Abstract
Myotonic dystrophy type 1 (DM1) is caused by a CTG trinucleotide expansion in the 3′ untranslated region (3′ UTR) of DM protein kinase (DMPK). The key feature of DM1 pathogenesis is nuclear accumulation of RNA, which causes aberrant alternative splicing of specific pre-mRNAs by altering the functions of CUG-binding proteins (CUGBPs). Cardiac involvement occurs in more than 80% of individuals with DM1 and is responsible for up to 30% of disease-related deaths. We have generated an inducible and heart-specific DM1 mouse model expressing expanded CUG RNA in the context of DMPK 3′ UTR that recapitulated pathological and molecular features of DM1 including dilated cardiomyopathy, arrhythmias, systolic and diastolic dysfunction, and misregulated alternative splicing. Combined in situ hybridization and immunofluorescent staining for CUGBP1 and CUGBP2, the 2 CUGBP1 and ETR-3 like factor (CELF) proteins expressed in heart, demonstrated elevated protein levels specifically in nuclei containing foci of CUG repeat RNA. A time-course study demonstrated that colocalization of MBNL1 with RNA foci and increased CUGBP1 occurred within hours of induced expression of CUG repeat RNA and coincided with reversion to embryonic splicing patterns. These results indicate that CUGBP1 upregulation is an early and primary response to expression of CUG repeat RNA.
Introduction
Myotonic dystrophy type 1 (DM1) is the most common form of adult-onset muscular dystrophy. The disease is dominantly inherited and affects multiple tissues with symptoms including muscle hyperexcitability (myotonia), progressive muscle wasting, cardiac conduction defects and cardiomyopathy, insulin resistance, and neuropsychiatric disturbances (1). DM1 is caused by a CTG trinucleotide expansion in the 3′ untranslated region (3′ UTR) of the DM protein kinase gene (DMPK). The primary mechanism of pathogenesis is a gain of function for the mutant DMPK mRNA, which contains long tracts of CUG repeats, accumulates in nuclear foci, and affects nuclear and cytoplasmic activities of RNA binding proteins such as muscleblind like (MBNL) as well as CUGBP1 and ETR-3 like factor (CELF) proteins (2). Sequestration of MBNL and increased steady-state levels of the CELF protein CUG-binding protein 1 (CUGBP1) result in the misregulated alternative splicing of specific pre-mRNAs in human DM1 tissues and cause disease features such as myotonia and insulin resistance (3–8). The molecular mechanism by which expression of CUG RNA repeats induces CUGBP1 overexpression in DM1 tissues is not clear.
Cardiac phenotypes occur in more than 80% of individuals with DM1; these include conduction defects, arrhythmias, and sudden cardiac death (9, 10). Two interrelated cardiac phenotypes are observed in individuals with DM1. The first is conduction defects, which are particularly prevalent and can progress to complete heart block or other potentially fatal arrhythmias. The second phenotype is mechanical, in which both diastolic and systolic dysfunction can progress to combined systolic and diastolic heart failure (11). Conduction defects include prolonged PR interval, altered QRS complex, and prolonged His to ventricle interval on ECG analysis (1, 12, 13). Light microscopy revealed infiltration of fatty tissue and fibrosis in the myocardium, whereas electron microscopy revealed vacuolation and disorganization of sarcoplasmic reticulum and accumulation of mitochondria (12, 14, 15). The molecular mechanisms causing abnormalities in electric conduction or contractility in DM1 have not yet been identified.
We used a Cre-loxP approach including tamoxifen-inducible Cre to generate an inducible mouse model for heart-specific expression of 960 CUG RNA repeats in the context of DMPK 3′ UTR. Adult mice in which high levels of expanded CUG RNA was induced developed severe cardiomyopathy and arrhythmias resulting in 100% mortality within 2 weeks of induction. Mice from lines that expressed more than 5-fold the level of the identical mRNA lacking repeats exhibited no phenotypic or molecular changes. Repeat-expressing mice exhibited diastolic and systolic dysfunction, arrhythmias, and a full set of molecular features observed in DM1 heart tissue such as RNA foci formation, colocalization of MBNL1 with RNA foci, elevated CELF protein expression, and misregulated alternative splicing. Combined immunofluorescence and in situ hybridization demonstrated elevated CUGBP1 and its paralog, CUGBP2 (ETR-3/NAPOR/BRUNO3), specifically in nuclei containing CUG repeat RNA foci. A time-course study of molecular changes following induction of DMPK-CUG repeat RNA expression demonstrated that splicing abnormalities were observed beginning at 12 hours following induction of Cre-mediated recombination. Importantly, RNA foci formation, MBNL1 colocalization with foci, and induction of CUGBP1 protein preceded splicing changes. These results demonstrate that an increased steady-state level of CUGBP1 is a specific and early event of DM1 pathogenesis.
Results
Inducible expression of expanded CUG RNA in cardiomyocytes.
To establish conditional mouse models for DM1 that reproduce the full array of symptoms, we used a Cre-loxP approach to induce the expression of expanded CUG RNA within the context of the DMPK 3′ UTR. The EpA960 transgene contains a ubiquitously expressed CMV promoter, a floxed concatemer of the SV40 polyadenylation site, and human DMPK exon 15 containing 960 copies of interrupted CTG repeats (Figure 1A). The SV40 polyadenylation sites prevent expression of RNA from downstream genomic segments (16), and their removal by Cre-mediated recombination results in transcription and splicing of DMPK exon 15 into the transgene mRNA. We also generated lines containing a transgene lacking CTG repeats (EpA0) to express an identical mRNA containing only the DMPK 3′ UTR following Cre-mediated recombination (Figure 1A). Three different EpA960 lines expressed different levels of EpA960 transgene mRNA from the nonrecombined allele, as determined by quantitative real-time RT-PCR (Figure 1B). Expression of one EpA0 line was comparable to that of the highest-expressing EpA960 line (Figure 1B). In addition, the 5 lines tested (3 EpA960 and 2 EpA0) expressed the transgene in all 3 tissues that were tested: heart, skeletal muscle, and brain (Figure 1B).
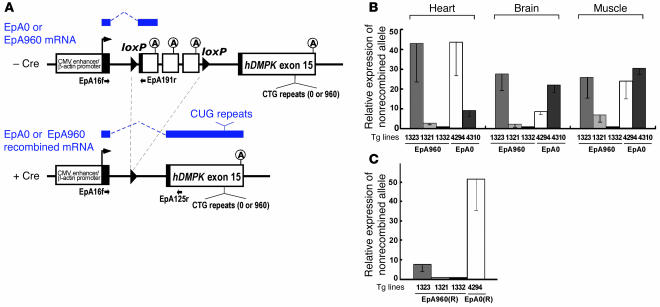
Figure 1. Generation of bitransgenic mouse expressing expanded CUG RNA in heart.
(A) EpA960 and EpA0 transgene constructs. The spliced mRNA transcripts from the nonrecombined (top) and recombined alleles (bottom) are indicated in blue above the gene diagrams. The mRNAs from the recombined EpA0 and EpA960 alleles are identical except for the presence or absence of the CUG repeats. Primer pairs used for RT-PCR analysis of nonrecombined and recombined alleles are shown. The CMV enhancer and β-actin promoter drive the transcription ubiquitously. Black boxes represent segments of different exons added for splicing. The cassette containing SV40 polyadenylation sites that prevent expression of RNA from downstream gene segments is located between 2 loxP sites. (B) Relative levels of the nonrecombined transgene mRNAs in different tissues from EpA960 and EpA0 transgenic mice using real-time RT-PCR. Results are from 3 mice from each line, with the exception of the muscle sample, taken from 1 mouse in line EpA960/MCM 1332. (C) Relative expression of EpA960(R) and EpA0(R) mRNAs from the recombined alleles after Cre-mediated recombination in heart.
