Abstract
The molecular bases of spectral tuning in the UV-, violet-, and blue-sensitive pigments are not well understood. Using the in vitro assay, here we show that the SWS1, SWS2-A, and SWS2-B pigments of bluefin killifish (Lucania goodei) have the wavelengths of maximal absorption (λmax’s) of 354, 448, and 397 nm, respectively. The spectral difference between the SWS2-A and SWS2-B pigments is largest among those of all currently known pairs of SWS2 pigments within a species. The SWS1 pigment contains no amino acid replacement at the currently known 25 critical sites and seems to have inherited its UV-sensitivity directly from the vertebrate ancestor. Mutagenesis analyses show that the amino acid differences at sites 44, 46, 94, 97, 109, 116, 118, 265, and 292 of the SWS2-A and SWS2-B pigments explain 80% of their spectral difference. Moreover, the larger the individual effects of amino acid changes on the λmax-shift are, the larger the synergistic effects tend to be generated, revealing a novel mechanism of spectral tuning of visual pigments.
Keywords: visual pigments, absorption spectra, mutagenesis, bluefin killifish
1. Introduction
The photosensitive molecules, visual pigments, consist of an opsin and a chromophore, either 11-cis-retinal or 11-cis-3, 4-dehydroretinal, and their major function can be characterized by the wavelength of maximal absorption (λmax). In the early stage of vertebrate evolution, our ancestor already possessed five paralogous visual pigments: 1) rhodopsin (RH1); 2) RH1-like (RH2); 3) short wavelength-sensitive type 1 (SWS1); 4) SWS type 2 (SWS2); and 5) middle and long wavelength-sensitive (M/LWS) pigments (e. g., Yokoyama and Yokoyama, 1996; Yokoyama, 2000a; Ebrey and Koutalos, 2001). To adapt to their diverse ecological and physiological environments, organisms have modified the absorption spectra of their visual pigments during vertebrate evolution (e.g., Lythgoe, 1979; Yokoyama and Yokoyama, 1996). Using different experimental methods, currently known RH1, RH2, SWS1, SWS2, and M/LWS pigments are shown to haveλmax’s of 480–520, 460–510, 360–440, 410–460, and 510–570 nm, respectively (e.g., Yokoyama, 2000a; Ebrey and Koutalos, 2001; Takahashi and Ebrey, 2003). These evolutionary characteristics and the availability of functional assays make visual pigments one of only handful molecules that allow us to conduct experimental analyses of adaptive evolution in vertebrates (Yokoyama and Yokoyama, 1996; Yokoyama, 1997; Yokoyama, 2000a; Carroll, 2006).
The bluefin killifish, Lucania goodei, live in freshwater in South Carolina, Georgia, and Florida. Using the microspectrophotometry (MSP), Fuller and her colleagues have identified cone pigments of bluefin killifish with λmax’s of 538 nm (RH2 pigment), 359 nm (SWS1 pigment), 454 nm (SWS2-A pigment), 405 nm (SWS2-B pigment), and 572 nm (LWS pigment) (Fuller et al., 2003). Thus, in addition to the UV-sensitive SWS1 pigment, bluefin killifish uses blue-sensitive SWS2-A and violet-sensitive SWS2-B pigments. More recently, medaka (Matusmoto et al., 2006), tilapia (Spady et al., 2006), and cichlids (e. g., Parry et al., 2005) are also shown to have two types of SWS2 pigments. Among these, bluefin killifish has the largest spectral difference between SWS2-A and -B pigments and provides an opportunity to explore the molecular basis of spectral tuning in SWS2 pigments. Therefore, we constructed the bluefin killifish SWS1, SWS2-A, and SWS2-B pigments and studied the evolutionary patterns of their functional differentiation. The results show that the λmax’s of the three pigments evaluated using the in vitro assay are consistently 5–8 nm shorter than the corresponding MSP estimates. Furthermore, the majority of the λmax difference between the two SWS2 pigments can be explained by nine amino acid differences; in particular, those at sites 94, 97, 118, and 265 significantly modulate the λmax’s of the SWS2 pigments individually as well as synergistically.
2. Materials and methods
2.1. In vitro assays of SWS1 and SWS2 pigments
Total retinal RNAs of bluefin killifish (Lucania goodei) are gift from Dr. R. C. Fuller at the University of Illinois at Urbana/Champaign. The full-length SWS1, SWS2-A, and SWS2-B opsin cDNAs were obtained by RT-PCR using primers based on the nucleotide sequences of the 5′ and 3′ end cDNA clones (SWS1: 5′–ATTGCAGAATTCCACCATGGGGAAACATTTTC-3′ (forward) and 5′–TCCCAAGTCGACGAAGCTGTGGACAC-3′ (reverse); SWS2-A: 5′–GCAGAAGAATTCCACCATGAGGTCCACCCGAGTCATAGAGC-3′ (forward) and 5′–CTGTTAGTCGACGCTGGGCCAACTTTGGAGA-3′ (reverse); and SWS2-B: 5′–AAACAAGAATTCCACCATGAAGATGAGGGTAAAC-3′ (forward) and 5′–GCCGAGAGTCGACGAGGGTCCAACTTTTGAG-3′ (reverse)). The opsin cDNAs of full length were subcloned into the EcoRI and SalI restriction sites of the expression vector pMT5 (e.g., Yokoyama, 2000b). To rule out spurious mutations, these DNA fragments were sequenced by cycle sequencing reactions using the Sequitherm Excel II long-read kits (Epicentre Technologies, Madison, WI) with dye-labeled M13 forward and reverse primers. Reactions were run on a LI-COR (Lincoln, NE) 4200LD automated DNA sequencer.
