively, PAR-5 may indeed form a complex with DAF-16::GFP in worms. Because our experiments indicate that PAR-5 does not affect daf-2-mediated dauer formation, DAF-16 subcellular localization or DAF-16 transcriptional activities, PAR-5 may be involved in other aspects of DAF-16 functions (Berdichevsky et al., 2006).
Our analyses demonstrate that specific knock down of ftt-2, but not par-5, affects dauer formation, DAF-16 localization, and DAF-16 transcriptional activities. Such specificity is consistent with the expression patterns of FTT-2 and PAR-5. Using northern blotting analyses, par-5 is detected to express highly in the embryos and its mRNA level drops drastically by the L1 stage and remains low through larval development. ftt-2 is also found to be the most highly expressed in embryos but its expression through larval stages only drops a little compared to that in embryos (Wang and Shakes, 1997). Also, whereas par-5 expression is highly germline enriched, ftt-2 is not (Wang and Shakes, 1997). Using GFP fusion strategies, PAR-5::GFP is shown to express strongly in the neurons and the intestine of transgenic larvae and FTT-2::GFP is shown to express most strongly in the pharynx and the nervous system, and weakly in the intestine of transgenic larvae (Wang et al., 2006). Based on the expression patterns of FTT-2 and PAR-5, it is not surprising that FTT-2 has important functions during C. elegans larval development. Furthermore, as FTT-2 and PAR-5 express in overlapping but distinct tissues, it is also expected that they will have different functions. Interestingly, in our RT-PCR experiments (Fig. 1), we noticed that specific knock down of par-5 led to ~2-fold increase in ftt-2 mRNA expression, probably due to a yet unknown compensation mechanism. Our results indicate that C. elegans has assigned different tasks for the closely related members of the 14-3-3 family, further supporting the notion that 14-3-3 proteins have isoform-specific functions (Roberts and de Bruxelles, 2002).
In summary, we report here that the 14-3-3 protein FTT-2 specifically regulates dauer formation, DAF-16 subcellular localization, and DAF-16 transcriptional activities in C. elegans. We provide evidence that FTT-2 forms a complex with DAF-16 in worm, similar to that in mammalian cultured cells. The complex formation between FTT-2 and DAF-16 likely results in cytoplasmic sequestration of DAF-16 and prevents it from regulating its transcriptional targets. Our results advance the understanding of the regulation of DAF-16, an important determinant of longevity, metabolism, and development, and further highlight the high degree of conservation between the C. elegans daf-2 and the mammalian insulin/IGF-1 signal transduction pathways.
Acknowledgments
We thank members of the Lee, Liu, and Kemphues labs (Cornell University, Ithaca NY) for helpful discussion. We especially thank Dr. Diane Morton (Cornell University, Ithaca NY) for providing the PCR primers for ftt-2 and par-5, and for insightful discussion. We thank the Lis lab (Cornell University, Ithaca NY) for usage of the real-time PCR machine and we are grateful to Behfar Ardehali for technical assistance. We are especially grateful to Dr. Andy Golden (NIDDK, National Institutes of Health, Bethesda, MD) for providing the anti-FTT-2 and anti-PAR-5 antibodies. We thank Philippe Vaglio for assistance with the bioinformatics. We also thank CGC for providing strains. The RNAi resource used here was supported by a NCI grant to MV. This work was supported by a New Scholar Award in Aging from the Ellison Medical Foundation and a R01 grant AG024425-01 from the NIA awarded to SSL.