The 3 EpA960 lines and the highest-expressing EpA0 control line, 4294, were crossed with MerCreMer (MCM) mice, in which heart-specific expression of a tamoxifen-inducible Cre is driven by the α–myosin heavy chain (α-MHC) promoter (17). EpA960/MCM and EpA0/MCM bitransgenic mice were treated with tamoxifen at 8–9 weeks of age to induce Cre-mediated recombination, and RNA was extracted from heart tissue after 1 week. Real-time RT-PCR using primers specific for mRNAs from the recombined allele demonstrated that line EpA960/MCM 1323 expressed the highest level of EpA960(R) mRNA from the recombined allele (Figure 1, B and C). All mice from line EpA960/MCM 1323 died within 2 weeks of tamoxifen injection, while the 2 lower-expressing lines remained viable and expressed no obvious phenotype 1 year after tamoxifen injection. Mice of the EpA0/MCM 4294 line expressed greater than 5-fold more mRNA from the recombined allele than did EpA960/MCM 1323 mice (Figure 1C) and showed no phenotype up to 6 months after tamoxifen injection. We conclude that CUG repeats are required for RNA toxicity. Unless otherwise indicated below, our subsequent experiments used the lines EpA960/MCM 1323 and EpA0/MCM 4294.
Mice expressing EpA960(R) RNA exhibit cardiomyopathy, arrhythmia, and systolic and diastolic dysfunction.
Histological analysis of heart tissues from EpA960/MCM mice revealed dilated cardiomyopathy characterized by dilation of the left ventricle, thinning of the left ventricular wall, and myocyte hypertrophy (Figure 2, A–F). The cardiomyocyte cytoplasm appeared pale in patchy areas and contained variably prominent numbers of pink granules (Figure 2E). Rare degenerating myocytes and focal areas of mild interstitial fibrosis were also observed with trichrome stain (data not shown). Ultrastructural analysis by electron microscopy revealed focally abundant mitochondria that correlated with the pink granules seen by light microscopy (Figure 2, E and F). The sarcomeres and Z-lines appeared normal. Electron microscopy also confirmed myocyte hypertrophy with enlarged, irregular myocyte nuclei. In some myocytes, aggregates of proliferating membranes of the sarcoplasmic reticulum were also noticeable. Notably, dilated cardiomyopathy, interstitial fibrosis, hypertrophic or degenerated myocardial cells, and mitochondriosis are observed in individuals with DM1 (14, 15, 18).
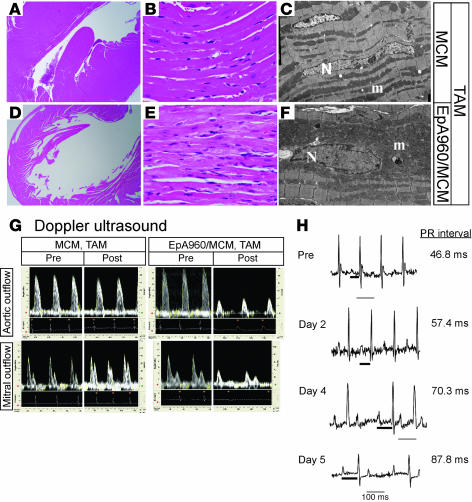
Figure 2. Induced EpA960/MCM mice reproduce functional and histopathological features of DM1.
(A–F) EpA960/MCM and MCM littermates were given tamoxifen (TAM) at the same time. (A and D) H&E stain revealed that EpA960/MCM hearts were dilated compared with hearts of tamoxifen-treated MCM mice. Original magnification, ×2. (B and E) H&E stain revealed that induced EpA960/MCM hearts exhibited hypertrophied cardiomyocytes that focally contained pale, granular cytoplasm compared with MCM hearts. Original magnification, ×40. (C and F) Electron microscopy revealed hypertrophied myocytes with irregular nuclei (N) and abundant mitochondria (m) in EpA960/MCM hearts compared with MCM hearts. Original magnification, ×4,000. (G) Cardiac functional abnormalities in mice expressing EpA960(R) mRNA. Doppler ultrasound was performed before (Pre) and after (Post) tamoxifen administration on EpA960/MCM and MCM littermates and revealed both systolic and diastolic dysfunction in hearts expressing EpA960(R) mRNA. (H) ECG telemetry revealed progressive arrhythmias following induction of expanded CUG RNA expression. The length of the PR interval (thick bar) is indicated; thin bars represent 100 ms.
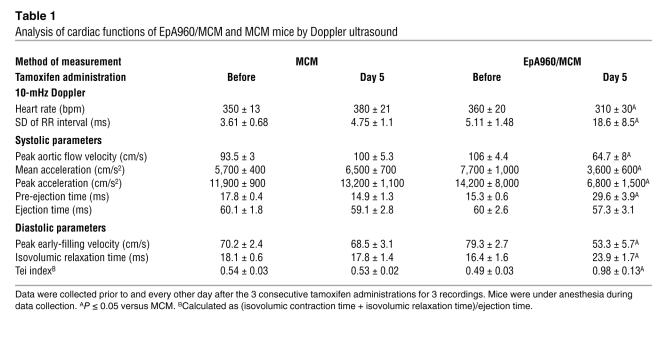
The effects of EpA960(R) mRNA expression on heart function were determined by 10-mHz Doppler ultrasound. The analysis was performed sequentially before and every 2 days after tamoxifen administration on MCM and EpA960/MCM mice. No differences in systolic or diastolic parameters were observed between MCM and EpA960/MCM mice prior to tamoxifen administration. However, following tamoxifen administration, EpA960/MCM but not MCM mice exhibited cardiac dysfunction in both systolic and diastolic parameters, similar to what has been documented in DM1 patients (19, 20). Systolic function exhibited a 50% or greater decrease in peak aortic flow velocity and both mean and peak acceleration (Table 1 and Figure 2G). Peak early diastolic filling velocity was decreased by more than 30% in EpA960/MCM mice, but was unchanged in the MCM mice, compared with mice before tamoxifen treatment (Table 1 and Figure 2G). Isovolumic relaxation time was prolonged by more than 50% in EpA960/MCM mice. Because there were some changes in heart rate, the Tei index, which normalizes isovolumic relaxation time plus isovolumic contraction time by ejection time, was calculated. Again, EpA960/MCM mice exhibited impaired function around 1 week following tamoxifen administration. There was not severe edema or audible respiration in these mice indicative of heart failure. We also examined the variation among RR intervals, which is used to measure the time between the R peaks in the electrocardiogram, to check the tendency toward arrhythmia. The standard deviation of RR intervals increased 5-fold after tamoxifen administration in EpA960/MCM mice but did not change in MCM mice. These results suggested that expression of expanded CUG RNA in heart prompted mice to develop arrhythmia.
Table 1 .
Analysis of cardiac functions of EpA960/MCM and MCM mice by Doppler ultrasound
ECG telemetry was used to analyze arrhythmias in unanesthetized mice. Analysis of the mouse with the most severe phenotype indicated that before tamoxifen administration, there was no spontaneous arrhythmia (Figure 2H). Progressive lengthening of the PR interval and widening of the QRS complex was observed beginning 2 days after the last day of tamoxifen administration (Figure 2H). At 4 days, this mouse showed 2:1 atrioventricular block. These conduction defects recapitulated those observed in DM1 patients (11). Thus, mice expressing EpA960(R) RNA exhibit cardiomyopathy, arrhythmia, and diastolic and systolic dysfunction.