These opsins were expressed in COS1 cells by transient transfection. The pigments were regenerated by incubating the opsins with 11-cis-retinal (Storm Eye Institute, Medical University of South Carolina) and were purified using immobilized 1D4 (The Culture Center, Minneapolis, MN) in buffer W1 (50 mM N-(2-hydroxyethyl) piperazine-N’-2-ethanesulfonic acid (HEPES) (pH 6.6), 140 mM NaCl, 3mM MgCl2, 20% (w/v) glycerol and 0.1% dodecyl maltoside) (for more details, see Yokoyama, 2000b). UV visible spectra were recorded at 20° C using a Hitachi U-3000 dual beam spectrophotometer. Visual pigments were bleached for 3 min using a 60 W standard light bulb equipped with a Kodak Wratten #3 filter at a distance of 20 cm. Data were analyzed using Sigmaplot software (Jandel Scientific, San Rafael, CA).
2.2. Site-directed mutagenesis
Mutant opsins were generated by using QuickChange site-directed mutagenesis kit (Stratagene, La Jolla, CA). All DNA fragments that were subjected to mutagenesis were sequenced to rule out spurious mutations.
2.3. Sequence data analyses
We considered a total of 38 representative fish pigments from the five pigment groups: RH1 pigments (goldfish 1, GenBank accession no. L11863; zebrafish 1, AF109368; cavefish 1, U12328; medaka 1, AB001606; and bluefin killifish, AY296738), RH2 pigments (goldfish 2-A, L11865; goldfish 2-B, L11866; zebrafish 2–1, AB087805; zebrafish 2–2, AB087806; zebrafish 2–3, AB087807; zebrafish 2–4, AB087808; cavefish 2; S75255; medaka 2-A, AB223053; medaka 2-B, AB223054; medaka 2-C, AB223055; and bluefin killifish 2, AY296739), SWS1 pigments (goldfish S1, D85863; zebrafish S1, AB087810; medaka S1, AB223058; tilapia S1, AF191221; and bluefin killifish S1, AY296735), SWS2 pigments (goldfish S2, L11864; zebrafish S2, AB087809; cavefish S2, AF134762; medaka S2-A, AB223056; medaka S2-B, AB223057; tilapia S2-A, AF247116; tilapia S2-B, AF247120; bluefin killifish S2-A, AY296737; and bluefin killifish S2-B, AY296736), and M/LWS pigments (goldfish L, L11867; cavefish L, M38625; cavefish M, M38619; zebrafish L-1, AB087803; zebrafish L-2, AB087804; medaka L-A, AB223051; medaka L-B, AB223052; and bluefin killifish L, AY296740). The phylogenetic tree of these visual pigments has been constructed by applying the neighbor-joining (NJ) method (Saitou and Nei, 1987) to the nucleotide sequences between codon positions 9 and 320. On the basis of this tree topology, we inferred the ancestral amino acid sequences of visual pigments by using a computer program, PAML, based on a likelihood-based Bayesian method (Yang, 1997).
3. Results
3.1. Absorption spectra of the SWS1 and SWS2 pigments of bluefin killifish
The SWS1, SWS2-A, and SWS2-B opsins were expressed in the COS1 cells transiently and were reconstituted with the 11-cis-retinal. The in vitro assays show that the SWS1, SWS2-A, and SWS2-B pigments have λmax’s of 354 nm ± 0 (referred to as bfin killifish S1 (P354)), 448 ± 1 nm (bfin killifish S2-A (P448)), and 397 ± 0 nm (bfin killifish S2-B (P397)), respectively (Fig. 1). These values are close to the corresponding MSP estimates of 359 ± 2, 454 ± 3, and 405 ± 2 (Fuller et al., 2003), but our estimates are consistently 5–8 nm shorter than the MSP values. In particular, the λmax of bfin killifish S2-B (P397) is lowest among all currently known SWS2 pigments; in fact, it is almost identical to the λmax of 393 nm of the pigeon violet pigment, which is lowest among those of the currently known violet pigments in the paralogous SWS1 pigment group (Yokoyama, 2000a). Consequently, the observed λmax difference of 51 nm between the SWS2-A and SWS2-B pigments of bluefin killifish is largest among those of the currently known pairs of SWS2 pigments within a species.
Fig. 1.
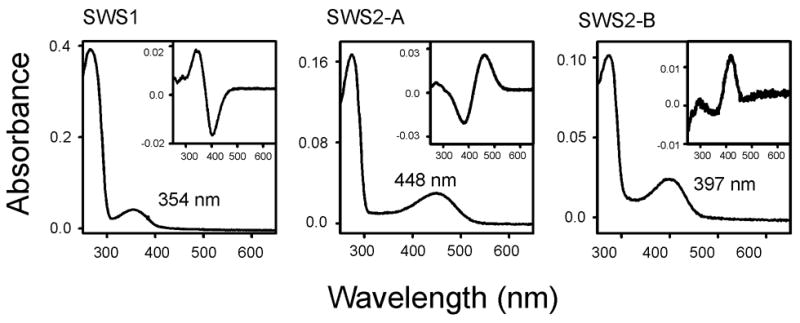
The absorption spectra of the bluefin killifish SWS1, SWS2-A, and SWS2-B pigments evaluated by the in vitro assay. The dark-light difference spectra are shown in the inset.
