Abstract
A neuron’s dendrites typically do not cross one another. This intrinsic self-avoidance mechanism ensures unambiguous processing of sensory or synaptic inputs. Moreover, some neurons respect the territory of others of the same type, a phenomenon known as tiling. Different types of neurons, however, often have overlapping dendritic fields. We found that Down’s syndrome Cell Adhesion Molecule (Dscam) is required for dendritic self-avoidance of all four classes of Drosophila dendritic arborization (da) neurons. However, neighboring mutant class IV da neurons still exhibited tiling suggesting that self-avoidance and tiling differ in their recognition and repulsion mechanisms. Introducing one of the 38,016 Dscam isoforms to da neurons in Dscam mutants was sufficient to significantly restore self-avoidance. Remarkably, expression of a common Dscam isoform in da neurons of different classes prevented their dendrites from sharing the same territory suggesting that co-existence of dendritic fields of different neuronal classes requires divergent expression of Dscam isoforms.
Keywords: dendrite development, dendrite morphogenesis, Dscam, self-avoidance, tiling, repulsion, self-recognition, co-existence
Introduction
Proper sampling of sensory or synaptic inputs critically depends on the morphogenesis and organization of dendrites. Several mechanisms contribute to the orderly organization of dendritic fields of various types of neurons in a common area.
First, the dendrites of an individual neuron typically do not fasciculate or cross one another (self-avoidance). Given the likely local processing and integration of inputs impinging onto a dendritic branch (Jan and Jan, 2003; Stuart et al., 1999), the absence of overlap of a neuron’s dendritic branches facilitates orderly projection of sensory or synaptic inputs for efficient and unambiguous signal processing. In addition, self-avoidance could also contribute to the maximal dispersion of dendritic branches for more uniform coverage of the entire dendritic field of a neuron.
Second, certain types of neurons exhibit a phenomenon known as tiling, which refers to the avoidance between dendrites of adjacent neurons of the same type. Tiling was first reported for the mammalian retinal ganglion cells (Perry and Linden, 1982; Wassle et al., 1981) and allows neurons of the same class to cover the entire dendritic field like tiles on the floor, completely but without redundancy.
Third, neurons of different types often have overlapping dendritic fields to allow different neuronal types to process different aspects of inputs. For example, different classes of retinal ganglion cells that selectively respond to motion in different directions need to cover the same visual field (Amthor and Oyster, 1995).
Drosophila dendritic arborization (da) neurons exhibit the full range of possible dendritic interactions encompassing self-avoidance, tiling, and co-existence. Larval da neurons are sensory neurons of the peripheral nervous system and fall into four different classes with different morphology (Grueber et al., 2002). All classes display self-avoidance, while only two of the four classes of da neurons exhibit tiling, namely, the class III da neurons with characteristic dendritic spikes and the class IV da neurons exhibiting the greatest dendritic complexity (Grueber et al., 2002; Grueber et al., 2003; Sugimura et al., 2003). The simpler class I and class II da neurons have smaller dendritic fields that normally do not overlap with those of the same class. However, when class I neurons are duplicated in hamlet mutants they occupy the same area and do not repel each other. In contrast, duplicated class IV neurons tile and hence subdivide the dendritic field (Grueber et al., 2003). Moreover, da neurons belonging to different classes extensively share their dendritic fields (Gao et al., 2000; Grueber et al., 2002), analogous to the repeated coverage of the retina by different classes of retinal ganglion cells tuned to different features of the visual field (Rockhill et al., 2000). Drosophila da neurons thus present an opportunity to examine the role of molecular interactions between dendrites and serve as a useful model to study the mechanism of dendritic field organization.
The tiling phenomenon and self-avoidance both involve recognition and repulsion of dendrites. In recent years, the NDR-family kinase Tricornered/Sax-1 (trc), its activator Furry/Sax-2 (fry), and the upstream Ste-20 family kinase Hippo (hpo) have been identified as important components of the intracellular signaling cascade regulating branching and tiling of da neurons (Emoto et al., 2004; Emoto et al., 2006). Sax-1 and Sax-2 have been found to serve a similar role in C. elegans mechanosensory neurons (Gallegos and Bargmann, 2004). In trc and fry mutants, all classes of da neurons display dendritic overbranching, while class IV da neurons additionally exhibit dendritic crossing of isoneuronal branches (one feature of self-avoidance defects) and between neighboring class IV neurons (tiling defects) (Emoto et al., 2004). It is not known whether self-avoidance and tiling employ the same dendrite recognition and avoidance mechanism.
One candidate for mediating dendritic interactions is Dscam, a type I membrane protein of the immunoglobulin superfamily (Schmucker et al., 2000). Alternative usage of twelve exon 4, forty-eight exon 6 and thirty-three exon 9 variants coding for parts of the extracellular Ig domains, as well as two alternative exons for the transmembrane segment, yields a total of 38,016 possible splice variants of Dscam—most of which have been detected in vivo (Neves et al., 2004; Watson et al., 2005; Zhan et al., 2004). Strong homophilic interactions are only observed between the same isoforms of Dscam (Wojtowicz et al., 2004), raising the possibility that isoform-specific interactions contribute to the intricate patterning within the nervous system. Interestingly, different combinations of Dscam isoforms are found in neighboring neurons of the same type (Neves et al., 2004; Zhan et al., 2004). Moreover, Dscam mutants display abnormal axonal targeting and axonal branch segregation (Hummel et al., 2003; Wang et al., 2002), and even a modest reduction of Dscam diversity alters mechanosensory neuronal projections (Chen et al., 2006). Loss of Dscam function also reduces the dendritic field of olfactory projection neurons leading to the suggestion that Dscam may have a role in mediating dendritic repulsion (Zhu et al., 2006). It is therefore of particular interest to analyze individual da neurons for Dscam loss of function and misexpression phenotypes to assess the role of Dscam in self-avoidance and tiling, and co-existence of overlapping dendritic fields.
In this study, we examined the cell-autonomous function of Dscam by employing MARCM (Mosaic Analysis with a Repressible Cell Marker). We found that loss of Dscam function caused self-avoidance defects in all four da neuron classes, resulting in crossing and bundling of dendrites in clones of individual Dscam mutant da neurons. In contrast, self-avoidance in class I–III da neurons did not require trc function, and loss of trc function caused dendritic crossing but not bundling in class IV neurons. All da neurons lacking Dscam also displayed abnormal coverage of the dendritic field, perhaps partly due to the failure in self-avoidance. However, adjacent class IV da neurons lacking Dscam function still exhibited tiling. Thus, Dscam is required for self-avoidance, but probably not tiling. Self-avoidance and dendritic field coverage in Dscam mutant animals were restored to a large extent in class I and IV da neurons expressing any of four arbitrarily chosen full length Dscam isoforms, but not Dscam lacking the intracellular domain. Remarkably, whereas duplicated class I neurons normally showed extensive overlap of their dendrites, overexpression of a single Dscam isoform restricted their sharing of dendritic fields. Moreover, expression of the same Dscam isoform in all da neurons led to dendritic avoidance between the different classes, thus strongly reducing their dendritic field overlap. We conclude that while a single Dscam isoform is sufficient for self-avoidance, Dscam diversity appears to be necessary to ensure co-existence of da neuron dendrites sharing the same territory.
Results
Dscam function is essential for self-avoidance and proper dendritic field coverage of da neurons
To test whether Dscam function in da neurons is important for dendrite morphogenesis, we first determined whether Dscam is expressed in da neurons by carrying out the following two experiments. First, we used a 4.5 kb Dscam promoter fragment fused to Gal4, which has been shown to reflect the endogenous Dscam expression pattern in the central nervous system (Wang et al., 2004), to drive expression of the reporter mCD8-GFP, and found reporter expression in all four classes of da neurons (Supplemental Figure 1A). Next, we confirmed Dscam expression in da neurons with immunofluorescence using an antibody against Dscam revealing a punctate pattern of endogenous Dscam in both axons and dendrites of all da neurons in wild type (Supplemental Figure 1B) but not Dscam mutant larvae (Supplemental Figure 1C).
To specifically remove Dscam function in individual da neurons, we used the MARCM technique (Lee et al., 2000) to generate da neuron clones homozygous for either of two mutant alleles known to yield no detectable Dscam protein, Dscam18 and DscamP1 (Schmucker et al., 2000; Wang et al., 2002). When mutant for Dscam, each class of da neurons displayed dendritic self-avoidance defects in third instar larvae.
Wild type MARCM clones of class I da neurons project stereotypical, well-spaced comb-like dendrites to the segmental boundary (Grueber et al., 2002); the ddaD neuron dendrites extend towards the anterior boundary (not shown) and the ddaE neuron dendrites to the posterior boundary (Figure 1 A). In contrast, the dendritic branches of individual Dscam mutant class I da neurons were often fasciculated (arrows, Figure 1 B) or crossed one another (arrowheads, Figure 1 B, G, Supplemental Figure 2 D). Thus, whereas the dendrites initially projected in the right direction, a failure of self-avoidance resulted in an uneven and abnormal coverage of the dendritic field.
Figure 1. MARCM analysis of dendritic morphogenesis of Dscam mutant da neurons.