Misregulated alternative splicing in mice expressing EpA960(R) RNA.
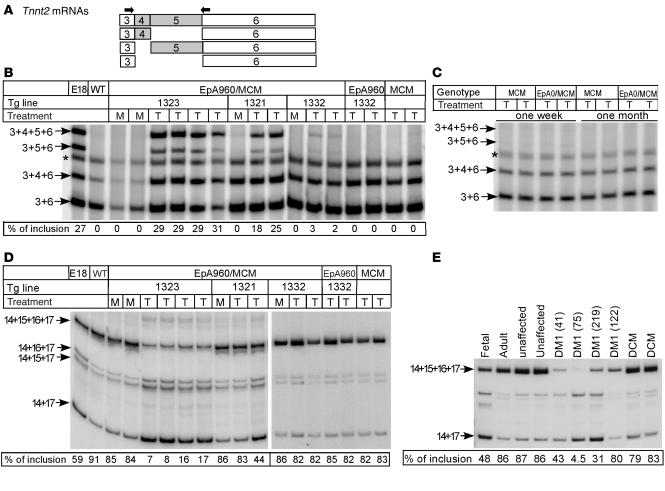
A characteristic molecular feature of DM1 is misregulation of developmental alternative splicing transitions. We found that Tnnt2 exhibited the embryonic splicing pattern of alternative exons 4 and 5 in tamoxifen-treated EpA960/MCM mice, whereas there was no inclusion of exons 4 and 5 in mock-treated EpA960/MCM mice (Figure 3, A and B). This pattern of exon inclusion recapitulates what was previously found in DM1 cardiac tissue (4, 21). Tamoxifen did not induce splicing transitions of exons 4 and 5 in EpA or MCM mice (Figure 3B). Importantly, the extent of Tnnt2 exon 5 inclusion in the 3 EpA960/MCM lines correlated with the level of expression of repeat-containing mRNA (29.5%, 21.5%, and 2.5% for EpA960/MCM lines 1323, 1321, and 1332, respectively), and the lethal bitransgenic line expressed a level of inclusion similar to that observed in embryonic heart (29.5% versus 27%; Figure 3B). Cardiac tissue from bitransgenic mice expressing high levels of EpA0(R) mRNA for 1 week or 1 month did not exhibit the embryonic splicing pattern of exons 4 and 5 (Figure 3C). Thus, overexpression of DMPK exon 15 containing repeats reproduces aberrant splicing of Tnnt2, as observed in individuals with DM1, while DMPK exon 15 lacking repeats in adult cardiac tissue does not affect Tnnt2 alternative splicing.
Figure 3. Misregulated alternative splicing of Tnnt2 and Fxr1h in heart expressing EpA960(R) mRNA but not EpA0(R) mRNA.
(A) Alternative splicing patterns of mouse Tnnt2 exons 4 and 5 and the location of the RT-PCR primers (arrows). (B) Mice expressing EpA960(R) mRNA reverted to the embryonic splicing pattern of Tnnt2. The EpA960/MCM bitransgenic mice from 3 EpA960 lines as well as transgenic EpA960 and MCM mice were given tamoxifen (T) at a dosage of 20 mg/kg/d for 5 consecutive days, and RNA was extracted 1 week after the last injection. EpA960/MCM bitransgenic littermates treated with oil served as mock (M) controls. E18 cardiac tissue exhibited the Tnnt2 embryonic alternative splicing pattern. Percent of mRNAs containing exons 4 and 5 is shown. Asterisks denote a hybrid band of the 2 smallest PCR products. (C) Mice expressing EpA0(R) mRNA exhibited no phenotype and retained adult splicing patterns of Tnnt2 splicing both 1 week and 1 month following tamoxifen injection. (D) Decreased inclusion of exons 15 and 16 of FXR1h in mouse heart expressing EpA960(R) mRNA. Percent of mRNAs containing exons 15 and 16 is shown. (E) Decreased inclusion of exons 15 and 16 of FXR1h was also observed in individuals with DM1 (patient identification number shown in parentheses). Splicing abnormalities were not observed in tissue from unaffected individuals or individuals with non-DM1 forms of dilated cardiomyopathy (DCM).
We then examined splicing of Fragile X–related protein 1 (Fxr1h) exons 15 and 16, which undergo developmentally regulated alternative splicing (22, 23). We found striking reversion to an embryonic pattern in the severe line (Figure 3D). However, misregulated splicing of Fxr1h was observed in 1 of 3 EpA960/MCM 1321 mice examined and in none of the lowest-expressing EpA960/MCM 1332 mice. Tamoxifen alone did not alter the splicing pattern of Fxr1h in EpA960 or MCM transgenic mice (Figure 3D). We then checked splicing of FXR1 exons 15 and 16 in heart samples of patients with DM1 and found a similar reversion to an embryonic pattern that was not observed in individuals without cardiac disease or affected by idiopathic dilated cardiomyopathy (Figure 3E). The results indicated that the EpA960 lines induced to express CUG repeat RNA reproduced characteristic splicing abnormalities observed in DM1 heart.
Nuclear RNA foci formation in mice expressing expanded CUG RNA.
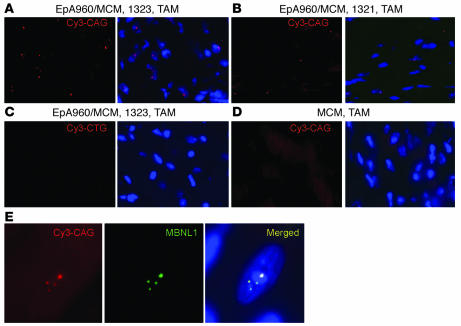
Expanded DMPK-CUG RNA forms nuclear foci in DM1 cardiac tissue (21). In situ hybridization using Cy3-labeled CAG–peptide nucleic acid (CAG-PNA) probes demonstrated nuclear RNA foci in cardiac tissue from tamoxifen-induced bitransgenic mice (Figure 4, A and B). Heart from the highest-expressing line, EpA960/MCM 1323, contained foci in a higher fraction of nuclei than that of the moderate-expressing line EpA960/MCM 1321 (Figure 4, A and B). In addition, multiple RNA foci were detected in the same nucleus in the higher-expressing line, whereas in the moderate line, the majority of foci-containing nuclei had single RNA foci. Foci were not detected in heart tissue from tamoxifen-treated EpA960/MCM 1323 mice using sense Cy3-labeled CTG-PNA probes (Figure 4C) or in tamoxifen-treated MCM mice (Figure 4D). MBNL1 was found in nuclear foci that colocalized with the RNA foci in bitransgenic mice induced to express repeat RNA (Figure 4E), as previously demonstrated in cardiac tissue from individuals with DM1 (21).
Figure 4. EpA960(R) mRNA forms foci that colocalize with MBNL1.
In situ hybridization using a Cy3-CAG PNA probe was used to detect RNA foci. DAPI was used for nuclear staining. Heart tissue from the higher-expressing line EpA960/MCM 1323 (A) showed 8- to 10-fold more nuclei containing RNA foci than the lower-expressing line EpA960/MCM 1321 (B). (C) No signal was detected in cardiac tissue from the highest-expressing line using a Cy3-CTG (sense) probe or (D) in MCM mice treated with tamoxifen. (E) Endogenous MBNL1 colocalized with nuclear RNA foci in heart cells expressing EpA960(R) mRNA in line EpA960/MCM 1323. Original magnification, ×40 (A–D); ×63 (E).
CELF protein steady-state levels are induced in hearts expressing expanded CUG RNA.