3.2. The phylogenetic tree of fish visual pigments
At present, the in vitro assays of visual pigments in fish have been performed for goldfish (Carassius auratus), zebrafish (Danio rerio), medaka (Oryzias latipes), cavefish (Astyanax fasciatus), tilapia (Oreochromis niloticus), and bluefin killifish. Applying the NJ method to the nucleotide sequences of 38 pigments (see Materials and methods), we have constructed the unrooted phylogenetic tree (Fig. 2). A majority of bootstrap values in Fig. 2 are > 0.95 and most bifurcation patterns are highly reliable. The NJ tree shows that the visual pigments are classified into RH1, RH2, SWS1, SWS2, and M/LWS groups and that in each group, the goldfish and zebrafish pigments form one cluster and medaka, tilapia, and bluefin killifish pigments form another cluster; the cavefish pigment seems to belong to the goldfish-zebrafish pigment cluster (see also Yokoyama, 2000a; Matsumoto et al., 2006; Spady et al., 2006). The exact phylogenetic relationships of medaka, tilapia, and bluefin killifish pigments, however, cannot be established. When the amino acid sequences are considered, the tree topologies of the SWS1 and SWS2 pigments are identical to those in Fig. 2. However, the cavefish 1 is most distantly related to the other RH1 pigments, and the medaka 2-A in the RH2 group is more closely related to the medaka-bluefin killifish cluster. Since the bootstrap values of these nodes in the amino acid tree are much lower than 0.95, the two tree topologies are consistent with each other. Because of its higher bootstrap values at different nodes, the tree topology in Fig. 2 seems to be more reliable than the amino acid tree.
Fig. 2.
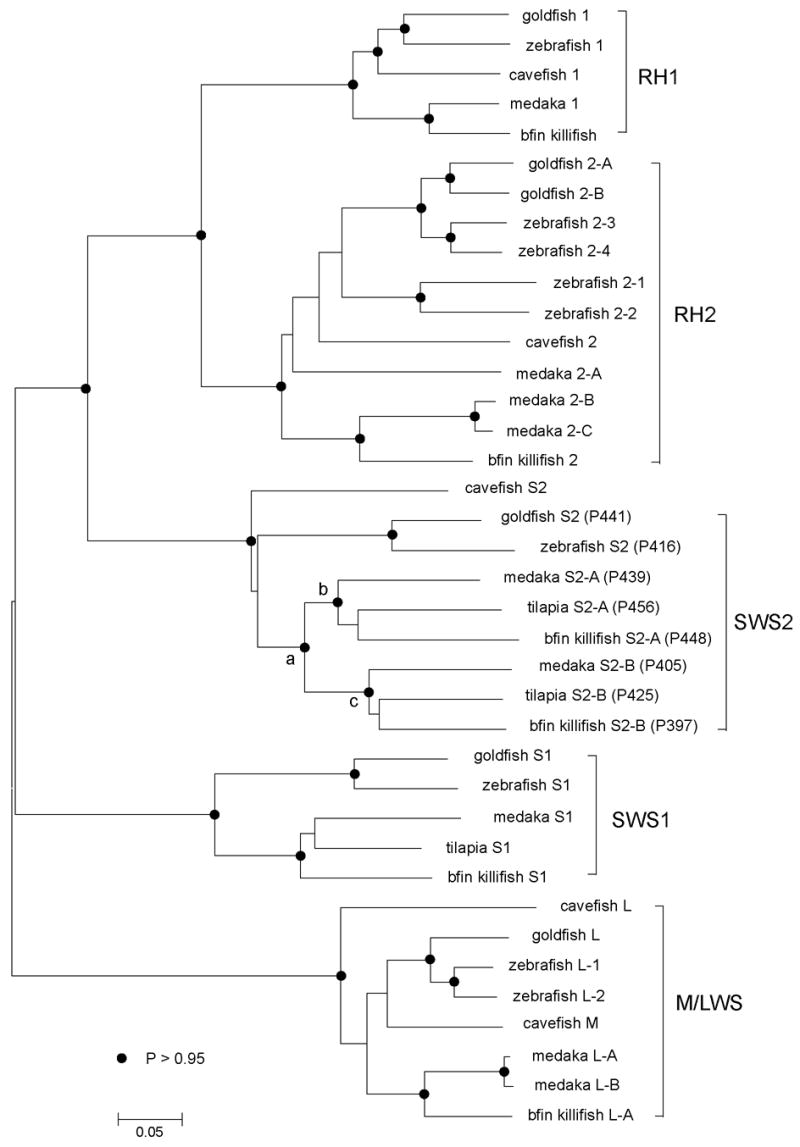
The phylogenetic tree of some representative fish visual pigments. The numbers after P in the SWS2 pigments refer to λmax’s evaluated by the in vitro assay. The filled circles at different nodes indicate that their bootstrap values are >0.95; otherwise, the bootstrap values are <0.95. The bar at the bottom indicates evolutionary distance measured as the number of nucleotide substitutions per site.
3.3. Molecular bases of spectral tuning in the SWS1 and SWS2 pigments of bluefin killifish
Based on the tree topology in Fig. 2, we inferred amino acid sequences of all ancestral pigments at all nodes using the JTT, Dayhoff, and WAG models of amino acid replacements (Yang, 1997). At present, certain amino acid replacements at 25 sites (46, 49, 52, 83, 86, 90, 91, 93, 94, 97, 109, 113, 114, 116, 118, 122, 164, 181, 207, 211, 261, 265, 269, 292, and 295) are known to have modified the λmax’s of various visual pigments in nature (see the Discussion section), where amino acid site numbers correspond to those of the bovine RH1 pigment. At these sites, no amino acid replacement has occurred in bfin killifish S1 (P354). Since the ancestral vertebrate SWS1 pigment had a λmax of ~360 nm (Shi and Yokoyama, 2003), like many other fishes, bfin killifish S1 (P354) must have inherited its UV-sensitivity directly from the vertebrate ancestor. Because of its small shift, we did not explore the exact molecular basis for the decrease in the λmax of bfin killifish S1 (P354).