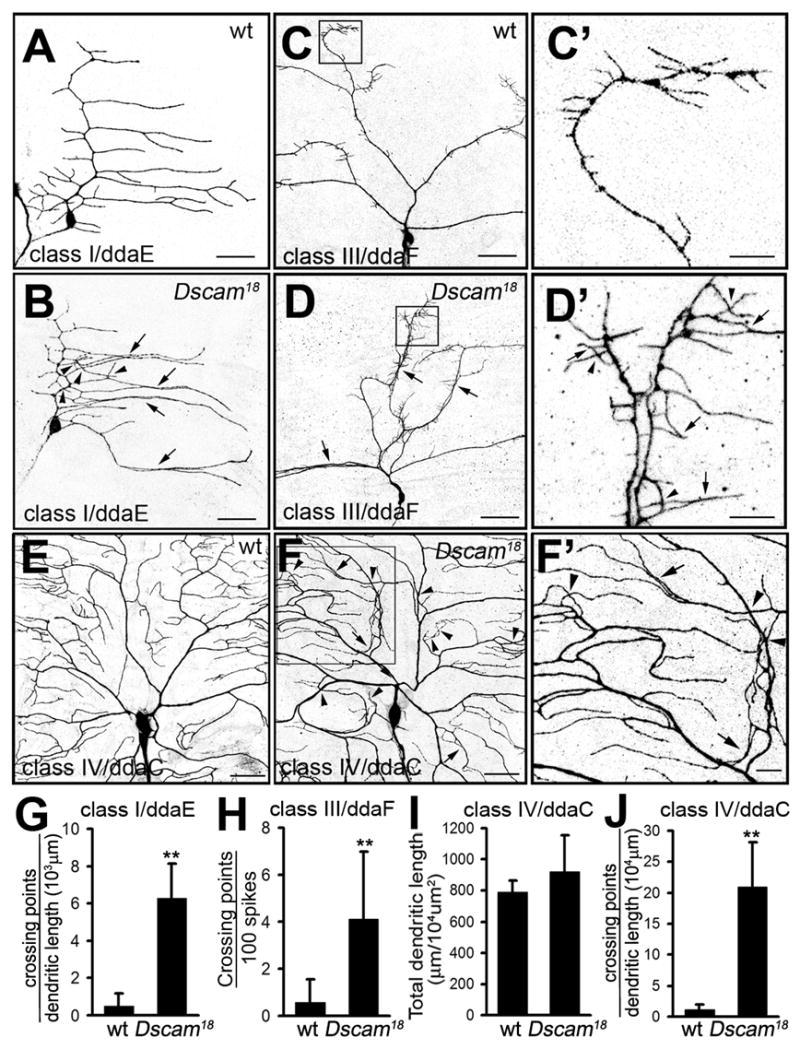
(A–F) Representative MARCM clones of class I ddaE (A–B), class III ddaF (C–D), and class IV ddaC (E–F) neurons. Wild type and Dscam18 mutant MARCM clones are shown as indicated. (C′), (D′), and (F′) are enlarged views of the highlighted areas in (C), (D), and (F). Arrows indicate dendritic bundling and arrowheads indicate crosses of dendritic branches (or spikes in ddaF). Scale bars, 30 μm in (A–F) or 10 μm in (C′, D′, and F′). (G–J) Quantitative analysis of the crossing points of class I ddaE dendrites (G, n=7 for each group), dendritic spikes of ddaF (H, n=7 for each group), total dendritic length (I) and the crossing points of dendritic branches (J) of ddaC neurons (n=6 for each group). All the data are mean ± SD in this and the following figures (**, P<0.01, student’s t-test).
Similar phenotypes were found in mutant clones of class II and class III da neurons. We observed partial bundling of primary and higher order dendritic branches (arrows, Figure 1 D, D′, Supplemental Figure 2 B, C, E). Moreover, whereas dendritic spikes (short, actin-rich dendritic protrusions found only in class III neurons (Andersen et al., 2005)) of control MARCM clones of class III da neurons did not cross (Figure 1 C, C′) as reported (Grueber et al., 2003), the spikes in Dscam mutant clones frequently crossed (arrowheads, Figure 1 D, D′, H) or bundled together (arrows, Figure 1 D′). This suggests that dendritic spikes as well as dendritic branches require Dscam function for self-avoidance.
The Dscam mutant phenotype was further analyzed in class IV da neurons, which display the most complex dendrite morphology. Compared to control MARCM clones that exhibited extensive dendritic field coverage but virtually no crossing between isoneuronal dendrites (Figure 1E), loss of Dscam function in class IV da neurons led to frequent crossing (arrowheads) and bundling (arrows) of dendritic branches (Figure 1 F, F′, Supplemental Figure 2F). There was a ten-fold increase in dendritic crossings (Figure 1 J) but no significant alteration in overall dendritic length (Figure 1 I). It thus appears that Dscam is required for self-avoidance of all classes of da neurons.
Dscam does not play a major role in tiling between dendrites of neighboring class IV da neurons
Given that avoidance of crossing between dendrites of the same or neighboring class IV da neurons both require the NDR-kinase Tricornered (Trc), its activator Furry (Fry) and its upstream kinase, the tumor suppressor Hippo (Hpo)(Emoto et al., 2004; Emoto et al., 2006), it is of interest to determine whether Dscam is required for tiling as well as self-avoidance.
Tiling mutants typically display abnormally enlarged dendritic fields, the result of a decrease in dendritic growth inhibition (Emoto et al., 2004). In class IV da neuron clones mutant for Dscam, however, despite abnormal field coverage, the dendritic field did not exhibit signs of overgrowth and its size was comparable to that of control MARCM clones (Figure 2 A, B, C).
Figure 2. Dscam does not play a major role in interneuronal recognition and tiling.
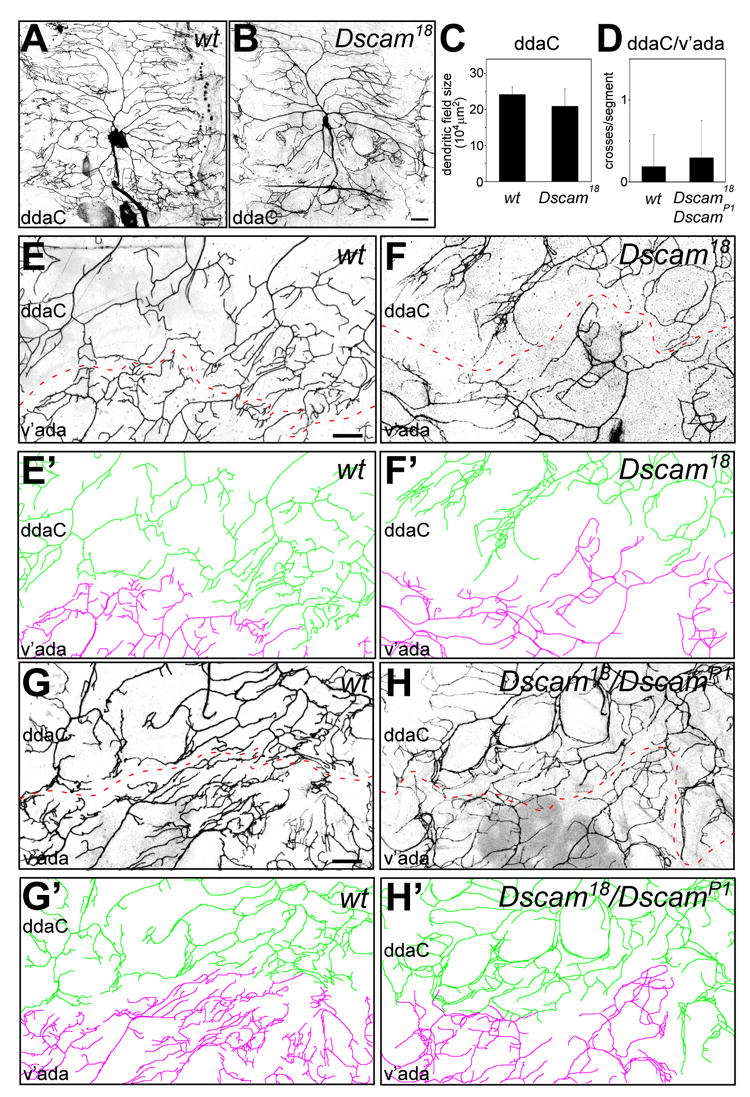
(A–B) MARCM clones of wild type (A) and Dscam18 mutant (B) class IV ddaC neurons. Note that the dendritic field size is largely unchanged in Dscam mutants (B) compared to the wild type (A) and does not show prominent overgrowth. (C) Quantitative analysis of total dendritic field size of wild type (n=4) and Dscam18 (n=5) ddaC MARCM clones. (D) Quantitative analysis of interneuronal dendritic crosses between adjacent ddaC and v’ada class IV da neurons in wild type (n=16) and Dscam18/DscamP1 (n=17) third instar larvae. (E–F) Dendrites of two adjacent MARCM clones of wild type (E and E′) and Dscam18 mutant (F and F′) ddaC (color coded in green in E′ and F′) and v’ada (color coded in magenta in E′ and F′) neurons form a boundary (indicated by dashed lines) without showing any overlap. (G–H) The boundary between dendrites of adjacent ddaC and v’ada neurons is shown for wild type (G and G′) and Dscam18/DscamP1 third instar larvae (H and H′). ddaC (color coded in green in G′ and H′) and v’ada (color coded in magenta in G′ and H′) neurons form a boundary (indicated by dashed lines) without showing major overlap or crossing of dendrites. Scale bar, 30 μm.