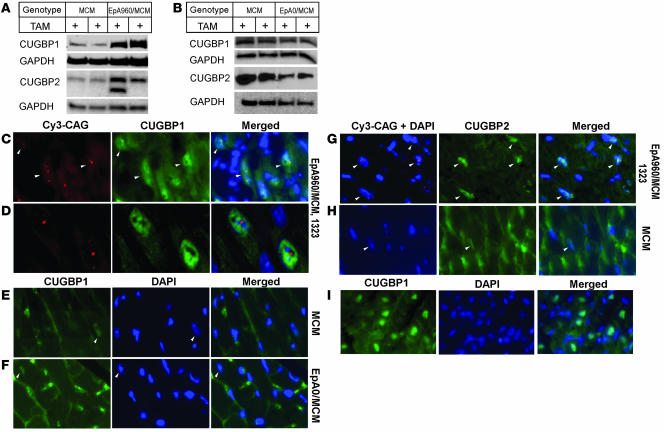
Heart tissue expresses 2 of the 6 CELF paralogs, CUGBP1 and CUGBP2. CUGBP1 and CUGBP2 proteins are normally downregulated during heart development to barely detectable levels in adult tissues (5, 24). CUGBP1 protein expression is increased in DM1 heart and skeletal muscle tissues as well as in DM1 cell cultures (7, 25–27). Reversion to an embryonic expression pattern of CUGBP1 has been proposed to be relevant to misregulation of alternative splicing and translation events that are regulated by this protein (2). Western blot analysis indicated that levels of CUGBP1 and CUGBP2 proteins were significantly increased in heart tissues from EpA960/MCM 1323 mice 1 week following tamoxifen administration (Figure 5A). In contrast, the tamoxifen-induced EpA0/MCM line did not show an increase in either CUGBP1 or CUGBP2 protein steady-state levels (Figure 5B).
Figure 5. CUGBP1 and CUGBP2 protein steady-state levels increase in cardiac tissue expressing EpA960(R) mRNA.
(A) Western blot analysis demonstrated elevated CUGBP1 and CUGBP2 protein expression in cardiac tissues expressing EpA960(R) mRNA (line EpA960/MCM 1323) 1 week following tamoxifen administration compared with tamoxifen-treated MCM littermates. An additional CUGBP2 band, possibly a splice variant, was expressed in some hearts expressing EpA960(R) mRNA. GAPDH was used as loading control. (B) Expression of CELF proteins remained unchanged in cardiac tissues expressing EpA0(R) mRNA (line 4294) 1 week following tamoxifen administration. (C) Nuclei expressing EpA960(R) mRNA foci exhibited elevated CUGBP1 protein expression, as shown by immunofluorescence staining.Arrowheads indicate nuclei containing RNA foci. (D) Higher magnification showed nuclei containing RNA foci also exhibited higher levels of CUGBP1. (E) Endogenous expression of CUGBP1 (arrowheads) in MCM mice given tamoxifen remained low. Nonspecific background in the cytoplasm was the result of mouse IgG. Nuclear CUGBP1 in cells expressing EpA0(R) mRNA (line 4294) remained low (F), as did nuclear CUGBP1 in MCM mice (E), 1 month following tamoxifen administration. (G) Nuclei containing EpA960(R) mRNA foci exhibited elevated expression of CUGBP2 protein. Arrowheads indicate nuclei containing RNA foci. (H) CUGBP2 expression (arrowheads) was low in tamoxifen-treated MCM hearts, with some areas of nonspecific staining in the cytoplasm. (I) Expression of CUGBP1 in the mouse heart at postnatal day 2. Images in E–I were taken using the same exposure time. Original magnification, ×40 (C and E–I); ×63 (D).
Combined immunofluorescence staining and in situ hybridization demonstrated that CUGBP1 expression was enhanced specifically in nuclei in which RNA foci were also detected (Figure 5, C and D). In contrast, nuclear CUGBP1 was low or undetectable in hearts from tamoxifen-treated MCM and EpA0/MCM mice (Figure 5, E and F). Consistent with several previous reports (21, 28, 29), CUGBP1 did not colocalize with RNA foci (Figure 5D). Nuclear CUGBP2 expression was also elevated in nuclei expressing RNA foci in tamoxifen-treated EpA960/MCM mice compared with tamoxifen-treated MCM control mice (Figure 5, G and H). EpA960(R) RNA expression should be limited to cardiomyocytes, which make up 20%–30% of the cell population within the myocardium (30). While control mice exhibited nonspecific cytoplasmic staining, control mice did not exhibit nuclei expressing levels of nuclear CELF proteins comparable to those of mice expressing EpA960(R) RNA. Cytoplasmic staining was likely to be nonspecific because it was removed when hearts were perfused (see Figure 6C, MCM and 0 hours). Thus, expression of the 2 CELF proteins that are normally downregulated during heart development was upregulated in response to expression of EpA960(R) but not EpA0(R) RNA. The nuclear expression pattern of CUGBP1 resembled its expression in mouse cardiac tissue at postnatal day 2 (Figure 5I).
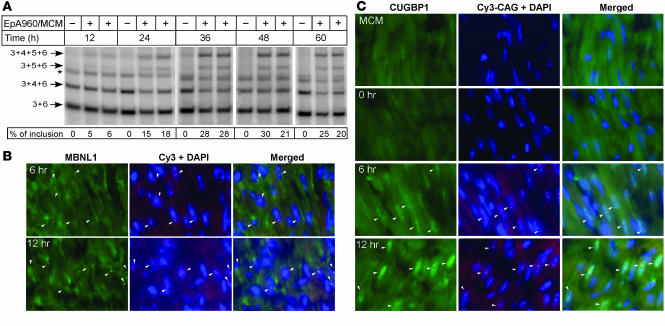
Figure 6. Time course demonstrates rapid molecular changes associated with induction of EpA960(R) mRNA.
(A) Tnnt2 splicing changes. Heart tissue was collected from mock and tamoxifen-treated littermates at the indicated time points following a single tamoxifen dose. Inclusion of Tnnt2 exons 4 and 5 was first detected at 12 hours, with increasing exon inclusion at subsequent time points. Asterisk indicates hybrid bands of the 2 smallest PCR products. (B) RNA foci and colocalized MBNL1 foci were detected by 6 hours and with increased intensity at 12 hours. (C) Elevated nuclear staining for CUGBP1 was detected 6 hours following tamoxifen administration compared with tamoxifen-treated MCM and untreated EpA960/MCM mice (0 hours). Arrowheads indicate nuclei containing RNA foci. Original magnification, ×40.
CELF protein induction is an early event following CUG RNA expression.
We took advantage of the inducible system to investigate the timing of RNA foci formation, MBNL colocalization with RNA foci, enhanced nuclear CUGBP1 expression, and Tnnt2 splicing changes following induction of EpA960(R) RNA expression. Reversion of Tnnt2 alternative splicing to the embryonic pattern was first detected 12 hours following tamoxifen treatment and, by 36 hours, reached the level observed at 1 week following tamoxifen administration and in E18 heart (compare Figure 6A and Figure 3B). Interestingly, we found that sparse RNA foci were detected as early as 3 hours following tamoxifen administration, and MBNL1 colocalization with RNA foci was weakly detected at these early time points (data not shown). MBNL colocalization with RNA foci became clearly detectable by 6 hours following tamoxifen administration and exhibited increased intensity at 12 hours (Figure 6B). We conclude that foci formation is extremely rapid following induction of EpA960(R) mRNA expression and that MBNL1 colocalizes with foci as soon as foci are detectable. We then traced the earliest time point at which elevated CUGBP1 expression could be detected. As noted above, staining for CUGBP1 was low in hearts from MCM and EpA960/MCM without tamoxifen injection (Figure 6C), which do not express EpA960(R) RNA. However, we found that CUGBP1 expression in foci-containing nuclei was consistently elevated by 6 hours and showed increased signal at 12 hours following tamoxifen administration (Figure 6C). The rapidity of the response indicates that induction of CUGBP1 steady-state levels is a primary response to expression of EpA960(R) RNA rather than a secondary response to cardiac cell injury.
