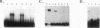Abstract
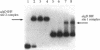
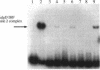
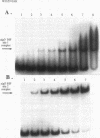
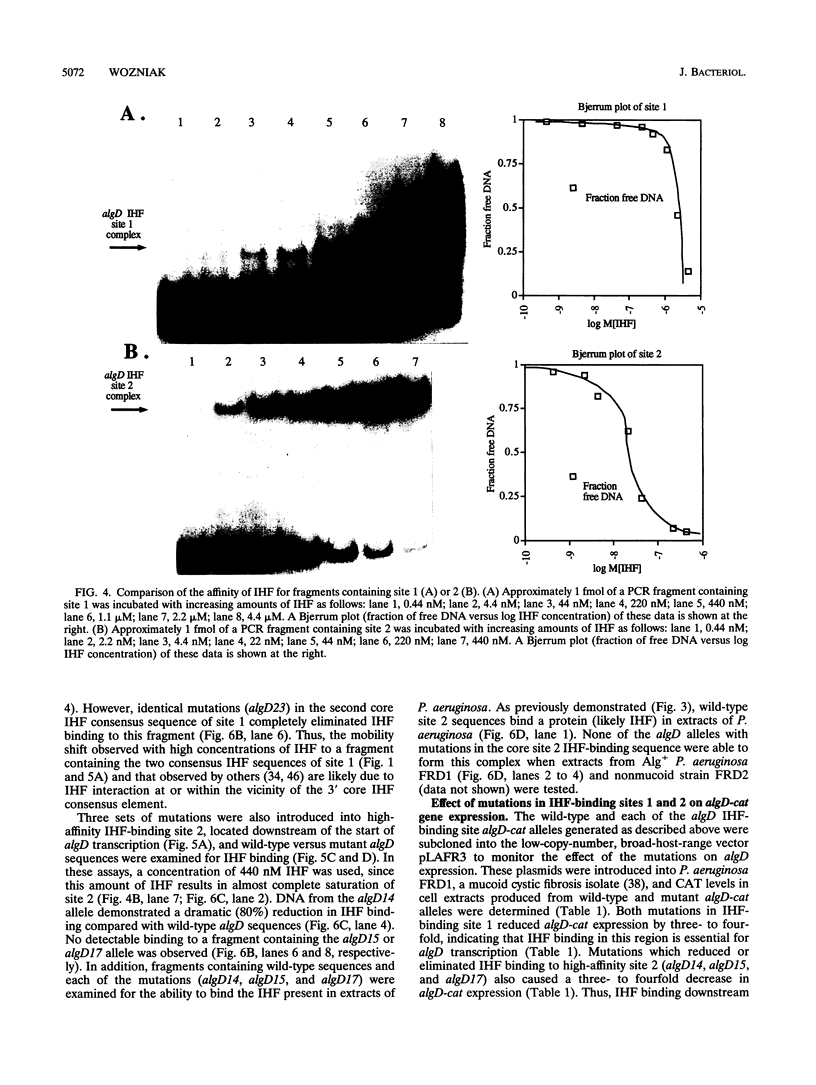
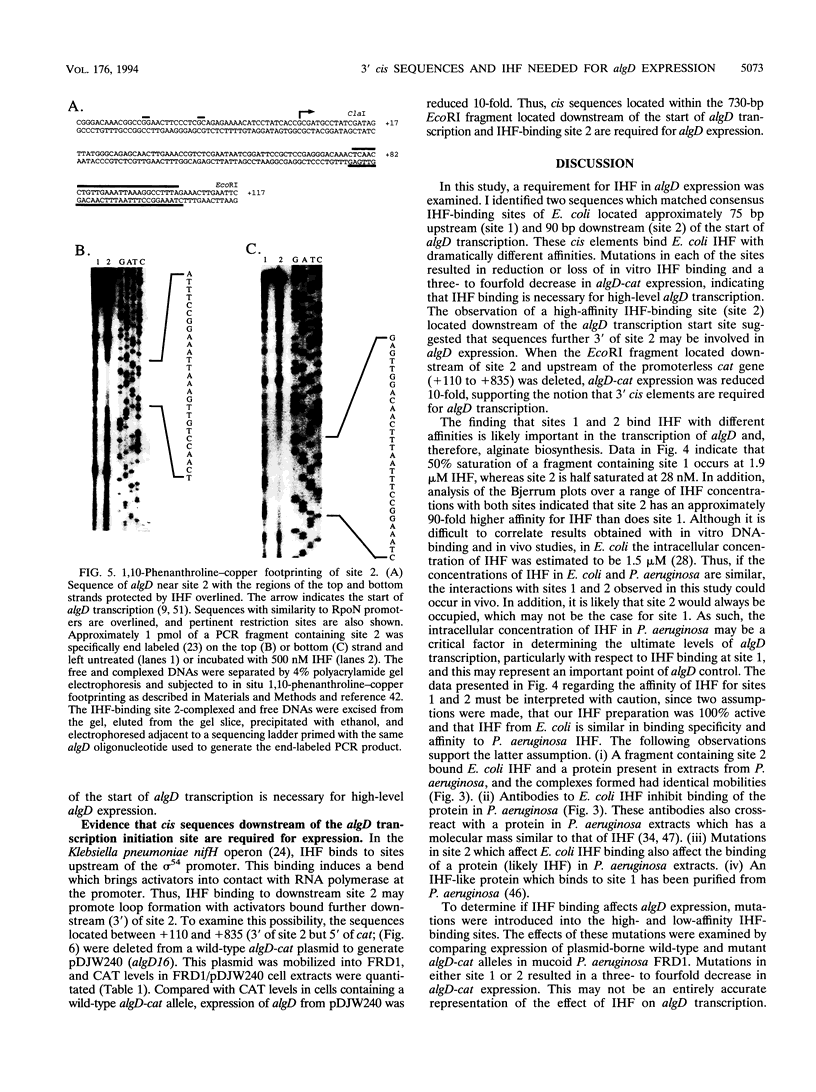
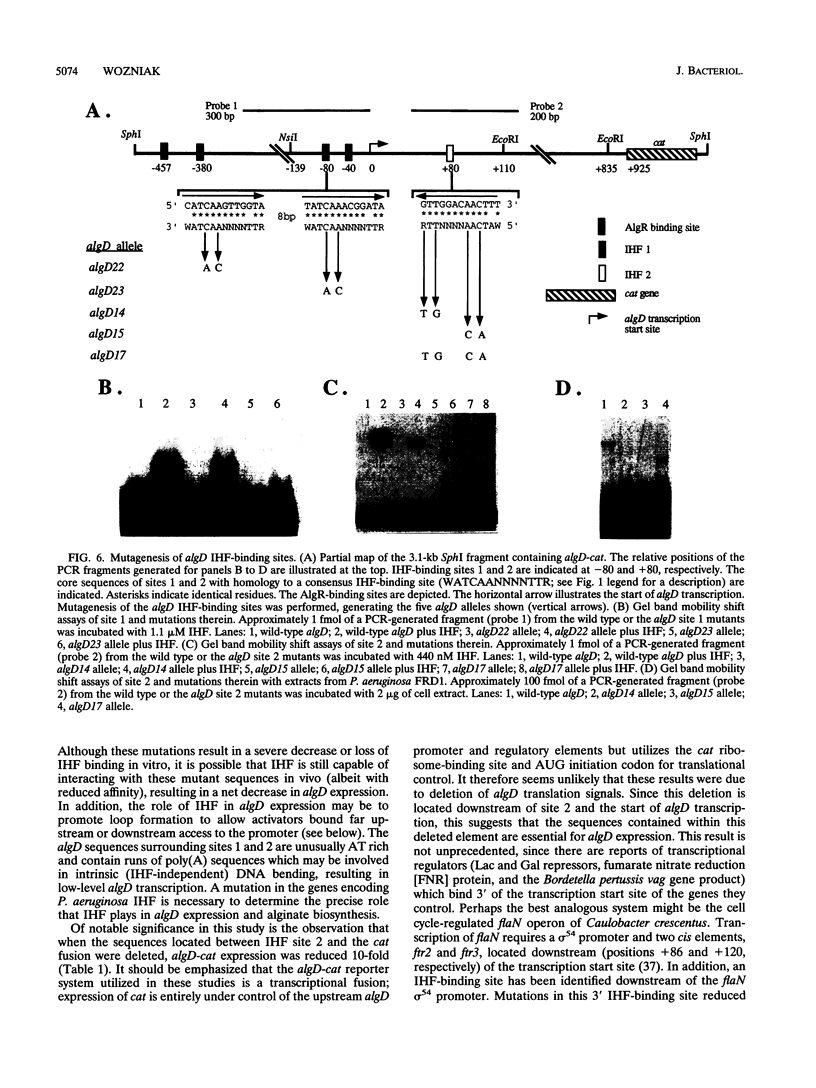
Pseudomonas aeruginosa is an extremely important opportunistic pathogen in immunocompromised individuals. Strains of P. aeruginosa isolated from chronic lung infections in patients with the genetic disease cystic fibrosis have a mucoid colony morphology. This phenotype is due to overproduction of the exopolysaccharide alginate, which is believed to confer a selective advantage on P. aeruginosa in cystic fibrosis lungs. Alginate biosynthesis is controlled by a complex regulatory mechanism. Genes located in the 34-min region of the P. aeruginosa chromosome form an operon which encodes most of the biosynthetic enzymes necessary for alginate production. algD, the first gene in the operon and a critical point for the transcriptional regulation of alginate biosynthesis, is controlled by several trans, cis, and environmental factors. In this study, the involvement of the histone-like protein integration host factor (IHF) in algD expression was examined. Sequences with similarity to consensus IHF-binding sites of Escherichia coli were identified 75 bp upstream (site 1) and 90 bp downstream (site 2) of the start of algD transcription. In gel band mobility shift assays, DNA fragments containing either site bind IHF but site 2 has an approximately 90-fold higher affinity for IHF. Mutations in each of the elements were generated, and they resulted in the reduction or loss of in vitro IHF binding and a three- to fourfold decrease in algD-cat expression. This indicates that IHF binding is necessary for high-level algD transcription. The presence of a high-affinity IHF-binding site located 3' of the algD transcription start site suggested that sequences further downstream of this element are involved in algD expression. When a fragment located downstream of site 2 and upstream of the promoterless cat gene (+110 to +835) was deleted, algD-cat expression was reduced 10-fold supporting the notion that 3' enhancer elements are required for algD transcription. This is the first direct evidence of a 3' element involved in the control of a P. aeruginosa gene. It is postulated that IHF mediates the formation of a higher-order looped structure which is necessary for efficient algD transcription.