Furthermore, we looked for the relatively rare events of two adjacent class IV da neuron MARCM clones homozygous for Dscam mutations. We found that neighboring ddaC and v’ada mutant neurons displayed fairly normal tiling (n=3, Figure 2 F and F′). Although we observed rare dendritic branch crossings (data not shown), overall the tiling of adjacent mutant clones resembled the tiling of neighboring control MARCM class IV da neuron clones (Figure 2 E and E′).
To allow for a more thorough quantitative analysis with a larger sample number we also analyzed tiling in larvae carrying the trans-heterozygous combination of Dscam18/DscamP1 alleles, since they can survive to the 3rd instar stage. We used ppk-Gal4, UAS-mCD8-GFP to specifically visualize class IV neurons and examined the adjacent dendritic fields of the v’ada and ddaC class IV neurons for interneuronal crossing defects. Similar to the MARCM analysis, virtually no crossing of dendrites from the neighboring neurons was observed in wild type (Fig. 2 D, G, G′). Likewise, in the Dscam mutant larvae, despite extensive isoneuronal dendrite crossing and bundling, no significant increase in interneuronal crossing of v’ada and ddaC dendrites was observed (Fig. 2 D, H, H′). The rare crossing or touching of inter-neuronal dendrites was often associated with bundling and therefore likely represents a secondary effect rather than a real tiling phenotype. These results support the notion that Dscam is required for self-avoidance, but not tiling of dendrites from adjacent class IV da neurons.
The Hpo/Trc/Fry pathway does not mediate self-avoidance in class I-III da neurons or prevention of class IV da neuron dendritic bundling
Dscam function in da neurons ensures self-avoidance by preventing dendritic crossing and bundling. To examine whether trc also contributes to self-avoidance, we compared the MARCM phenotypes of Dscam with trc mutants with respect to dendrite crossing and bundling in class I and IV da neurons.
Dscam mutant class I da neurons showed crossing and bundling of dendrites as described above (Figure 3 B). As expected of trc function in dendritic branching (Emoto et al., 2004), we observed overbranching proximal to the soma in trc mutant ddaE neurons (Figure 3 A). However, trc mutant class I da neurons exhibited no dendritic crosses or bundling (Figure 3 A, C, D). Moreover, we found no evidence for self-avoidance defects in class I da neurons in hpo or fry mutants (data not shown). This suggests that the Hpo/Trc/Fry signaling pathway is not required for self-avoidance of class I da neuron dendrites. Similarly, no self-avoidance defects were detected in trc or hpo mutant class II and III da neurons (data not shown).
Figure 3. trc function is not required for self-avoidance in class I da neurons and prevention of dendritic bundling in class IV da neurons.
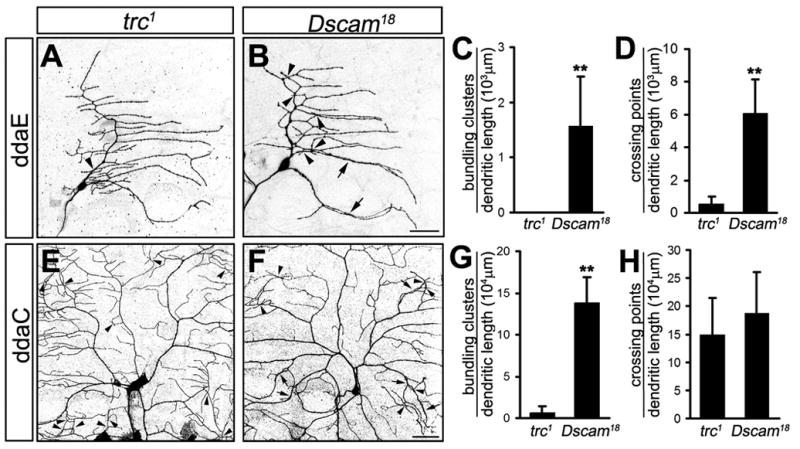
(A–D) MARCM clones of class I ddaE neurons mutant for trc1 (A) and Dscam18 (B) are shown and dendritic bundling (C) and crossing (D) was quantified for both genotypes. Dendritic bundling events were defined as the physical contact between two or more branches over a distance of more than 5 μm. Scale bar, 30 μm.
(E–H) MARCM clones of class IV ddaC neurons mutant for trc1 (E) and Dscam18 (F) are shown and dendritic bundling (G) and crossing (H) was quantified for both genotypes. While both trc and Dscam loss of function caused crossing defects, bundling of dendrites was only observed in the Dscam mutant. Scale bar, 30 μm. **, P<0.01, student’s t-test.
Class IV da neurons mutant for Dscam or trc showed iso-neuronal dendritic crossing as previously described (Figure 3 E, F, H) (Emoto et al., 2004). However, unlike in Dscam mutants, trc mutant class IV da neurons did not exhibit significant dendritic bundling (Figure 3 G). We also found no bundling defects in class IV da neurons in hpo or fry mutant animals (data not shown). Thus, the Hpo/Trc/Fry pathway is not required to prevent dendrite bundling of class IV da neurons, or self-avoidance of class I–III da neurons.
Expression of single Dscam isoforms significantly restored self-avoidance and normal dendritic projection
Neurons in wild type animals typically express multiple isoforms of Dscam (Neves et al., 2004; Zhan et al., 2004). While expression of a single Dscam isoform was sufficient to rescue axonal branch segregation in mushroom body neurons (Wang et al., 2002; Zhan et al., 2004), it proved insufficient to rescue the axonal projections of Drosophila mechanosensory neurons (Chen et al., 2006). However, given that self-avoidance involves dendritic interactions within a single neuron, we wondered whether the homophilic interaction of a single isoform of Dscam is sufficient to restore self-avoidance of Dscam mutant da neurons. We tested this possibility in several transgenic lines each carrying a randomly selected Dscam isoforms with or without a C-terminal GFP-tag.
First, we generated lines expressing Dscam isoforms carrying the alternative transmembrane exons 17.1 (TM1) and 17.2 (TM2) in class IV da neurons using the ppk-Gal4 driver. Although the ectopic expression might not completely reflect the endogenous localization pattern, previous studies have shown a distinct localization pattern of TM1 and TM2 isoforms in mushroom body neurons (Wang et al., 2004; Zhan et al., 2004). TM1 isoforms localize preferentially to the somatodendritic compartment, while TM2 isoforms localize to both dendritic and axonal compartments. In class IV da neurons, the TM1 isoform (3.36.25.1GFP) was primarily localized to the soma, but also present in punctuate structures in dendrites (Figure 4 A. B, E, and F). This TM1 isoform was also found at low levels in the class IV axon terminals (Figure 4 I). The same TM1 isoform without a GFP-tag (3.36.25.1) showed a similar localization pattern in dendrites and axons as revealed by Dscam antibody staining (Supplemental Figure 3). The TM2 isoform (3.36.25.2) localized to a similar extent to both axons and dendrites of class IV da neurons (Figure 4 C, D, and J), although in dendrites, it was present in larger puncta (Figure 4 G, H) as compared to the TM1 isoform (Figure 4 E, F).
Figure 4. Subcellular localization of TM1 and TM2 Dscam isoforms.
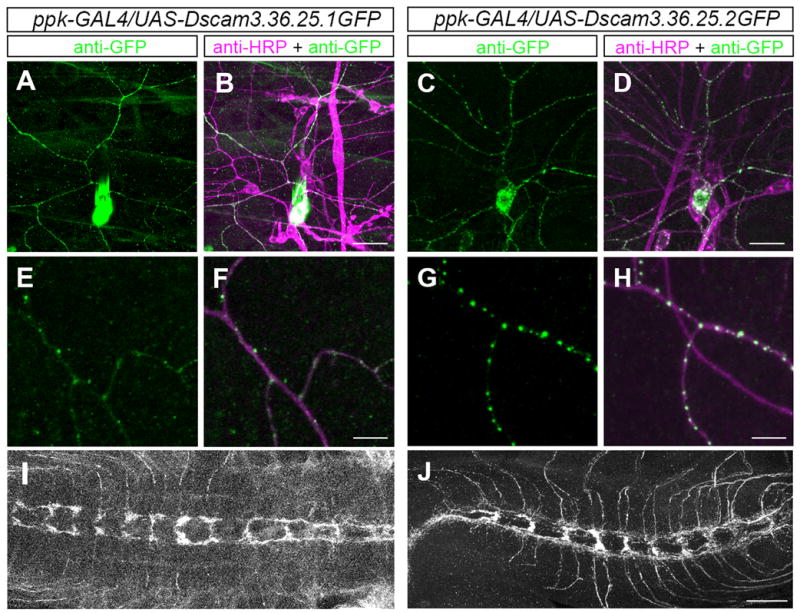
(A–J) The transgenic GFP-tagged Dscam isoforms TM1 (3.36.25.1) and TM2 (3.36.25.2) were expressed in class IV da neurons using ppk-Gal4. The somatodendritic distribution pattern of GFP-labeled TM1 (A) and TM2 (C) isoforms in ddaC neurons is shown, and overlaid with staining using the anti-HRP antibody, a pan-neuronal membrane marker (Jan and Jan, 1982) (B and D). Scale bar, 30 μm. (E–H) Enlarged view of a ddaC dendritic segment showing a punctate pattern of TM1 (E) and TM 2 (G) Dscam isoforms and merged with the anti-HRP staining (F, H). Scale bar, 30 μm. TM1 (I) and TM2 (J) isoform expression was also detected in class IV axonal projections in the VNC. Scale bar, 30 μm.