Discussion
In the present study, an inducible and heart-specific DM1 mouse model expressing expanded CUG RNA in the context of DMPK 3′ UTR exhibited the functional, pathological, electrophysiological, and molecular features of DM1. These features include prolonged PR intervals and QRS duration, decreased diastolic and systolic velocities, dilated cardiomyopathy, hypertrophy of cardiomyocytes, and proliferation of mitochondria. Importantly, the EpA960/MCM model reproduced a feature of DM1 that has not to our knowledge been previously reproduced in heart tissue of a DM1 mouse model: increased steady-state levels of CUGBP1 protein. Combined in situ hybridization and immunofluorescent staining for CUGBP1 and its paralog, CUGBP2, demonstrated elevated levels of both proteins specifically in nuclei containing RNA foci. A time course of molecular events following induction of EpA960(R) mRNA expression demonstrated that colocalization of MBNL1 with RNA foci and increased CUGBP1 expression occurred early and coincided with reversion to an embryonic splicing pattern for Tnnt2. CUGBP1 and CUGBP2 are the only CELF proteins expressed in heart, and both are normally downregulated by 3 weeks postnatally (ref. 24 and our unpublished observations). Our analysis in EpA960/MCM mice demonstrated that a rapid increase in CELF protein expression, recapitulating the embryonic expression patterns, was an early response to the mutant RNA.
Altered activities of MBNL and CELF proteins mediate the disrupted alternative splicing observed in DM1 (2, 31). In addition, altered CUGBP1 cytoplasmic function is proposed to alter regulated translation of key myogenic proteins in skeletal muscle contributing to muscle degeneration (32). MBNL1 is sequestered by CUG repeat RNA nuclear foci, resulting in a loss of nuclear activity, while steady-state levels of CUGBP1 increase in DM1 heart and skeletal muscle (6, 7, 21, 33). Here we demonstrate that RNA foci formed and MBNL1 colocalized with foci within 6 hours — even as soon as 3 hours — following tamoxifen administration. Elevated CUGBP1 protein expression was detected by immunofluorescence within 6 hours of tamoxifen administration. The time between RNA accumulation and changes in MBNL and CELF expression must be substantially less than 6 hours, because time is required for tamoxifen uptake, Cre-mediated recombination, and accumulation of EpA960(R) RNA from the recombined allele. The timing of CUGBP1 nuclear accumulation and MBNL colocalization directly correlates with Tnnt2 splicing changes, suggesting that both events could contribute to the disease.
Increased CUGBP1 was previously demonstrated to be pathogenic in CUGBP1-overexpressing mice in which increased levels of CUGBP1 in heart and skeletal muscle or in skeletal muscle alone resulted in neonatal lethality (32, 34). A pathogenic effect of CUGBP1 was also demonstrated in a fly model for DM1 (35). While loss of MBNL activity can be explained by sequestration on nuclear CUG repeat RNA, the mechanism of increased CUGBP1 expression in cells containing RNA foci is less straightforward. Our present results confirm results from other labs indicating that neither CUGBP1 nor CUGBP2 is sequestered by RNA foci (21, 36); however, this does not preclude a transient interaction between CUGBP1 and RNA foci or transient or even stable interactions of CUGBP1 with a form of CUG repeat RNA not associated with foci. Our recent results from EpA960/MCM mice and DM1 cells indicate that steady-state levels of CUGBP1 protein increase as a result of hyperphosphorylation and increased half-life. Furthermore, CUG repeat RNA mediates this effect via activation of PKC, which was found to be activated in heart tissue from EpA960/MCM mice treated with tamoxifen as well as in heart tissue and cultured cells from individuals with DM1 (37).
A comparison of our results with results from individuals with DM1 or DM2 and with other mouse models strongly suggests that the context of the pathogenic repeats determines whether CUGBP1 protein levels are affected. CUGBP1 levels are not elevated by repeat-containing RNAs that lack the DMPK 3′ UTR, that is, in the HSALR mouse model in which CUG repeats are expressed in the context of the human skeletal α-actin mRNA or in DM2 skeletal muscle in which a CCUG repeat within ZNF9 intron 1 causes disease. In both cases, MBNL1 colocalizes with RNA foci and splicing is altered, but CUGBP1 levels are not affected (5). In contrast, CUGBP1 protein is elevated in heart and skeletal muscle tissues and in cultured myoblasts and fibroblasts from DM1 patients (7, 26, 27, 32). Recently, high expression of RNA in mice containing the DMPK 3′ UTR with only 5 CTG repeats, GFP, and a segment of DMPK intron 1 exhibited histological changes, myotonia, and splicing changes in skeletal muscle that were characteristic of DM1 (38). These mice also exhibited arrhythmias. The absence of RNA foci and lack of obvious MBNL sequestration suggested that elevated CUGBP1 promotes the splicing changes (39); however, an alternative explanation is that MBNL is effectively inactivated by binding to RNA containing 5 CUGs without forming foci. Mice expressing repeat RNA exhibited elevated CUGBP1 in skeletal muscle but not in heart, correlating with higher levels of RNA expression in muscle than in heart (38). The absence of overt cardiomyopathy and the limitation of the cardiac phenotype to the conduction system suggest that the effects of the repeat RNA is limited to cells involved in conduction. It is possible that elevated CUGBP1 expression was not sufficiently widespread to be detected by Western blot analysis. We find that low-level induction of CUGBP1 protein expression, particularly when limited to specific cell types, is detectable by immunofluorescence but not Western blot analysis (our unpublished observations). It would be of interest to use immunofluorescence to examine heart tissue for cell-specific elevated CUGBP1 expression.
Expression of EpA960(R) mRNA in the EpA960/MCM 1323 line was lethal within 2 weeks of tamoxifen induction, while an identical mRNA lacking repeats had no noticeable phenotype even when more than 5-fold the amount of RNA was expressed. These results are consistent with results from the mouse C2C12 myoblast cell line, in which only CTG repeats in the context of the DMPK 3′ UTR inhibited skeletal muscle differentiation, while either CTG or DMPK alone had no effect (40, 41). Similarly, expression of RNA containing the DMPK 3′ UTR with 960 CUG repeats induced hyperphosphorylation of CUGBP1 and increased protein stability, while comparable expression of the 3′ UTR alone had no effect (our unpublished observations). These results indicate that one or more cis-acting elements within DMPK exon 15 act in combination with the CUG repeat RNA to induce alterations of CUGBP1 expression.
Expression of expanded CUG RNA in the heart affected both electric activity and contractility in EpA960/MCM mice. The prominent arrhythmias observed in DM1 (increased PR interval and heart block) are consistent with the observed degeneration of the conduction system (11, 42) and suggest a mechanism in which cell loss is caused by toxicity of CUG repeat RNA. This does not rule out the possibility that arrhythmias result from misregulated alternative splicing of genes critical for proper electrophysiological properties of the heart. Several genes have been shown to be misspliced in DM1 cardiac tissues, such as TNNT2, KCNAB1, Titin, and ALP (21) as well as FXR1h, identified in the present study. Mutations within a large number of ion channels have been shown to cause familial forms of arrhythmias, and these genes often express multiple isoforms by alternative splicing (43, 44). The effect of EpA960(R) RNA expression on cardiac function is independent of the arrhythmias and indicates that CUG repeat RNA induces a functional defect within the myocardium. It remains to be determined whether this reflects an effect on alternative splicing or other functions of MBNL, CELF, or other proteins.