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Balke V. L., Gralla J. D. Changes in the linking number of supercoiled DNA accompany growth transitions in Escherichia coli. J Bacteriol. 1987 Oct;169(10):4499–4506. doi: 10.1128/jb.169.10.4499-4506.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayer A. S., Eftekhar F., Tu J., Nast C. C., Speert D. P. Oxygen-dependent up-regulation of mucoid exopolysaccharide (alginate) production in Pseudomonas aeruginosa. Infect Immun. 1990 May;58(5):1344–1349. doi: 10.1128/iai.58.5.1344-1349.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourret R. B., Fox M. S. Lysogenization of Escherichia coli him+, himA, and himD hosts by bacteriophage Mu. J Bacteriol. 1988 Apr;170(4):1672–1682. doi: 10.1128/jb.170.4.1672-1682.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford M. M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976 May 7;72:248–254. doi: 10.1006/abio.1976.9999. [DOI] [PubMed] [Google Scholar]
- Darzins A., Chakrabarty A. M. Cloning of genes controlling alginate biosynthesis from a mucoid cystic fibrosis isolate of Pseudomonas aeruginosa. J Bacteriol. 1984 Jul;159(1):9–18. doi: 10.1128/jb.159.1.9-18.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeVault J. D., Hendrickson W., Kato J., Chakrabarty A. M. Environmentally regulated algD promoter is responsive to the cAMP receptor protein in Escherichia coli. Mol Microbiol. 1991 Oct;5(10):2503–2509. doi: 10.1111/j.1365-2958.1991.tb02096.x. [DOI] [PubMed] [Google Scholar]
- Deretic V., Dikshit R., Konyecsni W. M., Chakrabarty A. M., Misra T. K. The algR gene, which regulates mucoidy in Pseudomonas aeruginosa, belongs to a class of environmentally responsive genes. J Bacteriol. 1989 Mar;171(3):1278–1283. doi: 10.1128/jb.171.3.1278-1283.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deretic V., Gill J. F., Chakrabarty A. M. Gene algD coding for GDPmannose dehydrogenase is transcriptionally activated in mucoid Pseudomonas aeruginosa. J Bacteriol. 1987 Jan;169(1):351–358. doi: 10.1128/jb.169.1.351-358.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deretic V., Govan J. R., Konyecsni W. M., Martin D. W. Mucoid Pseudomonas aeruginosa in cystic fibrosis: mutations in the muc loci affect transcription of the algR and algD genes in response to environmental stimuli. Mol Microbiol. 1990 Feb;4(2):189–196. doi: 10.1111/j.1365-2958.1990.tb00586.x. [DOI] [PubMed] [Google Scholar]