Besides the two isoforms with identical extracellular domains, we generated additional lines expressing Dscam isoforms with different extracellular domains and tested their ability to rescue the self-avoidance defects in class I and class IV da neurons as well. Using Gal42-21, UAS-mCD8-GFP to visualize class I da neurons, or ppk-Gal4, UAS-mCD8-GFP to delineate class IV neurons, we observed dendritic bundling and dendrite crossing phenotypes in Dscam18/DscamP1 larvae (Figure 5B, 6B), similar to the self-avoidance defects observed in MARCM clones of Dscam mutant da neurons. In Dscam mutant class I neurons, expression of a single Dscam isoform under the control of Gal42-21 largely restored wild type morphology of class I ddaE neurons (Figure 5C–E). The number of isoneuronal dendritic crosses (Figure 5 G) and bundling defects (Figure 5 H) was significantly reduced upon expression of each of five Dscam transgenes tested in our analysis. Notably, dendritic self-avoidance was restored to a similar extent by Dscam isoforms with three different extracellular domains and TM1 or TM2 transmembrane domains (Figure 5 G, H). In contrast, Dscam lacking the intracellular domain (Dscam 3.36.25.1ΔC) was not able to rescue dendrite crossing and bundling (Figure 5 F, G, H) indicating that intracellular signaling via the Dscam C-terminal domain is required for self-avoidance.
Figure 5. Introduction of single Dscam isoforms into the Dscam mutant background substantially rescues class I dendrite crossing and bundling.
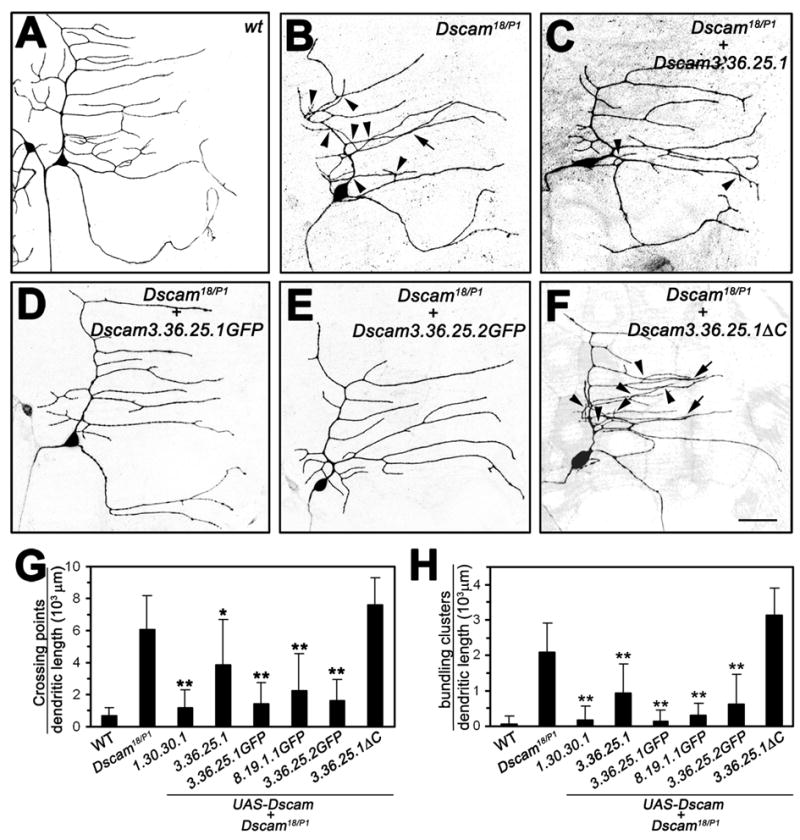
(A–F) Class I ddaE neurons visualized by the expression of UAS-mCD8-GFP driven by the class I specific Gal42-21 in third instar larvae. Wild type (A, n=12), Dscam18/DscamP1 (B, n=16), and Dscam18/DscamP1 expressing UAS-Dscam (3.36.25.1) (C, n=9), UAS-Dscam (3.36.25.1GFP) (D, n=16), UAS-Dscam (3.36.25.2GFP) (E, n=10), or UAS-Dscam (3.36.25.1ΔC) (F, n=13) under the control of Gal42-21. Arrowheads indicate dendritic branch crossing and arrows indicate bundling events. (G–H) Quantitative analysis of dendritic crosses (G) and bundling (H) of the indicated genotypes (sample number as indicated above, and for UAS-Dscam (1.30.30.1) (n=8) and UAS-Dscam (8.19.1.1GFP) (n=10)). Statistical significance of the rescue of self-avoidance defects was compared to Dscam mutant ddaE neurons. mean±SD; *, P<0.05, **, P<0.01, one-way ANOVA.
Figure 6. Introduction of single Dscam isoforms into the Dscam mutant background substantially rescues class IV dendrite crossing and bundling.
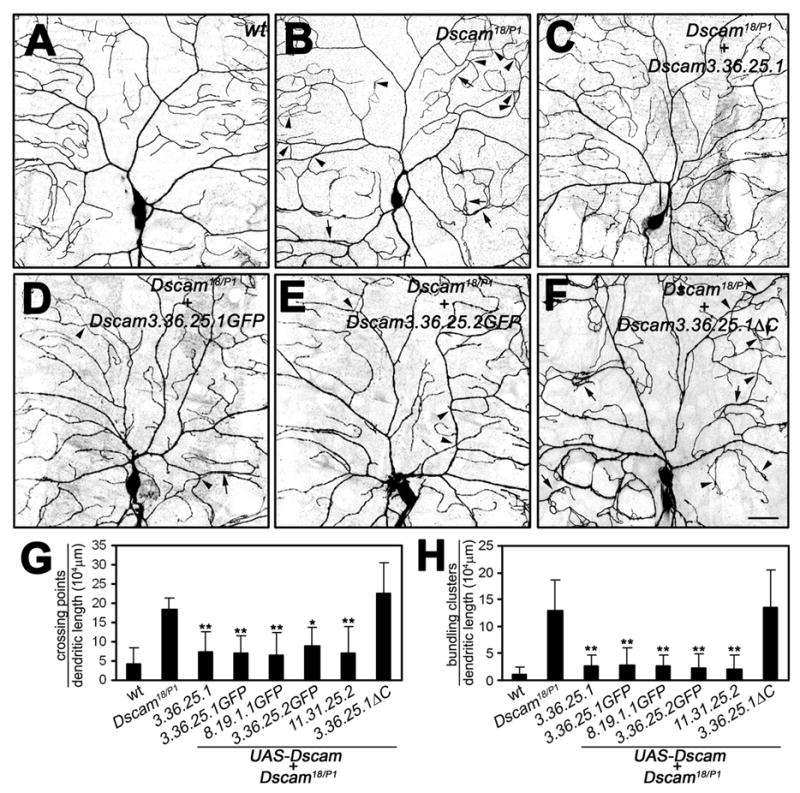
(A–F) Class IV ddaC neurons visualized via the expression of UAS-mCD8-GFP driven by class IV specific ppk-Gal4. Wild type (A, n=6), Dscam18/DscamP1 (B, n=6), and Dscam18/DscamP1 expressing UAS-Dscam (3.36.25.1) (C, n=8), UAS-Dscam (3.36.25.1GFP) (D, n=9), UAS-Dscam (3.36.25.2GFP) (E, n=6), or UAS-Dscam (3.36.25.1ΔC) (F, n=8). Arrowheads indicate dendritic branch crossing and arrows indicate bundling events. Scale bars, 30 μm. Note that expression of a single Dscam isoform in class IV neurons significantly rescued the Dscam loss of function phenotype with regard to dendritic crossing and bundling in ddaC neurons.
(G–H) Quantitative analysis of crossing points (G) and bundling events (H) of class IV ddaC neurons of the indicated genotypes (sample number as indicated above, and for UAS-Dscam (11.31.325.2) (n=8)). Statistical significance of the rescue of self-avoidance defects was compared to Dscam mutant ddaC neurons. mean±SD; **, P<0.01, one-way ANOVA.
Similarly, in class IV da neurons, the expression of a single isoform of Dscam rescued dendritic self-avoidance and bundling to a large extent (Figure 6 G, H); in rescued Dscam mutant larvae (Figure 6 C–E), the number of dendritic crosses and bundling defects in ddaC neurons expressing each of the tested Dscam isoforms was comparable to wild type (Figure 6 A) and significantly lower than that in Dscam mutant larvae (Figure 6 B). Furthermore, class IV da neurons expressing a single Dscam isoform displayed an overall dendritic field coverage resembling wild type with little or no dendrite bundling compared to Dscam mutants (Figure 6 C–E). In contrast, expression of Dscam lacking the intracellular domain (Dscam 3.36.25.1ΔC) did not rescue dendritic crosses or bundling (Figure 6 F, G, H), even though it showed a comparable expression and localization pattern (data not shown). Instead, it led to aberrant targeting especially of terminal dendrites, with the dendritic tips frequently touching and adhering to neighboring branches (Figure 6 F). These results indicate that while a single full length isoform of Dscam is sufficient for self-avoidance of class IV da neurons, the lack of the Dscam intracellular domain results in a loss of signaling required for repulsion.