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Agarwal-Mawal A, Qureshi HY, Cafferty PW, Yuan Z, Han D, Lin R, Paudel HK. 14-3-3 connects glycogen synthase kinase-3 beta to tau within a brain microtubule-associated tau phosphorylation complex. J Biol Chem. 2003;278:12722–8. doi: 10.1074/jbc.M211491200. [DOI] [PubMed] [Google Scholar]
- Berdichevsky A, Viswanathan M, Horvitz HR, Guarente L. C. elegans SIR-2.1 interacts with 14-3-3 proteins to activate DAF-16 and extend life span. Cell. 2006;125:1165–77. doi: 10.1016/j.cell.2006.04.036. [DOI] [PubMed] [Google Scholar]
- Birkenkamp KU, Coffer PJ. Regulation of cell survival and proliferation by the FOXO (Forkhead box, class O) subfamily of Forkhead transcription factors. Biochem Soc Trans. 2003;31:292–7. doi: 10.1042/bst0310292. [DOI] [PubMed] [Google Scholar]
- Brunet A, Bonni A, Zigmond MJ, Lin MZ, Juo P, Hu LS, Anderson MJ, Arden KC, Blenis J, Greenberg ME. Akt promotes cell survival by phosphorylating and inhibiting a Forkhead transcription factor. Cell. 1999;96:857–68. doi: 10.1016/s0092-8674(00)80595-4. [DOI] [PubMed] [Google Scholar]
- Cahill CM, Tzivion G, Nasrin N, Ogg S, Dore J, Ruvkun G, Alexander-Bridges M. Phosphatidylinositol 3-kinase signaling inhibits DAF-16 DNA binding and function via 14-3-3-dependent and 14-3-3-independent pathways. J Biol Chem. 2001;276:13402–10. doi: 10.1074/jbc.M010042200. [DOI] [PubMed] [Google Scholar]
- Chen MS, Ryan CE, Piwnica-Worms H. Chk1 kinase negatively regulates mitotic function of Cdc25A phosphatase through 14-3-3 binding. Mol Cell Biol. 2003;23:7488–97. doi: 10.1128/MCB.23.21.7488-7497.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clancy DJ, Gems D, Harshman LG, Oldham S, Stocker H, Hafen E, Leevers SJ, Partridge L. Extension of life-span by loss of CHICO, a Drosophila insulin receptor substrate protein. Science. 2001;292:104–6. doi: 10.1126/science.1057991. [DOI] [PubMed] [Google Scholar]
- Datta SR, Katsov A, Hu L, Petros A, Fesik SW, Yaffe MB, Greenberg ME. 14-3-3 proteins and survival kinases cooperate to inactivate BAD by BH3 domain phosphorylation. Mol Cell. 2000;6:41–51. [PubMed] [Google Scholar]
- DeLille JM, Sehnke PC, Ferl RJ. The arabidopsis 14-3-3 family of signaling regulators. Plant Physiol. 2001;126:35–8. doi: 10.1104/pp.126.1.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forrest A, Gabrielli B. Cdc25B activity is regulated by 14-3-3. Oncogene. 2001;20:4393–401. doi: 10.1038/sj.onc.1204574. [DOI] [PubMed] [Google Scholar]
- Fu H, Subramanian RR, Masters SC. 14-3-3 proteins: structure, function, and regulation. Annu Rev Pharmacol Toxicol. 2000;40:617–47. doi: 10.1146/annurev.pharmtox.40.1.617. [DOI] [PubMed] [Google Scholar]
- Fu H, Xia K, Pallas DC, Cui C, Conroy K, Narsimhan RP, Mamon H, Collier RJ, Roberts TM. Interaction of the protein kinase Raf-1 with 14-3-3 proteins. Science. 1994;266:126–9. doi: 10.1126/science.7939632. [DOI] [PubMed] [Google Scholar]