- Deretic V., Hibler N. S., Holt S. C. Immunocytochemical analysis of AlgP (Hp1), a histonelike element participating in control of mucoidy in Pseudomonas aeruginosa. J Bacteriol. 1992 Feb;174(3):824–831. doi: 10.1128/jb.174.3.824-831.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deretic V., Konyecsni W. M. A procaryotic regulatory factor with a histone H1-like carboxy-terminal domain: clonal variation of repeats within algP, a gene involved in regulation of mucoidy in Pseudomonas aeruginosa. J Bacteriol. 1990 Oct;172(10):5544–5554. doi: 10.1128/jb.172.10.5544-5554.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorman C. J., Barr G. C., Ni Bhriain N., Higgins C. F. DNA supercoiling and the anaerobic and growth phase regulation of tonB gene expression. J Bacteriol. 1988 Jun;170(6):2816–2826. doi: 10.1128/jb.170.6.2816-2826.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freundlich M., Ramani N., Mathew E., Sirko A., Tsui P. The role of integration host factor in gene expression in Escherichia coli. Mol Microbiol. 1992 Sep;6(18):2557–2563. doi: 10.1111/j.1365-2958.1992.tb01432.x. [DOI] [PubMed] [Google Scholar]
- Friedman D. I. Integration host factor: a protein for all reasons. Cell. 1988 Nov 18;55(4):545–554. doi: 10.1016/0092-8674(88)90213-9. [DOI] [PubMed] [Google Scholar]
- Gober J. W., Shapiro L. A developmentally regulated Caulobacter flagellar promoter is activated by 3' enhancer and IHF binding elements. Mol Biol Cell. 1992 Aug;3(8):913–926. doi: 10.1091/mbc.3.8.913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldberg J. B., Dahnke T. Pseudomonas aeruginosa AlgB, which modulates the expression of alginate, is a member of the NtrC subclass of prokaryotic regulators. Mol Microbiol. 1992 Jan;6(1):59–66. doi: 10.1111/j.1365-2958.1992.tb00837.x. [DOI] [PubMed] [Google Scholar]
- Goldberg J. B., Ohman D. E. Cloning and expression in Pseudomonas aeruginosa of a gene involved in the production of alginate. J Bacteriol. 1984 Jun;158(3):1115–1121. doi: 10.1128/jb.158.3.1115-1121.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Govan J. R., Harris G. S. Pseudomonas aeruginosa and cystic fibrosis: unusual bacterial adaptation and pathogenesis. Microbiol Sci. 1986 Oct;3(10):302–308. [PubMed] [Google Scholar]