Loss of Dscam function also affected the axonal projections of class IV da neurons in the ventral nerve cord (VNC), which could be partially restored by expression of a single Dscam isoform (Supplemental Figure 4).
Avoidance of dendrites of duplicated class I da neurons overexpressing the same Dscam isoform
Having found that a single isoform of Dscam could largely restore dendritic self-avoidance, we tested whether expression of the same Dscam isoform in neurons that normally do not avoid one another could induce avoidance of their dendrites. It was previously found that duplicated class I da neurons in hamlet mutants show no avoidance towards each other, but rather exhibit similar dendritic projection patterns with extensive overlap (Grueber et al., 2003). We obtained similar results by shifting a temperature sensitive Notch (Nts) mutant to the restrictive temperature for 1–2 hours during early embryonic development resulting in duplication of about one in ten vpda neurons. As in hamlet mutants, duplicated vpda neurons in Nts larvae extended dendritic arbors of normal shape and size that overlapped extensively (Figure 7 A, A′, B, B′). Non-duplicated vpda neurons in the same animals had identical morphology as wild type vpda neurons in control animals (Supplemental Figure 5). Thus, the transient inactivation of Notch did not affect vpda dendrite development, but provided an opportunity to examine dendritic interactions between the duplicated class I neurons. In contrast to the extensive overlap between duplicated control neuron dendrites, expression of a single Dscam isoform (3.36.25.1GFP) in the duplicated vpda neurons led to separation of their dendritic fields with dendrites of the neighboring neurons avoiding each other (Figure 7 C, C′, D, D′).
Figure 7. Ectopic expression of a single Dscam isoform induces recognition and repulsion of duplicated vpda neurons.
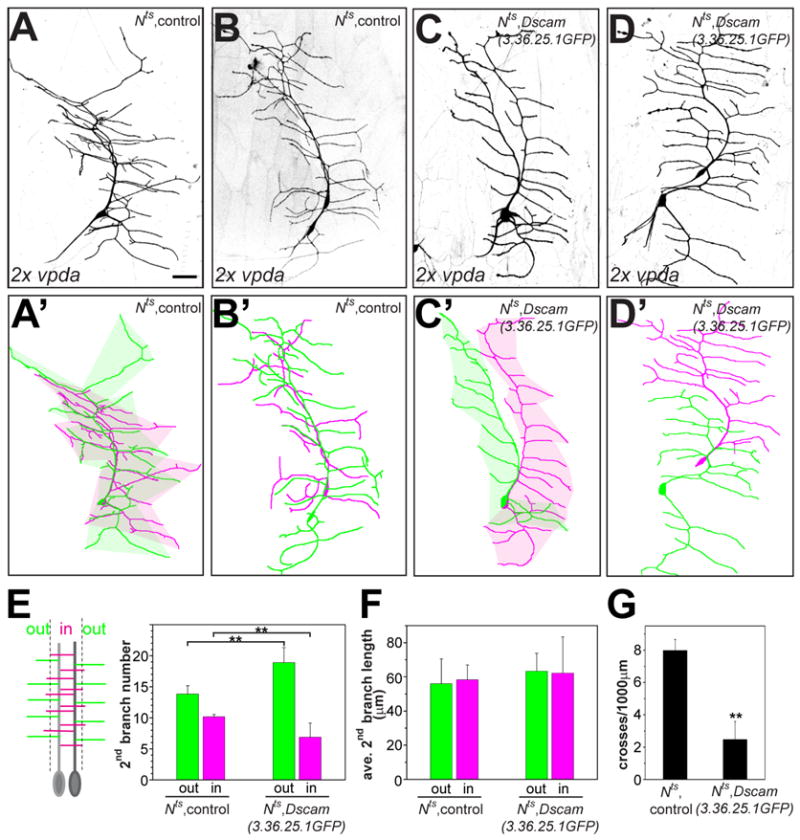
(A–D) Class I vpda neurons were duplicated using the temperature sensitive Nts allele with a 1–2 h shift to the restrictive temperature during embryonic development and visualized in third instar larvae with Gal42-21 driven UAS-mCD8-GFP. Two examples are shown each for Nts, control (A and B) and Nts, UAS-Dscam (3.36.25.1GFP) (C and D) are shown together with their corresponding color-coded tracings (A′–D′). Dendritic fields of duplicated vpda neurons expressing the same Dscam isoform show avoidance and highly reduced dendritic field overlap, as indicated by the shaded colored areas representing each individual dendritic field (A′ and C′). Scale bar, 30 μm. (E–G) Quantitative analysis of Dscam induced repulsion. Secondary branches of duplicated vpda neurons were classified for those branching away (“out“) or towards each other (“in”). The number (E) and length (F) of secondary “in” and “out” branches and the number of interneuronal dendritic crosses (G) were determined for each vpda pair of the indicated genotype (n=8 for each group). **, P<0.01, student’s t-test.
We quantified the length and number of secondary branches of duplicated vpda neurons growing either towards (“in”) or away (“out”) from each other (Figure 7 E, F) and the number of branch crosses between the two neurons (Figure 7 G). Duplicated control vpda neurons readily grew towards each other and their dendrites crossed extensively. In contrast, duplicated vpda neurons expressing the same Dscam isoform preferentially branched away from each and exhibited greatly reduced dendrite crossing, while the average length of secondary branches remained unaltered (Figure 7 F, G). In the same animals, non-duplicated vpda neurons expressing Dscam did not exhibit a preference in the direction of branching (Suplementary Figure 5). It thus appears that the presence of the same isoform of Dscam in the duplicated neurons is sufficient for their dendrites to avoid one another and force a switch in branching directionality.
Expression of the same Dscam isoform in different classes of da neurons prevents the overlap of their dendritic fields
Having found that expression of a common Dscam isoform prevented duplicated class I da neurons from sharing the same dendritic field, we wondered whether such avoidance behavior could be conferred to neurons of different classes that normally overlap in their dendritic field coverage. If expression of the same Dscam isoform in different classes of da neurons prevented them from innervating the same region of the body wall, this undesirable outcome would provide one rationale for the existence of a very large number of Dscam splice variants, namely, to ensure self-avoidance without significant risk of neighboring neurons inadvertently expressing a common Dscam isoform.
To address this question, we used the pan-da Gal421-7 driver to specifically express each of four different Dscam isoforms in all da neurons. During development, the class I vpda neuron is the first ventral da neuron to project dendrites dorsally, followed by the dendritic growth of neighboring class II, III and IV neurons, whose dendrites substantially overlap those of the vpda neuron (Figure 8 A and A′). Remarkably, pan-da neuronal overexpression of a single Dscam isoform with either the TM1 or Tm2 transmembrane domain greatly reduced the overlap and crossing of dendrites from different classes of da neurons (Figure 8 B, B′, C, and C′). The dendritic field established early on by the vpda neuron remained nearly void of other neuronal dendrites, acting as a barrier for ventral neurons projecting to the posterior segment boundary. In addition, some of the neighboring neurons showed enlarged dendritic terminals (Figure 8 B′, arrows) indicative of stalled dendritic growth. In da neurons expressing a common Dscam isoform, the number of heteroneuronal dendritic crosses with the vpda neuron was consistently reduced. Comparable results were obtained for each of the four Dscam isoforms tested (Figure 8D).
Figure 8. Ectopic expression of single Dscam isoforms induces heteroneuronal recognition and repulsion.
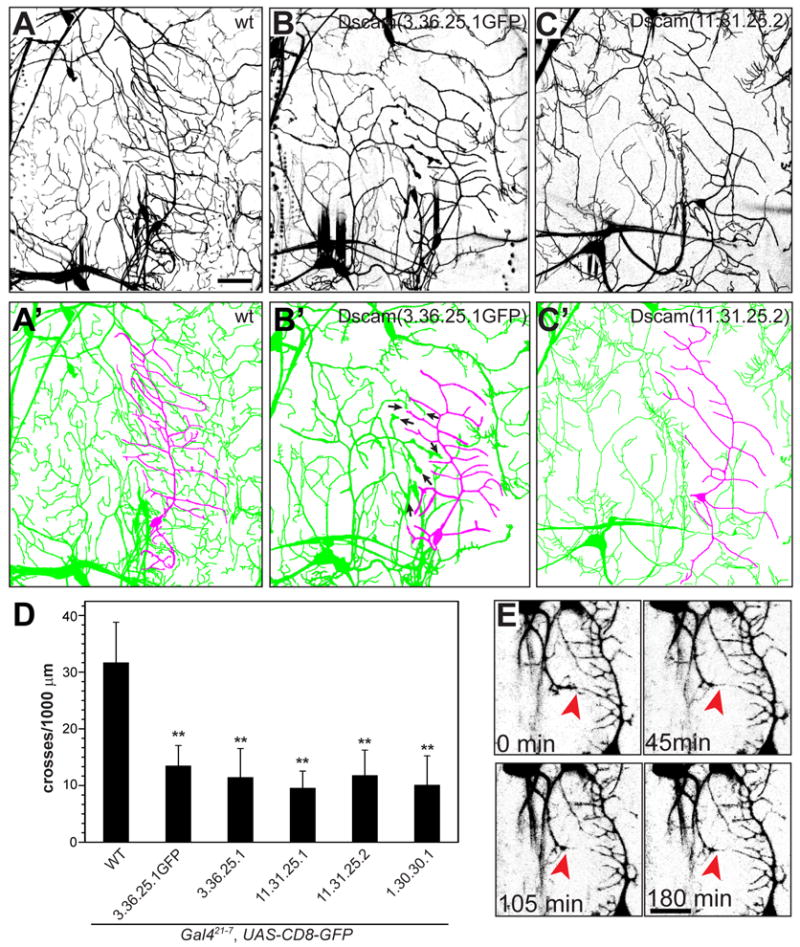
(A–C) The pan-da neuronal driver Gal421-7 driving UAS-mCD8-GFP allowed visualization of da neurons at the third instar larval stage. The wild type (A), UAS-Dscam (3.36.25.1GFP) (B), and UAS-Dscam (11.31.25.2) (C) overexpressing ventral and ventral’ da neuron clusters and their corresponding tracings (A′, B′ and C′) are shown. The vpda class I neuron is traced in magenta and the ventral and ventral’ class II, II and IV neurons are shown in green. Arrows in (B′) mark enlarged dendritic terminals indicative of stalled dendritic growth. Scale bar, 30 μm.