- Gems D, Sutton AJ, Sundermeyer ML, Albert PS, King KV, Edgley ML, Larsen PL, Riddle DL. Two pleiotropic classes of daf-2 mutation affect larval arrest, adult behavior, reproduction and longevity in Caenorhabditis elegans. Genetics. 1998;150:129–55. doi: 10.1093/genetics/150.1.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottlieb S, Ruvkun G. daf-2, daf-16 and daf-23: genetically interacting genes controlling Dauer formation in Caenorhabditis elegans. Genetics. 1994;137:107–20. doi: 10.1093/genetics/137.1.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grozinger CM, Schreiber SL. Regulation of histone deacetylase 4 and 5 and transcriptional activity by 14-3-3-dependent cellular localization. Proc Natl Acad Sci U S A. 2000;97:7835–40. doi: 10.1073/pnas.140199597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halaschek-Wiener J, Khattra JS, McKay S, Pouzyrev A, Stott JM, Yang GS, Holt RA, Jones SJ, Marra MA, Brooks-Wilson AR, Riddle DL. Analysis of long-lived C. elegans daf-2 mutants using serial analysis of gene expression. Genome Res. 2005;15:603–15. doi: 10.1101/gr.3274805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holzenberger M, Dupont J, Ducos B, Leneuve P, Geloen A, Even PC, Cervera P, Le Bouc Y. IGF-1 receptor regulates lifespan and resistance to oxidative stress in mice. Nature. 2003;421:182–7. doi: 10.1038/nature01298. [DOI] [PubMed] [Google Scholar]
- Honda Y, Honda S. The daf-2 gene network for longevity regulates oxidative stress resistance and Mn-superoxide dismutase gene expression in Caenorhabditis elegans. Faseb J. 1999;13:1385–93. [PubMed] [Google Scholar]
- Hu PJ, Xu J, Ruvkun G. Two membrane-associated tyrosine phosphatase homologs potentiate C. elegans AKT-1/PKB signaling. PLoS Genet. 2006;2:e99. doi: 10.1371/journal.pgen.0020099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ichimura T, Isobe T, Okuyama T, Takahashi N, Araki K, Kuwano R, Takahashi Y. Molecular cloning of cDNA coding for brain-specific 14-3-3 protein, a protein kinase-dependent activator of tyrosine and tryptophan hydroxylases. Proc Natl Acad Sci U S A. 1988;85:7084–8. doi: 10.1073/pnas.85.19.7084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irie K, Gotoh Y, Yashar BM, Errede B, Nishida E, Matsumoto K. Stimulatory effects of yeast and mammalian 14-3-3 proteins on the Raf protein kinase. Science. 1994;265:1716–9. doi: 10.1126/science.8085159. [DOI] [PubMed] [Google Scholar]
- Jones DH, Ley S, Aitken A. Isoforms of 14-3-3 protein can form homo- and heterodimers in vivo and in vitro: implications for function as adapter proteins. FEBS Lett. 1995;368:55–8. doi: 10.1016/0014-5793(95)00598-4. [DOI] [PubMed] [Google Scholar]
- Kenyon C, Chang J, Gensch E, Rudner A, Tabtiang R. A C. elegans mutant that lives twice as long as wild type. Nature. 1993;366:461–4. doi: 10.1038/366461a0. [DOI] [PubMed] [Google Scholar]
- Kimura KD, Tissenbaum HA, Liu Y, Ruvkun G. daf-2, an insulin receptor-like gene that regulates longevity and diapause in Caenorhabditis elegans. Science. 1997;277:942–6. doi: 10.1126/science.277.5328.942. [DOI] [PubMed] [Google Scholar]
- Larsen PL, Albert PS, Riddle DL. Genes that regulate both development and longevity in Caenorhabditis elegans. Genetics. 1995;139:1567–83. doi: 10.1093/genetics/139.4.1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee RY, Hench J, Ruvkun G. Regulation of C. elegans DAF-16 and its human ortholog FKHRL1 by the daf-2 insulin-like signaling pathway. Curr Biol. 2001;11:1950–7. doi: 10.1016/s0960-9822(01)00595-4. [DOI] [PubMed] [Google Scholar]