- Hassett D. J., Woodruff W. A., Wozniak D. J., Vasil M. L., Cohen M. S., Ohman D. E. Cloning and characterization of the Pseudomonas aeruginosa sodA and sodB genes encoding manganese- and iron-cofactored superoxide dismutase: demonstration of increased manganese superoxide dismutase activity in alginate-producing bacteria. J Bacteriol. 1993 Dec;175(23):7658–7665. doi: 10.1128/jb.175.23.7658-7665.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgins C. F., Dorman C. J., Stirling D. A., Waddell L., Booth I. R., May G., Bremer E. A physiological role for DNA supercoiling in the osmotic regulation of gene expression in S. typhimurium and E. coli. Cell. 1988 Feb 26;52(4):569–584. doi: 10.1016/0092-8674(88)90470-9. [DOI] [PubMed] [Google Scholar]
- Hooft van Huijsduijnen R. A. PCR-generated probes for the study of DNA-protein interactions. Biotechniques. 1992 Jun;12(6):830–832. [PubMed] [Google Scholar]
- Hoover T. R., Santero E., Porter S., Kustu S. The integration host factor stimulates interaction of RNA polymerase with NIFA, the transcriptional activator for nitrogen fixation operons. Cell. 1990 Oct 5;63(1):11–22. doi: 10.1016/0092-8674(90)90284-l. [DOI] [PubMed] [Google Scholar]
- Kato J., Chakrabarty A. M. Purification of the regulatory protein AlgR1 and its binding in the far upstream region of the algD promoter in Pseudomonas aeruginosa. Proc Natl Acad Sci U S A. 1991 Mar 1;88(5):1760–1764. doi: 10.1073/pnas.88.5.1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato J., Misra T. K., Chakrabarty A. M. AlgR3, a protein resembling eukaryotic histone H1, regulates alginate synthesis in Pseudomonas aeruginosa. Proc Natl Acad Sci U S A. 1990 Apr;87(8):2887–2891. doi: 10.1073/pnas.87.8.2887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konyecsni W. M., Deretic V. DNA sequence and expression analysis of algP and algQ, components of the multigene system transcriptionally regulating mucoidy in Pseudomonas aeruginosa: algP contains multiple direct repeats. J Bacteriol. 1990 May;172(5):2511–2520. doi: 10.1128/jb.172.5.2511-2520.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosturko L. D., Daub E., Murialdo H. The interaction of E. coli integration host factor and lambda cos DNA: multiple complex formation and protein-induced bending. Nucleic Acids Res. 1989 Jan 11;17(1):317–334. doi: 10.1093/nar/17.1.317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kur J., Hasan N., Szybalski W. Physical and biological consequences of interactions between integration host factor (IHF) and coliphage lambda late p'R promoter and its mutants. Gene. 1989 Sep 1;81(1):1–15. doi: 10.1016/0378-1119(89)90331-4. [DOI] [PubMed] [Google Scholar]