(D) Quantitative analysis of Dscam induced repulsion between ventral da neurons. Heteroneuronal dendritic crossing points between vpda dendrites and other ventral da neurons were counted and normalized to the total dendritic length of the vpda neuron for each genotype as indicated. (n=13 in each group, mean±SD). **, P<0.01, one-way ANOVA.
(E) Time lapse images of stage 17 embryos overexpressing GFP-tagged Dscam(8.19.1.1) in all da neurons under the control of Gal480-G2. Growing dendrites from the v’ada class IV neuron are repelled by vpda class I neuron dendrites and fail to penetrate the vpda dendritic field (indicated by red arrowheads). Scale bar equals 20 μm.
To look more closely into the behavior of da neuronal dendrites growing towards the vpda class I neuronal dendrites, we carried out time lapse imaging of da neurons expressing GFP-tagged Dscam (8.19.1.1) driven by the the pan-da Gal480-G2 driver in embryos, starting at stage 17, when the class I vpda neuron has established its dendritic field and neighboring dendrites begin to extend (Figure 8 E, 0 min). In these animals, the dendrites of neighboring neurons growing towards the vpda field retracted once they came into close proximity (Figure 8 E, 45 and 105 min). Moreover, even after reinitiation of growth they failed to extend into the vpda dendritic field (Figure 8 E, 180 min), an avoidance behavior never observed in control embryos. These results show that expression of a common Dscam isoform induces avoidance and severely compromises the ability of different classes of da neurons to cover the same territory.
Co-existence of overlapping dendritic fields is impaired in animals expressing only a single Dscam isoform
To further confirm that a single common Dscam isoform prevents co-existence of overlapping da neuron dendritic fields, we reduced the multiplicity of Dscam isoforms by expressing only a single isoform in all da neurons in Dscam mutant animals. For this purpose, we used Dscam18/DscamP1 mutant animals carrying a 4.5 kb Dscam promoter fragment fused to Dscam (3.36.25.1), and ppk-Gal, UAS-CD8-GFP to mark class IV da neurons. The 4.5 kb Dscam promoter fragment has previously been shown to reflect the endogenous Dscam expression pattern (Wang et al., 2004) and to be specific for pan-da neuronal expression in the PNS (see Supplemental Figure 1). Immunostaining with anti-CD8/anti-GFP and anti-HRP antibodies allowed us to visualize class IV da neurons and all sensory neurons (Jan and Jan, 1982), respectively, in the periphery.
In wild type (Figure 9 A) and Dscam mutant animals (data not shown), the class IV vdaB and v’ada neuron dendrites together covered the entire ventral field. The dendritic fields of these class IV da neurons completely overlapped with the class I vpda dendritic field (Figure 9 A′, A″). Remarkably, expression of a single isoform under the control of the Dscam promoter in the Dscam mutant background resulted in avoidance and partial exclusion of class IV dendrites (Figure 9 B) from the vpda dendritic field (Figure 9 B′, B″). Although expression of a single common Dscam isoform also reduced self-avoidance defects in all da neurons, it concomitantly caused inter-class avoidance. Quantitative analysis of Dscam mutant animals expressing a single Dscam isoform revealed that dendritic crosses (Figure 9 C) and dendritic field overlap (Figure 9 D) between class IV (vdaB, v’ada) and class I (vpda) da neurons was significantly reduced compared to wild type, reflecting an inability of class I and class IV da neurons to cover the same territory.
Figure 9. Co-existence of overlapping da neuron dendritic fields is impaired in larvae expressing only a single Dscam isoform.
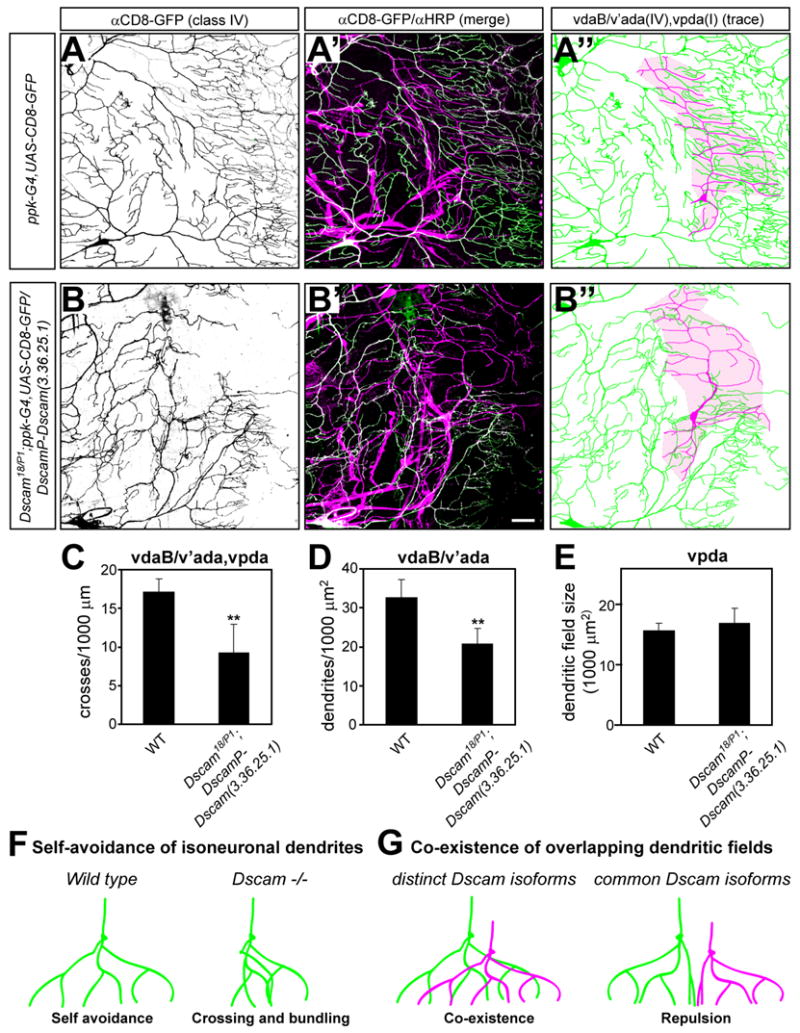
(A–E) Comparison of class IV (vdaB and v’ada) da neuron field coverage and overlap with the class I (vpda) neuron in wild type (A) and Dscam18/DscamP1; DscamP-Dscam (3.36.25.1) (B) third instar larvae carrying ppk-Gal, UAS-CD8-GFP. Larvae of both genotypes were immunostained with anti-CD8/anti-GFP and anti-HRP antibodies to visualize class IV da neurons (A and B) and all sensory neurons (merge with anti-CD8/GFP shown in A′ and B′), respectively. Corresponding tracings of the vpda class I neuron (shown in magenta) and class IV (vdaB. V’ada) neurons (shown in green) are shown for each genotype (A″ and B″). The vpda dendritic field is indicated by the shaded area (colored in magenta). Scale bar, 30 μm. Dendritic crosses (C, n=6) and dendritic field overlap between class IV (vdaB, v’ada) and class I (vpda) da neurons (D, n=6) and class I vpda dendritic field size (E) was quantified for each genotype as indicated. **, P<0.01, student’s t-test.
(F–G) Dscam plays a pivotal role in dendritic self avoidance and co-existence of overlapping dendritic fields. Self-avoidance of da neuron dendrites requires at least a single isoform of Dscam to ensure non-redundant and even target field coverage. The lack of Dscam leads to extensive crossing and bundling of dendrites due to a lack of self-recognition and repulsion (F). Dscam diversity is required to allow different types of neurons to innervate overlapping fields as observed for the different types of da neuron. Expression of a common Dscam isoform leads to dendrite recognition and avoidance resulting in non-overlapping dendritic fields (G).
Similarly, we observed avoidance between dendrites of the dorsal da neuron cluster using the same the Dscam promoter driven Dscam isoform in the Dscam mutant background as above (Supplemental Figure 6). Here, ddaC class IV neuron dendrites avoided other da neuron dendrites resulting in dendritic growth parallel to or around other da neuron dendrites. In severe cases, the failure of co-existence compromised the ability of ddaC neurons to cover the entire dorsal field (Supplemental Figure 6 C). Our results therefore show that in animals expressing only a single Dscam isoform, da neuron dendrites recognize and avoid each other. Thus, a lack of Dscam diversity severely impaired coexistence of da neuron dendritic fields in the same territory.