Of the 25 critical amino acid sites, bfin killifish S2-A (P448) and bfin killifish S2-B (P397) differ at eight positions: the former pigment contains amino acids L46, A94, S97, A109, I116, T118, W265, and A292 and the latter pigment contains F46, C94, C97, G109, T116, G118, Y265, and S292. The amino acid sequences of these pigments and their inferred ancestral pigments suggest that these amino acid differences might have been generated by F46L, M116I, and S292A (denoted as M46L/M116I/S292A), A94C/S97C/M116T, and A109G/T118G/W265Y at branches b - bfin killifish S2-A (P448), a – c, and c - bfin killifish S2-B (P397), respectively (Table 1). Thus, F46L/M116I/S292A and A94C/S97C/A109G/M116T/T118G/W265Y seem to have occurred in bfin killifish S2-A (P448) and bfin killifish S2-B (P397), respectively.
Table 1.
Amino acid replacements at the 25 critical sites that may cause λmax-shifts in the SWS2 pigments, which are inferred by three different amino acid replacement modelsa
| Models
|
|||
|---|---|---|---|
| Branch | JTT | Dayhoff | WAG |
| a – b | - | - | - |
| b – medaka S2-A (P439) | M0.58116A | M0.53116A | M0.77116A |
| b – tilapia S2-A (P456) | V0.9049I | V0.8849I | V0.9049I |
| b – bfin killifish S2-A (P448) | M1.044Tb | M1.044Tb | M1.044Tb |
| F1.046L | F1.046L | F1.046L | |
| M0.70116I | M0.63116I | M0.85116I | |
| S0.99292A | S0.99292A | S0.99292A | |
| a - c | A1.094C0.99 | A0.9994C1.0 | A0.9994C1.0 |
| S0.9997C0.99 | S1.097C1.0 | S1.097C1.0 | |
| M0.57116T1.0 | M0.52116T1.0 | M0.76116T1.0 | |
| c – medaka S2-B (P405) | F1.046V | F1.046V | F1.046V |
| A0.53109G | A0.54109G | A0.53109G | |
| T0.90118A | T0.89118A | T0.94118A | |
| W0.76265Y | W0.81265Y | W0.78265Y | |
| c – tilapia S2-B (P425) | - | - | - |
| c – bfin killifish S2-B (P397) | A0.53109G | A0.55109G | A0.53109G |
| T0.90118G | T0.89118G | T0.94118G | |
| W0.76265Y | W0.81265Y | W0.78265Y | |
Subscripts indicate the posterior probabilities; for nodes a, b, and c, see Fig. 1.
The effect of amino acid replacement on the λmax-shift has been identified by mutagenesis analyses.
In order to evaluate the effects of these amino acid replacements on the λmax-shift, we introduced mutations L46F, A94C, S97C, A109G, I116T, T118G, W265Y, and A292S into the bfin killifish S2-A (P448) individually, where the two independent events of M116I in bfin killifish S2-A (P448) and M116T in bfin killifish S2-B (P397) are combined into one step. If we follow the patterns of amino acid replacements in Table 1, then A94C, S97C, A109G, T118G, and W265Y are forward mutations and L46F and A292S are reverse mutations. The in vitro assays show that with the exception of A109G, these mutations decrease the λmax at least by 4 nm (Table 2). Although it is not included among the 25 critical sites, we also found that T44M decreases the λmax by 4 nm (Table 2).
Table 2.
The effects of amino acid changes on the λmax-shift
| Pigment | Mutation | λmax (nm) | δ λ max (nm) |
|---|---|---|---|
| bfin killifish S2-A (P448) | - | 448 ± 0 | |
| T44M | 444 ± 2 | -4 | |
| L46F | 442 ± 1 | -6 | |
| A94C | 432 ± 2 | -16 | |
| S97C | 431 ± 1 | -17 | |
| A109G | 446 ± 1 | -2 | |
| I116T | 441 ± 1 | -7 | |
| T118G | 433 ± 1 | -15 | |
| W265Y | 419 ± 1 | -29 | |
| A292S | 444 ± 0 | -4 | |
| A94C/W265Y | 416 ± 2 | -32 | |
| A94C/T118G/W265Y | 415 ± 1 | -33 | |
| A94C/T118G/W265Y/A292S | 422 ± 0 | -26 | |
| T44M/A94C/T118G/W265Y/A292S | 420 ± 1 | -28 | |
| T44M/L46F/A94C/S97C/A109G/I116T/ T118G/W265Y/A292S | 407 ± 0 | -41 | |
| bfin killifish S2-B (P397) | - | 397 ± 0 |
In theory, if the effects of these 9 amino acid changes on the λmax-shift were additive, then they would have decreased the λmax of bfin killifish S2-A (P448) by 100 nm, predicting that the bfin killifish S2-B (P397) should have a λmax of 348 nm, which is ~50 nm lower than the actual value. In practice, however, when the nine amino acid changes are introduced into bfin killifish S2-A (P448) together, they decrease the λmax by 41 nm, explaining a 80% of the entire λmax difference between the two SWS2 pigments (Table 2). We cannot explain the remaining 10 nm difference between the λmax’s of the two SWS2 pigments, but it is clear that the major portion of spectral difference between bfin killifish S2-A (P448) and bfin killifish S2-B (P397) is modulated by both individual and interactive amino acid replacements.