- Lee SS, Kennedy S, Tolonen AC, Ruvkun G. DAF-16 target genes that control C. elegans life-span and metabolism. Science. 2003a;300:644–7. doi: 10.1126/science.1083614. [DOI] [PubMed] [Google Scholar]
- Lee SS, Lee RY, Fraser AG, Kamath RS, Ahringer J, Ruvkun G. A systematic RNAi screen identifies a critical role for mitochondria in C. elegans longevity. Nat Genet. 2003b;33:40–8. doi: 10.1038/ng1056. [DOI] [PubMed] [Google Scholar]
- Lin K, Dorman JB, Rodan A, Kenyon C. daf-16: An HNF-3/forkhead family member that can function to double the life-span of Caenorhabditis elegans. Science. 1997;278:1319–22. doi: 10.1126/science.278.5341.1319. [DOI] [PubMed] [Google Scholar]
- Lin K, Hsin H, Libina N, Kenyon C. Regulation of the Caenorhabditis elegans longevity protein DAF-16 by insulin/IGF-1 and germline signaling. Nat Genet. 2001;28:139–45. doi: 10.1038/88850. [DOI] [PubMed] [Google Scholar]
- Martin H, Patel Y, Jones D, Howell S, Robinson K, Aitken A. Antibodies against the major brain isoforms of 14-3-3 protein. An antibody specific for the N-acetylated amino-terminus of a protein. FEBS Lett. 1993;331:296–303. doi: 10.1016/0014-5793(93)80356-y. [DOI] [PubMed] [Google Scholar]
- Masters SC, Pederson KJ, Zhang L, Barbieri JT, Fu H. Interaction of 14-3-3 with a nonphosphorylated protein ligand, exoenzyme S of Pseudomonas aeruginosa. Biochemistry. 1999;38:5216–21. doi: 10.1021/bi982492m. [DOI] [PubMed] [Google Scholar]
- McElwee J, Bubb K, Thomas JH. Transcriptional outputs of the Caenorhabditis elegans forkhead protein DAF-16. Aging Cell. 2003;2:111–21. doi: 10.1046/j.1474-9728.2003.00043.x. [DOI] [PubMed] [Google Scholar]
- Morton DG, Shakes DC, Nugent S, Dichoso D, Wang W, Golden A, Kemphues KJ. The Caenorhabditis elegans par-5 gene encodes a 14-3-3 protein required for cellular asymmetry in the early embryo. Dev Biol. 2002;241:47–58. doi: 10.1006/dbio.2001.0489. [DOI] [PubMed] [Google Scholar]
- Murphy CT, McCarroll SA, Bargmann CI, Fraser A, Kamath RS, Ahringer J, Li H, Kenyon C. Genes that act downstream of DAF-16 to influence the lifespan of Caenorhabditis elegans. Nature. 2003;424:277–83. doi: 10.1038/nature01789. [DOI] [PubMed] [Google Scholar]
- Obsil T, Ghirlando R, Klein DC, Ganguly S, Dyda F. Crystal structure of the 14-3-3zeta:serotonin N-acetyltransferase complex. a role for scaffolding in enzyme regulation. Cell. 2001;105:257–67. doi: 10.1016/s0092-8674(01)00316-6. [DOI] [PubMed] [Google Scholar]
- Ogg S, Paradis S, Gottlieb S, Patterson GI, Lee L, Tissenbaum HA, Ruvkun G. The Fork head transcription factor DAF-16 transduces insulin-like metabolic and longevity signals in C. elegans. Nature. 1997;389:994–9. doi: 10.1038/40194. [DOI] [PubMed] [Google Scholar]
- Peng CY, Graves PR, Thoma RS, Wu Z, Shaw AS, Piwnica-Worms H. Mitotic and G2 checkpoint control: regulation of 14-3-3 protein binding by phosphorylation of Cdc25C on serine-216. Science. 1997;277:1501–5. doi: 10.1126/science.277.5331.1501. [DOI] [PubMed] [Google Scholar]
- Pozuelo Rubio M, Geraghty KM, Wong BH, Wood NT, Campbell DG, Morrice N, Mackintosh C. 14-3-3-affinity purification of over 200 human phosphoproteins reveals new links to regulation of cellular metabolism, proliferation and trafficking. Biochem J. 2004;379:395–408. doi: 10.1042/BJ20031797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riddle D-L, Albert P-S. Genetic and environmental regulation of Dauer larva development. In: Riddle DL, Blumenthal T, Meyer BJ, Priess JR, editors. Cold Spring Harbor Monograph Series; C. elegans II. Vol. 33. Cold Spring Harbor Laboratory Press {a}, 10 Skyline Drive; Plainview, New York 11803, USA: 1997. 1997. pp. 739–768. [PubMed] [Google Scholar]