Discussion
We have identified Dscam as a key molecule for mediating dendritic self-avoidance of all four classes of da neurons. Loss of Dscam function resulted in excessive crossing and bundling of dendrites, accompanied by an uneven coverage of the dendritic field. These defects can be substantially corrected by the expression of each of four arbitrarily chosen Dscam isoforms. Our findings reveal that Dscam is critical for neurons to evenly extend their dendrites to cover the appropriate dendritic field, and to avoid the potential ambiguity arising from having different dendritic branches respond to the same input.
Given the necessity for different classes of da neurons to express at least one isoform of Dscam to ensure self avoidance, it is important to explore the possible consequences of having multiple da neurons in the same region express the same Dscam isoform. Different classes of da neurons presumably respond to different sensory inputs and hence their coverage of overlapping regions of the body wall will allow the animal to detect different types of sensory stimuli at the same physical locations. We found the expression of a common Dscam isoform made it almost impossible for different classes of da neurons to occupy the same territory, likely associated with deprivation of all but one type of sensory inputs at any one location. These considerations provide a plausible explanation for the existence of a vast number of Dscam splice variants (Neves et al., 2004; Zhan et al., 2004). For Dscam to mediate self-recognition manifested as dendritic self-avoidance in da neurons, it may be crucial to limit the expression of each of the Dscam isoforms to one or a small subset of neurons in any region of the nervous system.
This allows neighboring neurons to interact in ways distinct from the interactions that occur between isoneuronal processes of an individual neuron. Indeed, microarray analyses of individual Drosophila photoreceptor cells and mushroom body neurons have found distinct Dscam isoforms expressed in neighboring cells (Neves et al., 2004; Zhan et al., 2004). This supports the idea that an intricate splicing mechanism ensures that individual neurons can distinguish between self and non-self (Celotto and Graveley, 2001; Graveley, 2005; Neves and Chess, 2004). Given that divergent Dscam isoform expression is required for co-existence of da neurons in the same territory, it is likely that other mechanisms are involved in recognition of neighboring class IV da neurons for the purpose of tiling.
Intriguingly, even duplicated class I neurons most likely express distinct Dscam isoforms, since they occupy the same territory, but repel each other when expressing a common Dscam isoform. While the specific isoforms expressed by individual da neurons are currently unknown, the general principle of enabling dendrites of a single neuron to avoid one another without imposing recognition and avoidance of neighboring neurons underscores the importance of Dscam diversity. Since Dscam has been shown to primarily interact in an isoform specific manner (Wojtowicz et al., 2004), Dscam based self-recognition and avoidance depends on expression of the same isoforms. This is illustrated by the repulsive function of Dscam in vivo, where it induces branch retraction and avoidance of dendrites expressing identical isoforms (see Figure 8 B, C, E). The self avoidance phenomenon seems to be conserved in axonal development as well: in mushroom body neurons Dscam ensures the proper segregation and targeting of the two axonal processes to different lobes (Wang et al., 2002).
Our study reveals significant differences between self-avoidance, encompassing isoneuronal crossing and bundling of dendrites, and tiling. Dscam is essential for dendritic self-avoidance (dendritic crossing and bundling) of all classes of da neurons, but not for tiling of class IV da neurons. Thus, different recognition mechanisms are used for self-avoidance and tiling. The phenomenon of tiling, in which the dendrites of different neurons of the same class avoid one another, relies on components of the Hpo/Trc/Fry pathway (Emoto et al., 2004; Emoto et al., 2006). Whereas this signaling pathway also contributes to the avoidance of isoneuronal dendritic crossing of class IV da neurons, it is not required to prevent bundling of class IV da neuron dendrites, nor is it important for self-avoidance of the other three classes of da neurons. It is an interesting open question as to what machineries are employed to prevent the dendrites of each da neuron in class I-III from crossing one another. The involvement of the Hpo/Trc/Fry pathway in preventing crossing but not bundling of dendrites from the same class IV da neuron further suggests the possibility of multiple mechanisms for self-avoidance. While fasciculation of multiple processes, e.g. in axon guidance, is a widely used mechanism to ensure accurate projection to common target areas (Van Vactor, 1998), bundling of isoneuronal dendrites defeats the purpose of dendritic field coverage and non-redundant signal processing. Given the ample opportunities for neighboring dendritic branches of the same neuron to bundle, there is likely a specialized mechanism to repel branching dendrites from each other as they respond to cues for their extension.
Experimental Procedures
Fly stocks
Dscam18, DscamP1, Dscam21, Dscam23 (Hummel et al., 2003; Schmucker et al., 2000; Wang et al., 2002) and trc1, hpomgh4 (Emoto et al., 2006; Geng et al., 2000) mutant alleles have been described previously. The Dscam transgenic lines carrying the isoforms (3.36.25.1), (3.36.25.1GFP), (3.36.25.2GFP), (8.19.1.1GFP), (11.31.25.1), (11.31.25.2), (1.30.30.1), and (3.36.25.1ΔC) used in this study have been described (Wang et al., 2004; Zhu et al., 2006). Transgenic lines with the 4.5kb Dscam promoter fragment fused to Gal4 (DscamP-Gal4) or Dscam (3.36.25.1) (DscamP-Dscam (3.36.25.1)) have been described in (Wang et al., 2004). For visualization of class IV da neurons, a fusion of the pickpocket (ppk) enhancer (Grueber et al., 2003) to Gal4 was used to drive expression of mCD8-GFP. Gal42-21, UAS-mCD8-GFP was used to visualize class I da neurons (Parrish et al., 2006). The pan-da lines Gal421-7 (Song et al., 2007) and Gal480-G2 (Gao et al., 1999) were used to visualize all da neurons and for overexpression of Dscam isoforms. The notchts allele has been described in (Shellenbarger and Mohler, 1975).
MARCM analysis
MARCM analyses were essentially performed as described previously (Grueber et al., 2002). Briefly, FRTG13 Dscam18, UAS-mCD8GFP/CyO or FRTG13 DscamP1, UAS-mCD8GFP/CyO flies were mated with w; elav-Gal4, hs-Flp; FRTG13, tub-Gal80/CyO to generate mosaic clones. Analogously, FRT42D hpomgh4 and FRT80B trc1 flies were mated with the corresponding w; elav-Gal4, hs-Flp; FRT42D, tub-Gal80/CyO or w; elav-Gal4, hs-Flp; FRT80B, tub-Gal80/CyO, respectively. Embryos were collected for 2 h at 25 °C and allowed to develop for 3 h, then heat-shocked for 1h at 38 °C. Heat shocked embryos were kept at 25 °C and da neuron MARCM clones were analyzed in third instar larvae by immunohistochemistry.
Immunohistochemistry
Antibodies used in this study were: rat-anti-mCD8 antibody (1:100, Invitrogen, San Diego), mouse-anti-Dscam targeted against exon 18 (1:100), Cy5-mouse-anti-HRP (1:200, Jackson IR Labs, West Grove). Immunohistochemistry of Drosophila larvae was preformed essentially as described in (Grueber et al., 2002). Briefly, third instar larvae were dissected and fixed in 4% Formaldehyde/PBS and stained with the desired primary antibodies. Appropriate secondary antibodies coupled to Cy2, Rhodamine Red, or Cy5 were purchased from Jackson IR. Filets were mounted in Slowfade Gold (Invitrogen, San Diego) and analyzed by confocal microscopy on a Leica TCS SP2 microscope. Z-stacks of confocal sections were taken and maximized projections were used for quantitative analysis. For clarity and best reproduction of dendritic processes, images were inverted and contrast enhanced using Photoshop (Abobe Systems Inc., San Jose). Dendritic length and terminal branch numbers were quantified using the ImageJ (NIH, Bethesda) plug-in NeuronJ (Meijering et al., 2004).
Genetic duplication of class I neurons
Notchts females were crossed to Gal42-21, UAS-mCD8-GFP with or without UAS-Dscam-GFP(3.36.25.1). Embryos were collected for 1 h at 20 °C and allowed to develop for 3–4 h, and then transferred to the restrictive temperature of 29 °C for 1–2h. Embryos were allowed to develop at 20 °C and male third instar larvae (Nts/y; Gal42-21, UAS-mCD8-GFP/+ or Nts/y; UAS-Dscam-GFP(3.36.25.1)/+; Gal42-21, UAS-mCD8-GFP/+) were analyzed for duplication of class I neurons by confocal microscopy.