Methods
Transgenic mice.
The transgenes EpA960 and EpA0 contain a ubiquitously expressed CMV promoter (45), a floxed concatemer of the SV40 polyadenylation site (16), and human DMPK exon 15 containing 960 copies of interrupted CTG repeats (EpA960) or 0 repeats (EpA0). The interrupted CTG repeats were described previously (4) and were made by ligating double-stranded oligos containing 20 CTG repeats and overhangs for XhoI and SalI restriction sites. Digesting with SalI and XhoI selected for head-to-tail concatamers. The result was interrupted repeats containing CTCGA after every 20 CTG repeats. Pronuclei from FVB mice were used for microinjection to generate transgenic mice. Two primer sets were used to identify lines in which both the 5′ and 3′ ends of the transgene were intact. For the 5′ end, EpA5.3-GGGAGAGTGAAGCAGAACGTG (forward) and EpA5.5-CGGGGTCATTAGTTCATAGCC (reverse) were used. For the 3′ end, EpA3.3-CGGGGTCATTAGTTCATAGCC (forward) and EpA3.52-AAGCCGGGCCGTCCGTCTTC (reverse) were used. Transgenic MCM mice expressing tamoxifen-inducible MerCreMer under the control of the heart-specific α-MHC promoter were obtained from J. Molkentin (17). All mice reported were F1 progeny of EpA × MCM matings and were therefore hemizygous for 1 or both transgenes. Littermate bitransgenic, transgenic, and wild-type mice were used for all experiments whenever possible. All experiments were conducted in accordance with the NIH Guide for the Use and Care of Laboratory Animals and approved by the Institutional Animal Care and Use Committee of Baylor College of Medicine.
Tamoxifen administration.
To induce Cre-mediated recombination, bitransgenic mice, hemizygous for both transgenes (2–4 months of age) were treated with tamoxifen (Sigma-Aldrich) once a day for 5 consecutive days at a dosage of 20 mg/kg/d by intraperitoneal injection. Most bitransgenic mice from line EpA960/MCM 1323 died less than 1 week following tamoxifen administration, but by reducing the dosage to 10 mg/kg/d, the viability of mice could be extended to 2 weeks. In the time-course study (Figure 6), male EpA960/MCM 1323 bitransgenic mice were given 1 dose of tamoxifen (20 mg/kg). Cardiac tissues from all mice were collected for RNA extraction, protein extraction, and frozen sectioning at the indicated time points. To reduce the background generated from mouse IgG in the immunofluorescence staining, mice were anesthetized and perfused transcardially with PBS and then with 4% paraformaldehyde for some experiments. Hearts were then postfixed in 4% paraformaldehyde for overnight at 4°C before being subjected to frozen sectioning.
Real-time RT-PCR.
Total cellular RNA was extracted using TRI zol (Invitrogen) according to the manufacturer’s instructions. First-strand cDNA was synthesized using 1–2 μg total RNA, 100 ng random hexamer, 1.25 mM dNTP, and 6 U AMV reverse transcriptase. EpA16f (GCCCCGGCTCTGACTGA) and EpA191r (TGTGGTATGGCTGATTATGATCCT) were used for real-time PCR of RNA from the nonrecombined allele in EpA960 and EpA0 transgenic lines (Figure 1B). EpA16f and EpA125r (ACAGCACAATAACCAGCACGTT) were used for real-time RT-PCR to quantify mRNA from the recombined allele (Figure 1C). Primer pairs for 18S RNA were used as internal controls (ACCGCAGCTAGGAATAATGGA and GCCTCAGTTCCGAAAACCA). The PCR reaction was carried out on an ABI-Prism 7000 Sequence Detection System with SYBR-Green PCR master mix (Applied Biosystems). Cycle numbers of nonrecombined or recombined alleles were normalized first to cycle numbers of 18S. The relative expression level of nonrecombined and recombined alleles was determined using the formula 2–(cycle no., individual lines) – (cycle no., lowest-expressing EpA960/MCM 1332).
RT-PCR splicing analysis.
To assay splicing of endogenous Tnnt2, total RNA (5 μg) and 100 ng oligo(dT) primer were used for first-strand cDNA synthesis. PCR amplification was performed with primers as described previously (46, 47). To detect endogenous FXR1h mRNA, we used 1-step RT-PCR containing total RNA (1 μg), 1.5 mM MgCl2, 0.2 mM dNTP, 2.8 U AMV reverse transcriptase (Life Science), 0.4 U Platinum Taq (Invitrogen), 200 ng forward (GATAATACAGAATCCGATCAG) and reverse (CTGAAGGACCATGCTCTTCAATCAC) primers, and 1.5 ng 5′-P-end-labeled forward primer. These primers generate 4 fragments (195, 276, 287, and 368 bp). Reverse transcription was performed at 42°C for 1 hour, followed by 20 cycles of PCR amplification consisting of 45 seconds at 95°C, 45 seconds at 57°C, and 45 seconds at 72°C and a final 10-minute extension at 72°C. The products of PCR amplification were separated on 5% nondenaturing polyacrylamide gels. The radioactivity associated with each band was quantified using a PhosphorImager (Molecular Dynamics).
Patient samples.
Human tissue samples were obtained from tissue cooperatives including the University of Miami Tissue Bank and NDRI as well as from C. Thornton (University of Rochester, Rochester, New York, USA).
Fluorescence in situ hybridization and immunofluorescence.
Fluorescence in situ hybridization was performed on frozen sections (7 μm) using (CAG)5-Cy3– or (CTG)5-Cy3–labeled PNA probes (Applied Biosystems) as described previously (48). Cardiac tissues were fixed in 4% paraformaldehyde for 1–2 days, then soaked in 30% sucrose for 24–48 hours for frozen sectioning. After hybridization and wash with 1× SSC, sections were incubated with blocking solution (3% BSA and 0.2% Triton X-100 in 1× PBS) for 1 hour, incubated with primary antibodies [polyclonal anti-MBNL1 at 1:1,000 or monoclonal anti-CUGBP1 (3B1) at 2 μg/ml; Abcam; or monoclonal antiCUGBP2 (1H2) at 3 μg/ml] for 36–48 hours, washed with PBS, and then incubated with secondary antibodies (Alexa Fluor 488 goat anti-rabbit IgG at 1:500 or Alexa Fluor 488 goat anti-mouse IgG at 1:1,000) for 2 hours, washed, and mounted with mounting medium containing DAPI (1.5 mg/ml; Vector). The polyclonal anti-MBNL1 antibody was generated against the region of amino acids 1–255 of human MBNL1 (GenBank accession no. NM_207297).
Immunoblot.
Protein was extracted with a dounce homogenizer in 10 mM HEPES (pH 7.5), 0.32 M sucrose, 1% SDS, 5 μM MG132, and 5 mM EDTA with protease inhibitors. For Western blot, 50–70 μg total protein was used. The membrane was incubated with monoclonal anti–CUGBP1 antibody (3B1; Upstate) or CUGBP2 (1H2) conjugated with HRP (24).
Assessment of cardiac function.
The 10-mHz Doppler ultrasound was performed on EpA960/MCM mice and their MCM littermates of MCM × EpA960 (line 1323) crosses. Assessment was performed before tamoxifen administration and every other day after tamoxifen administration for 5 days. Mice were anesthetized with 1% isoflurane gas in oxygen (49).
ECG telemetry.