- Riddle DL, Swanson MM, Albert PS. Interacting genes in nematode dauer larva formation. Nature. 1981;290:668–71. doi: 10.1038/290668a0. [DOI] [PubMed] [Google Scholar]
- Roberts MR, de Bruxelles GL. Plant 14-3-3 protein families: evidence for isoform-specific functions? Biochem Soc Trans. 2002;30:373–8. doi: 10.1042/bst0300373. [DOI] [PubMed] [Google Scholar]
- Rual JF, Ceron J, Koreth J, Hao T, Nicot AS, Hirozane-Kishikawa T, Vandenhaute J, Orkin SH, Hill DE, van den Heuvel S, Vidal M. Toward improving Caenorhabditis elegans phenome mapping with an ORFeome-based RNAi library. Genome Res. 2004;14:2162–8. doi: 10.1101/gr.2505604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmer F, Tijsterman M, Parrish S, Koushika SP, Nonet ML, Fire A, Ahringer J, Plasterk RH. Loss of the putative RNA-directed RNA polymerase RRF-3 makes C. elegans hypersensitive to RNAi. Curr Biol. 2002;12:1317–9. doi: 10.1016/s0960-9822(02)01041-2. [DOI] [PubMed] [Google Scholar]
- Tatar M, Kopelman A, Epstein D, Tu MP, Yin CM, Garofalo RS. A mutant Drosophila insulin receptor homolog that extends life-span and impairs neuroendocrine function. Science. 2001;292:107–10. doi: 10.1126/science.1057987. [DOI] [PubMed] [Google Scholar]
- Tavernarakis N, Wang SL, Dorovkov M, Ryazanov A, Driscoll M. Heritable and inducible genetic interference by double-stranded RNA encoded by transgenes. Nat Genet. 2000;24:180–3. doi: 10.1038/72850. [DOI] [PubMed] [Google Scholar]
- Timmons L, Court DL, Fire A. Ingestion of bacterially expressed dsRNAs can produce specific and potent genetic interference in Caenorhabditis elegans. Gene. 2001;263:103–12. doi: 10.1016/s0378-1119(00)00579-5. [DOI] [PubMed] [Google Scholar]
- Wang W, Shakes DC. Molecular evolution of the 14-3-3 protein family. J Mol Evol. 1996;43:384–98. doi: 10.1007/BF02339012. [DOI] [PubMed] [Google Scholar]
- Wang W, Shakes DC. Expression patterns and transcript processing of ftt-1 and ftt-2, two C. elegans 14-3-3 homologues. J Mol Biol. 1997;268:619–30. doi: 10.1006/jmbi.1997.1002. [DOI] [PubMed] [Google Scholar]
- Wang Y, Oh SW, Deplancke B, Luo J, Walhout AJ, Tissenbaum HA. C. elegans 14-3-3 proteins regulate life span and interact with SIR-2.1 and DAF-16/FOXO. Mech Ageing Dev. 2006;127:741–7. doi: 10.1016/j.mad.2006.05.005. [DOI] [PubMed] [Google Scholar]
- Wolff S, Ma H, Burch D, Maciel GA, Hunter T, Dillin A. SMK-1, an essential regulator of DAF-16-mediated longevity. Cell. 2006;124:1039–53. doi: 10.1016/j.cell.2005.12.042. [DOI] [PubMed] [Google Scholar]
- Wook Oh S, Mukhopadhyay A, Dixit BL, Raha T, Green MR, Tissenbaum HA. Identification of direct DAF-16 targets controlling longevity, metabolism and diapause by chromatin immunoprecipitation. Nat Genet. 2006;38:251–7. doi: 10.1038/ng1723. [DOI] [PubMed] [Google Scholar]
- Yaffe MB. How do 14-3-3 proteins work?-- Gatekeeper phosphorylation and the molecular anvil hypothesis. FEBS Lett. 2002;513:53–7. doi: 10.1016/s0014-5793(01)03288-4. [DOI] [PubMed] [Google Scholar]
- Yaffe MB, Rittinger K, Volinia S, Caron PR, Aitken A, Leffers H, Gamblin SJ, Smerdon SJ, Cantley LC. The structural basis for 14-3-3:phosphopeptide binding specificity. Cell. 1997;91:961–71. doi: 10.1016/s0092-8674(00)80487-0. [DOI] [PubMed] [Google Scholar]