To see the extent of the synergistic effects of the critical amino acid replacements on the λmax-shift, we first considered A94C and W265Y, which reduced the λmax by 16 and 29 nm, respectively (Table 2). When both of these mutations were introduced into the bfin killifish S2-A (P448), they reduced the λmax by 32 nm. Thus, the interaction between A94C and W265 (denoted as A94CxW265) increases the λmax by 13 nm (Table 3). T118G decreases the λmax by 15 nm and the interactions between T118G and A94C/W265Y (the sum of T118G x A94C, T118G x W265Y, and T118G x A94C x W265Y, or T118GxA94C/W265) increase the λmax by 14 nm; similarly, the interactions between A292S and A94C/T118G/W265 (A292SxA94C/T118G/W265) increase the λmax by 11 nm (Table 3). On the other hand, the interactions between T44M and A94C/T118G/W265Y/A292S cause only a trivial λmax-shift of 2 nm (Table 3). These examples show that the larger the individual effects of amino acid changes on the λmax-shift are, the more the synergistic effects of these amino acid changes on the λmax-shift tend to be generated.
Table 3.
The effects of some amino acid interactions on the λmax-shift
| Interaction | λmax-shift (nm) |
|---|---|
| A94C x W265Y | 13 |
| T118G x A94C/W265Y | 14 |
| A292S x A94C/T118G /W265Y | 11 |
| T44M x A94C/T118G /W265Y/A292S | 2 |
4. Discussion
At present, certain amino acid changes at a total of 25 sites are known to have modified the λmax of various visual pigments during vertebrate evolution. They include sites 83, 122, 211, 261, 265, 292, and 295 in RH1 pigments (e. g. Yokoyama, 2000a), sites 49, 122 and 207 in RH2 pigments (Yokoyama et al., 1999; Chinen et al., 2005a), sites 46, 49, 52, 86, 90, 91, 93, 97, 109, 113, 114, 116, and 118 in SWS1 pigments (e. g. Yokoyama, 2000a; Babu et al., 2001; Shi and Yokoyama, 2003; Fasick et al., 2002; Takahashi and Yokoyama, 2005), sites 91, 94, 116, 122, 261, 292, and 295 in SWS2 pigments (Takahashi and Ebrey, 2003; Chinen et al., 2005b), and sites 164, 181, 261, 269, and 292 in M/LWS pigments, which correspond to sites180, 197, 277, 285, and 308 of the human red- and green-sensitive pigments (Yokoyama and Radlwimmer, 2001). Our analyses of the bluefin killifish SWS2 pigments show that site 44 is also involved in the spectral tuning of visual pigments, increasing the total number of critical amino acid sites to 26.
At present, the mechanisms of spectral tuning in visual pigments in nature can be distinguished into two major groups: the absorption spectra of visual pigments are determined largely either 1) by a small number of individual amino acid changes, each causing a significant λmax-shift, or 2) by interactions among different critical amino acids, each causing a relatively small or no λmax-shift. The extreme examples of the first group can be seen in several amino acid changes at sites 86 and 90 in SWS1 pigments. That is, Y86F in bovine (Fasick et al., 2002; Cowing et al., 2002) and flying squirrel (Carvalho et al., 2006) pigments as well as F86Y in mouse (Fasick et al., 2002) and goldfish (Cowing et al., 2002) pigments shift the λmax by 60–75 nm; V86F in guinea pig pigment (Parry et al., 2004) and S86F in elephant pigment (Yokoyama et al., 2005) decrease the λmax by 52–53 nm. S90C and C90S in several avian and African clawed frog pigments also shift the λmax by 35–45 nm (Yokoyama et al., 2000; Wilkie et al., 2000; Dukkipati et al., 2002). The interactions of amino acids at these sites with others seem to cause significantly smaller magnitudes of λmax-shifts (Yokoyama et al., 2005). Although the magnitudes of changes are much smaller, S164A, H181Y, Y261F, T269A, and A292S in the LWS pigments and the reverse changes in the MWS pigments also shift the λmax by 7–30 nm individually. These amino acids act mostly in an additive fashion, but a significant interaction seems to occur only between sites 164 and 181 in rodent lineages (Yokoyama and Radlwimmer, 2001). Amino acid replacements D83N, E122Q, M207L, and A292S in various RH1 (Nathans, 1990; DeCaluwe et al., 1995; Sugawara et al., 2005; Fasick and Robinson, 1998; Yokoyama, 2000b; Jans and Farrens, 2001; Sun et al., 1997; Lin et al., 1998), RH2 (Yokoyama et al., 1999; Chinen et al., 2005a; Imai et al., 1997), SWS2 (Takahashi and Ebrey, 2003), and M/LWS (Fasick and Robinson, 1998; Sun et al., 1997) pigments also belong to the same group.
The second group is represented by certain critical amino acid changes in RH2 (Chinen et al., 2005a), SWS1 (Shi et al., 2001), and SWS2 (Chinen et al., 2005b) pigments. The most dramatic example of this type of spectral tuning can be seen in the mouse UV pigment, where F46T, F49L, T52F, F86L, T93P, A114G, and S118T cause no λmax-shift individually, but they together increase the λmax by 52 nm (Shi et al., 2001).
In the bluefin killifish, the nine amino acid changes are involved in generating a major λmax difference between bfin killifish S2-A (P448) and bfin killifish S2-B (P397). Out of the nine critical amino acid changes, A94C, S97C, T118G, and W265Y decrease the λmax by >15 nm individually, but the overall λmax of the latter pigment is also modulated by equally strong interactions among them. Thus, the molecular basis of spectral tuning in bfin killifish S2-B (P397) differs from the two currently known mechanisms of spectral tuning in visual pigments. It’s generality, however, remains to be tested by additional mutagenesis analyses using the other SWS2 and paralogous pigments.