ECG telemetry in unanesthetized and unrestrained mice was performed according to previously published methods (50). A transmitter (Data Sciences International) with subcutaneous electrodes was in a lead I configuration. Telemetry was recorded 7 days after surgery in baseline conditions and after the injection of 20 mg/kg tamoxifen for 3 consecutive days. ECG data were collected for 5 minutes every hour for a total of 4 weeks. Data collection was performed using Dataquest software, and offline data analyses were performed using Physiostat ECG analysis software (version 3.1; Data Sciences International). Analyses were performed without knowledge of genotype.
Statistics.
All data are expressed as mean ± SEM. Statistical significance was determined using unpaired Student’s t test. A P value less than 0.05 was considered significant.
Acknowledgments
We thank Jeffery Molkentin for providing us the MHC-MerCreMer mice and Donnie Bundman, Thuy Pham, Keith Weiser, and Kamrun Quader for their technical assistance. We also thank Muge Kuyumcu-Martinez and Cheng-Chiu Huang for helpful discussions on the manuscript. This work was supported by NIH grant R01AR45653, the Hunter Research Fund, and the Muscular Dystrophy Association (to T.A. Cooper) and by NIH grant R01DA017173 (to M. De Biasi).
Footnotes
Nonstandard abbreviations used: CELF, CUGBP1- and ETR-3–like factor; CUGBP, CUG-binding protein; DM1, myotonic dystrophy type 1; MBNL, muscleblind like; MCM, MHC-MerCreMer; PNA, peptide nucleic acid; 3′ UTR, 3′ untranslated region.
Conflict of interest: The authors have declared that no conflict of interest exists.
Citation for this article: J. Clin. Invest. 117:2802–2811 (2007). doi:10.1172/JCI32308
References
- 1.
- 2.Ranum L.P., Cooper T.A. RNA-mediated neuromuscular disorders. Annu. Rev. Neurosci. 2006;29:259–277. doi: 10.1146/annurev.neuro.29.051605.113014. [DOI] [PubMed] [Google Scholar]
- 3.Ho T.H., et al. Muscleblind proteins regulate alternative splicing. EMBO J. 2004;23:3103–3112. doi: 10.1038/sj.emboj.7600300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Philips A.V., Timchenko L.T., Cooper T.A. Disruption of splicing regulated by a CUG-binding protein in myotonic dystrophy. Science. 1998;280:737–741. doi: 10.1126/science.280.5364.737. [DOI] [PubMed] [Google Scholar]
- 5.Lin X., et al. Failure of MBNL1-dependent postnatal splicing transitions in myotonic dystrophy. Hum. Mol. Genet. 2006;15:2087–2097. doi: 10.1093/hmg/ddl132. [DOI] [PubMed] [Google Scholar]
- 6.Charlet-B. N., et al. Loss of the muscle-specific chloride channel in type 1 myotonic dystrophy due to misregulated alternative splicing. Mol. Cell. 2002;10:45–53. doi: 10.1016/s1097-2765(02)00572-5. [DOI] [PubMed] [Google Scholar]
- 7.Savkur R.S., Philips A.V., Cooper T.A. Aberrant regulation of insulin receptor alternative splicing is associated with insulin resistance in myotonic dystrophy. Nat. Genet. 2001;29:40–47. doi: 10.1038/ng704. [DOI] [PubMed] [Google Scholar]
- 8.Timchenko N.A., et al. Molecular basis for impaired muscle differentiation in myotonic dystrophy. Mol. Cell. Biol. 2001;21:6927–6938. doi: 10.1128/MCB.21.20.6927-6938.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bushby K., Muntoni F., Bourke J.P. 107th ENMC international workshop: the management of cardiac involvement in muscular dystrophy and myotonic dystrophy. June 7th–9th 2002, Naarden, the Netherlands. Neuromuscul. Disord. 2003;13:166–172. doi: 10.1016/s0960-8966(02)00213-4. [DOI] [PubMed] [Google Scholar]
- 10.Vinereanu D., et al. Subclinical cardiac involvement in myotonic dystrophy manifesting as decreased myocardial Doppler velocities. Neuromuscul. Disord. 2004;14:188–194. doi: 10.1016/j.nmd.2003.11.005. [DOI] [PubMed] [Google Scholar]
- 11.Phillips M.F., Harper P.S. Cardiac disease in myotonic dystrophy. Cardiovasc. Res. 1997;33:13–22. doi: 10.1016/s0008-6363(96)00163-0. [DOI] [PubMed] [Google Scholar]
- 12.Uemura N., et al. Electrophysiological and histological abnormalities of the heart in myotonic dystrophy. Am. Heart J. 1973;86:616–624. doi: 10.1016/0002-8703(73)90338-4. [DOI] [PubMed] [Google Scholar]
- 13.Dello Russo A., et al. Widespread electroanatomic alterations of right cardiac chambers in patients with myotonic dystrophy type 1. J. Cardiovasc. Electrophysiol. 2006;17:34–40. doi: 10.1111/j.1540-8167.2005.00277.x. [DOI] [PubMed] [Google Scholar]
- 14.Bulloch R.T., Davis J.L., Hara M. Dystrophia myotonica with heart block. A light and electron microscopic study. Arch. Pathol. 1967;84:130–140. [PubMed] [Google Scholar]
- 15.Ludatscher R.M., Kerner H., Amikam S., Gellei B. Myotonia dystrophica with heart involvement: an electron microscopic study of skeletal, cardiac, and smooth muscle. J. Clin. Pathol. 1978;31:1057–1064. doi: 10.1136/jcp.31.11.1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Maxwell I.H., Harrison G.S., Wood W.M., Maxwell F. A DNA cassette containing a trimerized SV40 polyadenylation signal which efficiently blocks spurious plasmid-initiated transcription. Biotechniques. 1989;7:276–280. [PubMed] [Google Scholar]
- 17.Sohal D.S., et al. Temporally regulated and tissue-specific gene manipulations in the adult and embryonic heart using a tamoxifen-inducible Cre protein. Circ. Res. 2001;89:20–25. doi: 10.1161/hh1301.092687. [DOI] [PubMed] [Google Scholar]
- 18.Motta J., Guilleminault C., Billingham M., Barry W., Mason J. Cardiac abnormalities in myotonic dystrophy. Electrophysiologic and histopathologic studies. Am. J. Med. 1979;67:467–473. doi: 10.1016/0002-9343(79)90795-2. [DOI] [PubMed] [Google Scholar]
- 19.Tokgozoglu L.S., et al. Cardiac involvement in a large kindred with myotonic dystrophy. Quantitative assessment and relation to size of CTG repeat expansion. JAMA. 1995;274:813–819. [PubMed] [Google Scholar]
- 20.Fragola P.V., Calo L., Luzi M., Mammarella A., Antonini G. Doppler echocardiographic assessment of left ventricular diastolic function in myotonic dystrophy. Cardiology. 1997;88:498–502. doi: 10.1159/000177398. [DOI] [PubMed] [Google Scholar]
- 21.Mankodi A., Lin X., Blaxall B.C., Swanson M.S., Thornton C.A. Nuclear RNA foci in the heart in myotonic dystrophy. Circ. Res. 2005;97:1152–1155. doi: 10.1161/01.RES.0000193598.89753.e3. [DOI] [PubMed] [Google Scholar]
- 22.Khandjian E.W., et al. Novel isoforms of the fragile X related protein FXR1P are expressed during myogenesis. Hum. Mol. Genet. 1998;7:2121–2128. doi: 10.1093/hmg/7.13.2121. [DOI] [PubMed] [Google Scholar]