Supplementary Material
Acknowledgments
We thank members of the Jan lab for discussion; Jill Wildonger and Yi Zheng for comments on the manuscript; Hsiu-Hsiang Lee and Yang Xiang for Gal421-7, UAS-CD8-GFP; Jay Parrish for the Nts duplication procedure; Liqun Luo and Haitao Zhu for Dscam reagents. This work was supported by NIH grant RO1 NS40929. L.Y.J. and Y.-N.J. are Investigators of the Howard Hughes Medical Institute.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Amthor FR, Oyster CW. Spatial organization of retinal information about the direction of image motion. Proc Nat Acad Sci. 1995;92:4002–4005. doi: 10.1073/pnas.92.9.4002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen R, Li Y, Resseguie M, Brenman JE. Calcium/calmodulin-dependent protein kinase II alters structural plasticity and cytoskeletal dynamics in Drosophila. J Neurosci. 2005;25:8878–8888. doi: 10.1523/JNEUROSCI.2005-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Celotto AM, Graveley BR. Alternative splicing of the Drosophila Dscam pre-mRNA is both temporally and spatially regulated. Genetics. 2001;159:599–608. doi: 10.1093/genetics/159.2.599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen BE, Kondo M, Garnier A, Watson FL, Puettmann-Holgado R, Lamar DR, Schmucker D. The molecular diversity of Dscam is functionally required for neuronal wiring specificity in Drosophila. Cell. 2006;125:607–620. doi: 10.1016/j.cell.2006.03.034. [DOI] [PubMed] [Google Scholar]
- Emoto K, He Y, Ye B, Grueber WB, Adler PN, Jan LY, Jan YN. Control of dendritic branching and tiling by the Tricornered-kinase/Furry signaling pathway in Drosophila sensory neurons. Cell. 2004;119:245–256. doi: 10.1016/j.cell.2004.09.036. [DOI] [PubMed] [Google Scholar]
- Emoto K, Parrish JZ, Jan LY, Jan YN. The tumour suppressor Hippo acts with the NDR kinases in dendritic tiling and maintenance. Nature. 2006;443:210–213. doi: 10.1038/nature05090. [DOI] [PubMed] [Google Scholar]
- Gallegos ME, Bargmann CI. Mechanosensory neurite termination and tiling depend on SAX-2 and the SAX-1 kinase. Neuron. 2004;44:239–249. doi: 10.1016/j.neuron.2004.09.021. [DOI] [PubMed] [Google Scholar]
- Gao FB, Brenman JE, Jan LY, Jan YN. Genes regulating dendritic outgrowth, branching, and routing in Drosophila. Gen Dev. 1999;13:2549–2561. doi: 10.1101/gad.13.19.2549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao FB, Kohwi M, Brenman JE, Jan LY, Jan YN. Control of dendritic field formation in Drosophila: the roles of flamingo and competition between homologous neurons. Neuron. 2000;28:91–101. doi: 10.1016/s0896-6273(00)00088-x. [DOI] [PubMed] [Google Scholar]
- Geng W, He B, Wang M, Adler PN. The tricornered gene, which is required for the integrity of epidermal cell extensions, encodes the Drosophila nuclear DBF2-related kinase. Genetics. 2000;156:1817–1828. doi: 10.1093/genetics/156.4.1817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graveley BR. Mutually exclusive splicing of the insect Dscam pre-mRNA directed by competing intronic RNA secondary structures. Cell. 2005;123:65–73. doi: 10.1016/j.cell.2005.07.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grueber WB, Jan LY, Jan YN. Tiling of the Drosophila epidermis by multidendritic sensory neurons. Development. 2002;129:2867–2878. doi: 10.1242/dev.129.12.2867. [DOI] [PubMed] [Google Scholar]
- Grueber WB, Ye B, Moore AW, Jan LY, Jan YN. Dendrites of distinct classes of Drosophila sensory neurons show different capacities for homotypic repulsion. Curr Biol. 2003;13:618–626. doi: 10.1016/s0960-9822(03)00207-0. [DOI] [PubMed] [Google Scholar]
- Hummel T, Vasconcelos ML, Clemens JC, Fishilevich Y, Vosshall LB, Zipursky SL. Axonal targeting of olfactory receptor neurons in Drosophila is controlled by Dscam. Neuron. 2003;37:221–231. doi: 10.1016/s0896-6273(02)01183-2. [DOI] [PubMed] [Google Scholar]
- Jan LY, Jan YN. Antibodies to horseradish peroxidase as specific neuronal markers in Drosophila and in grasshopper embryos. Proc Nat Acad Sci. 1982;79:2700–2704. doi: 10.1073/pnas.79.8.2700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jan YN, Jan LY. The control of dendrite development. Neuron. 2003;40:229–242. doi: 10.1016/s0896-6273(03)00631-7. [DOI] [PubMed] [Google Scholar]
- Lee T, Winter C, Marticke SS, Lee A, Luo L. Essential roles of Drosophila RhoA in the regulation of neuroblast proliferation and dendritic but not axonal morphogenesis. Neuron. 2000;25:307–316. doi: 10.1016/s0896-6273(00)80896-x. [DOI] [PubMed] [Google Scholar]
- Meijering E, Jacob M, Sarria JC, Steiner P, Hirling H, Unser M. Design and validation of a tool for neurite tracing and analysis in fluorescence microscopy images. Cytometry A. 2004;58:167–176. doi: 10.1002/cyto.a.20022. [DOI] [PubMed] [Google Scholar]
- Neves G, Zucker J, Daly M, Chess A. Stochastic yet biased expression of multiple Dscam splice variants by individual cells. Nat Genetics. 2004;36:240–246. doi: 10.1038/ng1299. [DOI] [PubMed] [Google Scholar]
- Parrish JZ, Kim MD, Jan LY, Jan YN. Genome-wide analyses identify transcription factors required for proper morphogenesis of Drosophila sensory neuron dendrites. Gen Dev. 2006;20:820–835. doi: 10.1101/gad.1391006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry VH, Linden R. Evidence for dendritic competition in the developing retina. Nature. 1982;297:683–685. doi: 10.1038/297683a0. [DOI] [PubMed] [Google Scholar]
- Rockhill RL, Euler T, Masland RH. Spatial order within but not between types of retinal neurons. Proc Nat Acad Sci. 2000;97:2303–2307. doi: 10.1073/pnas.030413497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmucker D, Clemens JC, Shu H, Worby CA, Xiao J, Muda M, Dixon JE, Zipursky SL. Drosophila Dscam is an axon guidance receptor exhibiting extraordinary molecular diversity. Cell. 2000;101:671–684. doi: 10.1016/s0092-8674(00)80878-8. [DOI] [PubMed] [Google Scholar]
- Shellenbarger DL, Mohler JD. Temperature-sensitive mutations of the notch locus in Drosophila melanogaster. Genetics. 1975;81:143–162. doi: 10.1093/genetics/81.1.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song W, Onishi M, Jan LY, Jan YN. Peripheral multidendritic sensory neurons are necessary for rhythmic locomotion behavior in Drosophila larvae. Proc Nat Acad Sci. 2007;104:5199–5204. doi: 10.1073/pnas.0700895104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuart G, Spruston N, Ha□usser M. Dendrites. Oxford; New York: Oxford University Press; 1999. [Google Scholar]
- Sugimura K, Yamamoto M, Niwa R, Satoh D, Goto S, Taniguchi M, Hayashi S, Uemura T. Distinct developmental modes and lesion-induced reactions of dendrites of two classes of Drosophila sensory neurons. J Neurosci. 2003;23:3752–3760. doi: 10.1523/JNEUROSCI.23-09-03752.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Vactor D. Adhesion and signaling in axonal fasciculation. Curr Opin Neurobiol. 1998;8:80–86. doi: 10.1016/s0959-4388(98)80011-1. [DOI] [PubMed] [Google Scholar]
- Wang J, Ma X, Yang JS, Zheng X, Zugates CT, Lee CH, Lee T. Transmembrane/juxtamembrane domain-dependent Dscam distribution and function during mushroom body neuronal morphogenesis. Neuron. 2004;43:663–672. doi: 10.1016/j.neuron.2004.06.033. [DOI] [PubMed] [Google Scholar]
- Wang J, Zugates CT, Liang IH, Lee CH, Lee T. Drosophila Dscam is required for divergent segregation of sister branches and suppresses ectopic bifurcation of axons. Neuron. 2002;33:559–571. doi: 10.1016/s0896-6273(02)00570-6. [DOI] [PubMed] [Google Scholar]
- Wassle H, Peichl L, Boycott BB. Dendritic territories of cat retinal ganglion cells. Nature. 1981;292:344–345. doi: 10.1038/292344a0. [DOI] [PubMed] [Google Scholar]
- Watson FL, Puttmann-Holgado R, Thomas F, Lamar DL, Hughes M, Kondo M, Rebel VI, Schmucker D. Extensive diversity of Ig-superfamily proteins in the immune system of insects. Science. 2005;309:1874–1878. doi: 10.1126/science.1116887. [DOI] [PubMed] [Google Scholar]
- Wojtowicz WM, Flanagan JJ, Millard SS, Zipursky SL, Clemens JC. Alternative splicing of Drosophila Dscam generates axon guidance receptors that exhibit isoform-specific homophilic binding. Cell. 2004;118:619–633. doi: 10.1016/j.cell.2004.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhan XL, Clemens JC, Neves G, Hattori D, Flanagan JJ, Hummel T, Vasconcelos ML, Chess A, Zipursky SL. Analysis of Dscam diversity in regulating axon guidance in Drosophila mushroom bodies. Neuron. 2004;43:673–686. doi: 10.1016/j.neuron.2004.07.020. [DOI] [PubMed] [Google Scholar]
- Zhu H, Hummel T, Clemens JC, Berdnik D, Zipursky SL, Luo L. Dendritic patterning by Dscam and synaptic partner matching in the Drosophila antennal lobe. Nat Neurosci. 2006;9:349–355. doi: 10.1038/nn1652. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