We have seen that the medaka, tilapia, and bluefin killifish SWS2 pigments have different λmax’s (Fig. 2). The λmax’s of medaka (Matsumoto et al., 2006) and bluefin killifish pigments were evaluated from dark spectra and those of tilapia (Spady et al., 2006) from dark-light spectra. It should be noted that the λmax’s of bfin killifish S1 (P354), bfin killifish S2-A (P448), and bfin killifish S2-B (P397) evaluated from dark-light spectra are 343 ± 0, 462 ± 0, and 425 ± 0 nm, respectively. Thus, the λmax’s evaluated from dark and dark-light spectra can differ by more than 10 nm and should not be compared directly. Thus, only the corresponding λmax’s of SWS2 pigments of medaka and bluefin killifish are coparable. Since their divergence, medaka S2-A (P439) and bfin killifish S2- A (P448) seem to have accumulated M116A and M44T/F46L/M116I/S292A, respectively (Table 1). Although we don’t know the effects of M116A and M116I on the λmax-shift, M44T, F46L, and S292A are all expected to increase the λmax of the latter pigment (Table 2) and, therefore, bfin killifish S2-A (P448) should have a higher λmax than medaka S2-A (P439). This explains the observed spectral difference. Similarly, F46V/A109G/T118A/W265Y and A109G/T118G/W265Y occurred in medaka S2-B (P405) and bfin killifish S2-B (P397), respectively (Table 1). Since T118A decreases the λmax of bovine RH1 pigment by 16 nm (Jans and Farrens, 2001), it is likely that A109G/T118AW265Y in medaka S2-B (P405) and A109G/T118G/W265Y in bfin killifish S2-B (P397) decrease the λmax by the same extent. Therefore, the spectral difference between medaka S2-B (P405) and bfin killifish S2-B (P397) might have been generated by F46V in medaka S2-B (P405) and/or other unidentified amino acid replacements.
In 1990, we proposed that the combination between the evolutionary prediction of amino acid changes which might have been involved in the functional adaptation of visual pigments, followed by experimental tests of such evolutionary hypotheses, would be useful in studying adaptive evolution of color vision in vertebrates (Yokoyama and Yokoyama, 1990; see also Yokoyama and Yokoyama, 1996; Yokoyama, 1997). Indeed, this approach has contributed significantly in elucidating not only the molecular mechanisms of the adaptation of organisms to various color environments but also the molecular bases of spectral tuning of visual pigments. As noted earlier, it has been established that specific amino acid differences at sites 164, 181, 261, 269, and 292 modulate the λmax’s of M/LWS pigments (e.g. Yokoyama and Yokoyama, 1990; Neitz et al., 1991; Asenjo et al., 1994; Sun et al., 1997; Yokoyama and Radlwimer, 2001). This genetic information has been used to establish a seemingly contradictory observation that red and green color vision as well as color blindness have undergone adaptive evolution in different lineages (Yokoyama and Takenaka, 2005). At the other end of the spectrum, using the evolutionary arguments, followed by mutagenesis analyses, it has been established that amino acid differences at more than 13 sites are involved in the differentiation of the absorption spectra of certain UV- and violet-sensitive pigments (e. g., Takahashi and Yokoyama, 2005). The evolution of these SWS1 pigments also reflects the ecological and physiological requirements for UV vision (Shi and Yokoyama, 2003). Furthermore, certain fish species have adapted to their deeper habitats by modifying a relatively small number of amino acids, such as E122Q, E122M, M207L, and A292S (Yokoyama et al., 1999; Yokoyama and Takenaka, 2004; Hunt et al., 2001; Sugawara et al., 2005). Indeed, we may claim that these analyses of dim-light and color vision have produced “the deepest body of knowledge linking differences in specific genes to differences in ecology and to the evolution of species (Carroll, 2006).”
To further deepen our understanding of the molecular bases of spectral tuning in visual pigments, it seems most effective to consider macroevolutionary changes of distantly related visual pigments, where a significant number of critical amino acid changes have accumulated, providing solid grounds in the molecular genetic analyses of dim-light and color vision in different organisms. The microevolutionary results of the visual pigments in the bluefin killifish by Fuller and her colleagues add an additional important dimension to the evolutionary analyses of organismal adaptation; that is, adaptive changes in color vision within a species can arise through differential expression of the same sets of opsin genes (Fuller et al. 2003, 2004). In the future, therefore, the molecular evolutionary genetic analyses of adaptive evolution of dim-light and color vision have to be understood by studying not only the coding regions of opsin genes but also the regulatory elements that modulate the expression of various opsin genes.
Acknowledgments
We thank Ruth Yokoyama for her comments and Dr. Rosalie Crouch of the Storm Eye Institute, Medical University of South Carolina, for the 11-cis-retinal. This work was supported by a grant from the National Institutes of Health.