- Lee E. C., Hales L. M., Gumport R. I., Gardner J. F. The isolation and characterization of mutants of the integration host factor (IHF) of Escherichia coli with altered, expanded DNA-binding specificities. EMBO J. 1992 Jan;11(1):305–313. doi: 10.1002/j.1460-2075.1992.tb05053.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee E. C., MacWilliams M. P., Gumport R. I., Gardner J. F. Genetic analysis of Escherichia coli integration host factor interactions with its bacteriophage lambda H' recognition site. J Bacteriol. 1991 Jan;173(2):609–617. doi: 10.1128/jb.173.2.609-617.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahajna J., Oppenheim A. B., Rattray A., Gottesman M. Translation initiation of bacteriophage lambda gene cII requires integration host factor. J Bacteriol. 1986 Jan;165(1):167–174. doi: 10.1128/jb.165.1.167-174.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin D. W., Holloway B. W., Deretic V. Characterization of a locus determining the mucoid status of Pseudomonas aeruginosa: AlgU shows sequence similarities with a Bacillus sigma factor. J Bacteriol. 1993 Feb;175(4):1153–1164. doi: 10.1128/jb.175.4.1153-1164.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohr C. D., Deretic V. In vitro interactions of the histone-like protein IHF with the algD promoter, a critical site for control of mucoidy in Pseudomonas aeruginosa. Biochem Biophys Res Commun. 1992 Dec 15;189(2):837–844. doi: 10.1016/0006-291x(92)92279-7. [DOI] [PubMed] [Google Scholar]
- Mohr C. D., Hibler N. S., Deretic V. AlgR, a response regulator controlling mucoidy in Pseudomonas aeruginosa, binds to the FUS sites of the algD promoter located unusually far upstream from the mRNA start site. J Bacteriol. 1991 Aug;173(16):5136–5143. doi: 10.1128/jb.173.16.5136-5143.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohr C. D., Leveau J. H., Krieg D. P., Hibler N. S., Deretic V. AlgR-binding sites within the algD promoter make up a set of inverted repeats separated by a large intervening segment of DNA. J Bacteriol. 1992 Oct;174(20):6624–6633. doi: 10.1128/jb.174.20.6624-6633.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullin D. A., Newton A. A sigma 54 promoter and downstream sequence elements ftr2 and ftr3 are required for regulated expression of divergent transcription units flaN and flbG in Caulobacter crescentus. J Bacteriol. 1993 Apr;175(7):2067–2076. doi: 10.1128/jb.175.7.2067-2076.