- 23.Kirkpatrick L.L., McIlwain K.A., Nelson D.L. Alternative splicing in the murine and human FXR1 genes. Genomics. 1999;59:193–202. doi: 10.1006/geno.1999.5868. [DOI] [PubMed] [Google Scholar]
- 24.Ladd A.N., Stenberg M.G., Swanson M.S., Cooper T.A. Dynamic balance between activation and repression regulates pre-mRNA alternative splicing during heart development. Dev. Dyn. 2005;233:783–793. doi: 10.1002/dvdy.20382. [DOI] [PubMed] [Google Scholar]
- 25.Timchenko L.T., et al. Identification of a (CUG)n triplet repeat RNA-binding protein and its expression in myotonic dystrophy. Nucleic Acids Res. 1996;24:4407–4414. doi: 10.1093/nar/24.22.4407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Timchenko N.A., et al. RNA CUG repeats sequester CUGBP1 and alter protein levels and activity of CUGBP1. J. Biol. Chem. 2001;276:7820–7826. doi: 10.1074/jbc.M005960200. [DOI] [PubMed] [Google Scholar]
- 27.Dansithong W., Paul S., Comai L., Reddy S. MBNL1 is the primary determinant of focus formation and aberrant insulin receptor splicing in DM1. J. Biol. Chem. 2005;280:5773–5780. doi: 10.1074/jbc.M410781200. [DOI] [PubMed] [Google Scholar]
- 28.Michalowski S., et al. Visualization of double-stranded RNAs from the myotonic dystrophy protein kinase gene and interactions with CUG-binding protein. Nucleic Acids Res. 1999;27:3534–3542. doi: 10.1093/nar/27.17.3534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fardaei M., Larkin K., Brook J.D., Hamshere M.G. In vivo co-localisation of MBNL protein with DMPK expanded-repeat transcripts. Nucleic Acids Res. 2001;29:2766–2771. doi: 10.1093/nar/29.13.2766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Oh H., et al. Cardiac muscle plasticity in adult and embryo by heart-derived progenitor cells. Ann. N. Y. Acad. Sci. 2004;1015:182–189. doi: 10.1196/annals.1302.015. [DOI] [PubMed] [Google Scholar]
- 31.Kuyumcu-Martinez N.M., Cooper T.A. Misregulation of alternative splicing causes pathogenesis in myotonic dystrophy. Prog. Mol. Subcell. Biol. 2006;44:133–159. doi: 10.1007/978-3-540-34449-0_7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Timchenko N.A., et al. Overexpression of CUG triplet repeat-binding protein, CUGBP1, in mice inhibits myogenesis. J. Biol. Chem. 2004;279:13129–13139. doi: 10.1074/jbc.M312923200. [DOI] [PubMed] [Google Scholar]
- 33.Miller J.W., et al. Recruitment of human muscleblind proteins to (CUG)n expansions associated with myotonic dystrophy. EMBO J. 2000;19:4439–4448. doi: 10.1093/emboj/19.17.4439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ho T.H., Bundman D., Armstrong D.L., Cooper T.A. Transgenic mice expressing CUGBP1 reproduce splicing mis-regulation observed in myotonic dystrophy. Hum. Mol. Genet. 2005;14:1539–1547. doi: 10.1093/hmg/ddi162. [DOI] [PubMed] [Google Scholar]
- 35.de Haro M., et al. MBNL1 and CUGBP1 modify expanded CUG-induced toxicity in a Drosophila model of myotonic dystrophy type 1. Hum. Mol. Genet. 2006;15:2138–2145. doi: 10.1093/hmg/ddl137. [DOI] [PubMed] [Google Scholar]
- 36.Jiang H., Mankodi A., Swanson M.S., Moxley R.T., Thornton C.A. Myotonic dystrophy type 1 is associated with nuclear foci of mutant RNA, sequestration of muscleblind proteins and deregulated alternative splicing in neurons. Hum. Mol. Genet. 2004;13:3079–3088. doi: 10.1093/hmg/ddh327. [DOI] [PubMed] [Google Scholar]
- 37.Kuyumcu-Martinez N.M., Wang G.S., Cooper T.A. Increased steady state levels of CUGBP1 in Myotonic Dystrophy 1 are due to PKC-mediated hyperphosphorylation. Mol. Cell. 2007 doi: 10.1016/j.molcel.2007.07.027. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mahadevan M.S., et al. Reversible model of RNA toxicity and cardiac conduction defects in myotonic dystrophy. Nat. Genet. 2006;38:1066–1070. doi: 10.1038/ng1857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Timchenko L. Reversal of fortune. Nat. Genet. 2006;38:976–977. doi: 10.1038/ng0906-976. [DOI] [PubMed] [Google Scholar]
- 40.Amack J.D., Mahadevan M.S. The myotonic dystrophy expanded CUG repeat tract is necessary but not sufficient to disrupt C2C12 myoblast differentiation. Hum. Mol. Genet. 2001;10:1879–1887. doi: 10.1093/hmg/10.18.1879. [DOI] [PubMed] [Google Scholar]
- 41.Amack J.D., Reagan S.R., Mahadevan M.S. Mutant DMPK 3′-UTR transcripts disrupt C2C12 myogenic differentiation by compromising MyoD. J. Cell Biol. 2002;159:419–429. doi: 10.1083/jcb.200206020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rakocevic-Stojanovic V., et al. Pathohistological changes in endomyocardial biopsy specimens in patients with myotonic dystrophy. Panminerva Med. 1999;41:27–30. [PubMed] [Google Scholar]
- 43.Snyders D.J. Structure and function of cardiac potassium channels. Cardiovasc Res. 1999;42:377–390. doi: 10.1016/s0008-6363(99)00071-1. [DOI] [PubMed] [Google Scholar]
- 44.Liao P., Yong T.F., Liang M.C., Yue D.T., Soong T.W. Splicing for alternative structures of Cav1.2 Ca2+ channels in cardiac and smooth muscles. Cardiovasc. Res. 2005;68:197–203. doi: 10.1016/j.cardiores.2005.06.024. [DOI] [PubMed] [Google Scholar]
- 45.Ikawa M., Kominami K., Yoshimura Y., Nishimune Y., Okabe M. Green fluorescent protein as a marker in transgenic mice. Dev. Growth Differ. 1995;37:455–459. doi: 10.1046/j.1440-169X.1995.t01-2-00012.x. [DOI] [PubMed] [Google Scholar]
- 46.Xu X., et al. ASF/SF2 regulated CaMKIIdelta alternative splicing temporally reprograms excitation-contraction coupling in cardiac muscle. Cell. 2005;120:59–72. doi: 10.1016/j.cell.2004.11.036. [DOI] [PubMed] [Google Scholar]
- 47.Charlet-B. N., Logan P., Singh G., Cooper T.A. Dynamic antagonism between ETR-3 and PTB regulates cell type-specific alternative splicing. Mol. Cell. 2002;9:649–658. doi: 10.1016/s1097-2765(02)00479-3. [DOI] [PubMed] [Google Scholar]
- 48.Mankodi A., et al. Muscleblind localizes to nuclear foci of aberrant RNA in myotonic dystrophy types 1 and 2. Hum. Mol. Genet. 2001;10:2165–2170. doi: 10.1093/hmg/10.19.2165. [DOI] [PubMed] [Google Scholar]
- 49.Appleton G.O., et al. Determinants of cardiac electrophysiological properties in mice. J. Interv. Card. Electrophysiol. 2004;11:5–14. doi: 10.1023/B:JICE.0000035922.14870.56. [DOI] [PubMed] [Google Scholar]
- 50.Salas R., Pieri F., Fung B., Dani J.A., De Biasi M. Altered anxiety-related responses in mutant mice lacking the beta4 subunit of the nicotinic receptor. J. Neurosci. 2003;23:6255–6263. doi: 10.1523/JNEUROSCI.23-15-06255.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]