Abbreviations
- λmax
wavelength of maximal absorption
- SWS2
short wavelength-sensitive type 2
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Asenjo AB, Rim J, Oprian DD. Molecular determination of human red/green color discrimination. Neuron. 1994;12:1131–1138. doi: 10.1016/0896-6273(94)90320-4. [DOI] [PubMed] [Google Scholar]
- Babu KR, Dukkipati A, Birge RR, Knox BE. Regulation of phototransduction in short-wavelength cone visual pigments via the retinylidene Schiff base counterion. Biochemistry. 2001;40:13760–13766. doi: 10.1021/bi015584b. [DOI] [PubMed] [Google Scholar]
- Carroll SB. The Making of the Fittest. W. W. Norton; New York: 2006. [Google Scholar]
- Carvalho LDS, Cowing JA, Wilkie SE, Bowmaker JK, Hunt DM. Shortwave visual sensitivity in tree and flying squirrels reflects changes in lifestyle. Current Biol. 2006;16:R81–R83. doi: 10.1016/j.cub.2006.01.045. [DOI] [PubMed] [Google Scholar]
- Chinen A, Matsumoto Y, Kawamura S. Reconstruction of ancestral green visual pigments of zebrafish and molecular mechanism of their spectral differentiation. Mol Biol Evol. 2005a;22:1001–1010. doi: 10.1093/molbev/msi086. [DOI] [PubMed] [Google Scholar]
- Chinen A, Matsumoto Y, Kawamura S. Spectral differentiation of blue opsins between phylogenetically close but ecologically distant goldfish and zebrafish. J Biol Chem. 2005b;280:9460–9466. doi: 10.1074/jbc.M413001200. [DOI] [PubMed] [Google Scholar]
- Cowing JA, Poopalasundaram S, Wilkie SR, Robinson PR, Bowmaker JK, Hunt DM. The molecular mechanism for the spectral shifts between vertebrate ultraviolet- and violet-sensitive cone visual pigments. Biochem J. 2002;367:129–135. doi: 10.1042/BJ20020483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeCaluwe GLJ, Bovee-Geurts PHM, Rath P, Rothschild KJ, De Grip WJ. Effect of carboxyl mutations on functional properties of bovine rhodopsin. Biophys Chem. 1995;56:79–87. doi: 10.1016/0301-4622(95)00018-s. [DOI] [PubMed] [Google Scholar]
- Dukkipati A, Kusnetzow A, Babu KR, Ramos L, Singh D, Knox BE, Birge RR. Phototransduction by vertebrate ultraviolet visual pigments: Protonation of the retinylidene Schiff base following photobleaching. Biochemistry. 2002;41:9842–9851. doi: 10.1021/bi025883g. [DOI] [PubMed] [Google Scholar]
- Ebrey T, Koutalos Y. Vertebrate photoreceptors. Prog Retin Eye Res. 2001;20:49–94. doi: 10.1016/s1350-9462(00)00014-8. [DOI] [PubMed] [Google Scholar]
- Fasick JI, Applebury ML, Oprian DD. Spectral tuning in the mammalian short wavelength sensitive cone pigments. Biochemistry. 2002;41:6860–6865. doi: 10.1021/bi0200413. [DOI] [PubMed] [Google Scholar]
- Fasick JI, Robinson PR. Mechanism of spectral tuning in the dolphin visual pigments. Biochemistry. 1998;37:432–438. doi: 10.1021/bi972500j. [DOI] [PubMed] [Google Scholar]
- Fuller RC, Fleishman LJ, Leal M, Travis J, Loew E. Intraspecific variation in retinal cone distribution in the bluefin killifish, Lucania goodie. J Comp Physiol A. 2003;189:609–616. doi: 10.1007/s00359-003-0435-x. [DOI] [PubMed] [Google Scholar]
- Fuller RC, Carleton KL, Fadool JM, Spady TC, Travis J. Population variation in opsin expression in the bluefin killifish, Lucania goodei: a real-time PCR study. J Comp Physiol A. 2004;190:147–154. doi: 10.1007/s00359-003-0478-z. [DOI] [PubMed] [Google Scholar]
- Hunt DM, Dulai KS, Partridge JC, Cottrill P, Bowmaker JK. The molecular basis for spectral tuning of rod visual pigments in deep-sea fish. J Exp Biol. 2001;204:3333–3344. doi: 10.1242/jeb.204.19.3333. [DOI] [PubMed] [Google Scholar]
- Imai H, Kojima D, Oura T, Tachibanaki S, Terakita A, Shichida Y. Single amino acid residue as a functional determinant of rod and cone visual pigments. Proc Natl Acad Sci USA. 1997;94:2322–2326. doi: 10.1073/pnas.94.6.2322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jans JM, Farrens DI. Engineering a functional blue-wavelength-shifted rhodopsin mutant. Biochemistry. 2001;40:7219–7227. doi: 10.1021/bi002937i. [DOI] [PubMed] [Google Scholar]
- Lin SW, Kochendoerfer CKS, Wang D, Mathies RA, Sakmar TP. Mechanisms of spectral tuning in blue cone visual pigments: visible and raman spectroscopy of blue-shifted rhodopsin mutants. J Biol Chem. 1998;273:24583–24591. doi: 10.1074/jbc.273.38.24583. [DOI] [PubMed] [Google Scholar]
- Lythgoe JN. The Ecology of Vision. Clarendon Press; Oxford: 1979. [Google Scholar]
- Matsumoto Y, Fukamachi S, Mitani H, Kawamura S. Functional characterization of visual opsin repertoire in medaka (Oryzias latipes) Gene. 2006;371:268–278. doi: 10.1016/j.gene.2005.12.005. [DOI] [PubMed] [Google Scholar]
- Nathans J. Determinations of visual pigment absorbance: role of charged amino acids in the putative transmembrane segments. Biochemistry. 1990;29:937–942. doi: 10.1021/bi00456a013. [DOI] [PubMed] [Google Scholar]
- Neitz M, Neitz J, Jacobs GH. Spectral tuning of pigments underlying red-green color vision. Science. 1991;252:971–974. doi: 10.1126/science.1903559. [DOI] [PubMed] [Google Scholar]