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohman D. E., Chakrabarty A. M. Utilization of human respiratory secretions by mucoid Pseudomonas aeruginosa of cystic fibrosis origin. Infect Immun. 1982 Aug;37(2):662–669. doi: 10.1128/iai.37.2.662-669.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parkinson J. S. Signal transduction schemes of bacteria. Cell. 1993 Jun 4;73(5):857–871. doi: 10.1016/0092-8674(93)90267-t. [DOI] [PubMed] [Google Scholar]
- Schleif R. DNA looping. Annu Rev Biochem. 1992;61:199–223. doi: 10.1146/annurev.bi.61.070192.001215. [DOI] [PubMed] [Google Scholar]
- Sigman D. S., Kuwabara M. D., Chen C. H., Bruice T. W. Nuclease activity of 1,10-phenanthroline-copper in study of protein-DNA interactions. Methods Enzymol. 1991;208:414–433. doi: 10.1016/0076-6879(91)08022-a. [DOI] [PubMed] [Google Scholar]
- Speert D. P., Farmer S. W., Campbell M. E., Musser J. M., Selander R. K., Kuo S. Conversion of Pseudomonas aeruginosa to the phenotype characteristic of strains from patients with cystic fibrosis. J Clin Microbiol. 1990 Feb;28(2):188–194. doi: 10.1128/jcm.28.2.188-194.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staskawicz B., Dahlbeck D., Keen N., Napoli C. Molecular characterization of cloned avirulence genes from race 0 and race 1 of Pseudomonas syringae pv. glycinea. J Bacteriol. 1987 Dec;169(12):5789–5794. doi: 10.1128/jb.169.12.5789-5794.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terry J. M., Piña S. E., Mattingly S. J. Role of energy metabolism in conversion of nonmucoid Pseudomonas aeruginosa to the mucoid phenotype. Infect Immun. 1992 Apr;60(4):1329–1335. doi: 10.1128/iai.60.4.1329-1335.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toussaint B., Delic-Attree I., Vignais P. M. Pseudomonas aeruginosa contains an IHF-like protein that binds to the algD promoter. Biochem Biophys Res Commun. 1993 Oct 15;196(1):416–421. doi: 10.1006/bbrc.1993.2265. [DOI] [PubMed] [Google Scholar]
- Wozniak D. J., Ohman D. E. Involvement of the alginate algT gene and integration host factor in the regulation of the Pseudomonas aeruginosa algB gene. J Bacteriol. 1993 Jul;175(13):4145–4153. doi: 10.1128/jb.175.13.4145-4153.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wozniak D. J., Ohman D. E. Pseudomonas aeruginosa AlgB, a two-component response regulator of the NtrC family, is required for algD transcription. J Bacteriol. 1991 Feb;173(4):1406–1413. doi: 10.1128/jb.173.4.1406-1413.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]