- Parry JWL, Carlton KL, Spady T, Carboo A, Hunt DM. Mix and match color vision: tuning spectral sensitivity by differential opsin gene expression in lake Malawi cichlids. Curr Biol. 2005;15:1734–1739. doi: 10.1016/j.cub.2005.08.010. [DOI] [PubMed] [Google Scholar]
- Parry JWL, Poopalasundaram S, BowmakeR JK, Hunt DM DM. A novel amino acid substitution is responsible for spectral tuning in a rodent violet-sensitive visual pigment. Biochemistry. 2004;43:8014–8020. doi: 10.1021/bi049478w. [DOI] [PubMed] [Google Scholar]
- Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic tree. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- Shi Y, Radlwimmer FB, Yokoyama S. Molecular genetics and the evolution of ultraviolet vision in vertebrates. Proc Natl Acad Sci USA. 2001;98:11731–11736. doi: 10.1073/pnas.201257398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y, Yokoyama S. Molecular analysis of the evolutionary significance of ultraviolet vision in vertebrates. Proc Natl Acad Sci USA. 2003;100:8308–8313. doi: 10.1073/pnas.1532535100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spady TC, Parry JWL, Robinson PR, Hunt DM, Bowmaker JK, Carleton KL. Evolution of the cihlid visual palette through ontogenetic subfunctionalization of the opsin gene arrays. Mol Biol Evol. 2006;23:1538–1547. doi: 10.1093/molbev/msl014. [DOI] [PubMed] [Google Scholar]
- Sugawara T, Terai Y, Imai H, Turner GF, Koblmuller S, Shichida Y, Okada N. Parallelism of amino acid changes at the RH1 affecting spectral sensitivity among deep-water cichlids from Lakes Tanganyika and Malawi. Proc Natl Acad sci USA. 2005;102:5448–5453. doi: 10.1073/pnas.0405302102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun H, Macke JP, Nathans J. Mechanisms of spectral tuning in the mouse green cone pigment. Proc Natl Acad Sci USA. 1997;94:8860–8865. doi: 10.1073/pnas.94.16.8860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi Y, Ebrey TG. Molecular basis of spectral tuning in the newt short wavelength sensitive visual pigment. Biochemistry. 2003;42:6025–6034. doi: 10.1021/bi020629+. [DOI] [PubMed] [Google Scholar]
- Takahashi Y, Yokoyama S. Genetic basis of spectral tuning in the violet-sensitive visual pigment of African clawed frog, Xenopus laevis. Genetics. 2005;171:1153–1160. doi: 10.1534/genetics.105.045849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkie SE, Robinson PR, Cronin TW, Poopalasundaram S, Bowmaker J, Hunt DM. Spectral tuning of avian violet-a dn ultraviolet-sensitive visual pigments. Biochemistry. 2000;39:7895–7901. doi: 10.1021/bi992776m. [DOI] [PubMed] [Google Scholar]
- Yang Z. PAML: a program package for phylogenetic analysis by maximum likelihood. Comp Appl Biosci. 1997;13:555–556. doi: 10.1093/bioinformatics/13.5.555. [DOI] [PubMed] [Google Scholar]
- Yokoyama R, Yokoyama S. Convergent evolution of the red- and green-like visual pigment genes in fish, Astyanax fasciatus, and human. Proc Natl Acad Sci USA. 1990;87:9315–9318. doi: 10.1073/pnas.87.23.9315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokoyama S. Molecular genetic basis of adaptive selection: examples from color vision in vertebrates. Annu Rev Genet. 1997;31:311–332. doi: 10.1146/annurev.genet.31.1.315. [DOI] [PubMed] [Google Scholar]
- Yokoyama S. Molecular evolution of vertebrate visual pigments. Prog Retin Eye Res. 2000a;19:385–419. doi: 10.1016/s1350-9462(00)00002-1. [DOI] [PubMed] [Google Scholar]
- Yokoyama S. Phylogenetic analysis and experimental approaches to study color vision in vertebrates. Methods Enzymol. 2000b;315:312–325. doi: 10.1016/s0076-6879(00)15851-3. [DOI] [PubMed] [Google Scholar]
- Yokoyama S, Radlwimmer FB. The molecular genetics of red and green color vision in vertebrates. Genetics. 2001;158:1697–1710. doi: 10.1093/genetics/158.4.1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokoyama S, Radlwimmer FB, Blow NS. Ultraviolet pigments in birds evolved from violet pigments by a single amino acid change. Proc Natl Acad Sci USA. 2000;97:7366–7371. doi: 10.1073/pnas.97.13.7366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokoyama S, Takenaka N. The molecular basis of adaptive evolution of squirrelfish rhodopsins. Mol Biol Evol. 2004;21:2071–2078. doi: 10.1093/molbev/msh217. [DOI] [PubMed] [Google Scholar]
- Yokoyama S, Takenaka N. Statistical and molecular analyses of evolutionary significance of red-green color vision and color blindness in vertebrates. Mol Biol Evol. 2005;22:968–975. doi: 10.1093/molbev/msi080. [DOI] [PubMed] [Google Scholar]
- Yokoyama S, Takenaka N, Agnew DW, Shoshani J. Elephants and human color-blind deuteranopes have identical sets of visual pigments. Genetics. 2005;170:335–344. doi: 10.1534/genetics.104.039511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokoyama S, Yokoyama R. Adaptive evolution of photoreceptors and visual pigments in vertebrates. Annu Rev Ecol Syst. 1996;27:543–567. [Google Scholar]
- Yokoyama S, Zhang H, Radlwimmer FB, Blow NS. Adaptive evolution of color vision of the Comoran coelacanth (Latimeria chalumnae) Proc Natl Acad Sci USA. 1999;96:6279–6284. doi: 10.1073/pnas.96.11.6279. [DOI] [PMC free article] [PubMed] [Google Scholar]


