Abstract
Human DNA polymerase κ (hPolκ) is a proficient extender of mispaired primer termini on undamaged DNA, wherein it extends directly by incorporating the next correct nucleotide, generating single-base substitutions in the process. Biochemical and genetic studies, however, have indicated that, in addition to single-base substitutions, Polκ generates single-base deletions. Here we show that hPolκ is very adept at using template–primer misalignment as a novel means for extending mispaired termini and for generating single-base deletions. The proficient ability of hPolκ to extend mispaired primer termini either directly or by misalignment could be important for the continued and efficient progression of the replication fork when mismatches introduced by the replicative polymerase are not proofread. In extending from nucleotides opposite DNA lesions, hPolκ uses the direct and misalignment modes of mispair extension to different extents, depending on whether the template base is present or not at the primer terminus; thus, although hPolκ can extend directly from nucleotides opposite damaged bases, it can use only the misalignment mechanism to extend from nucleotides opposite an abasic site. A particularly unconstrained active site at the template–primer junction could afford hPolκ the ability to tolerate the geometric distortions of mismatched base pairs or those resulting from template–primer misalignment, thereby enabling it to use both of these modes of mispair extension.
Keywords: DNA polymerase κ, template–primer misalignment, mispair extension, base deletions, translesion DNA synthesis
DNA lesions often block replicative DNA polymerases (Pols), and replication through such lesions requires the participation of specialized translesion synthesis (TLS) DNA polymerases. In both prokaryotes and eukaryotes, DNA polymerases belonging to the Y family are able to promote replication through DNA lesions. DNA polymerase η (Polη), from both yeast and humans, replicates through a cis–syn thymine–thymine (TT) dimer (Johnson et al. 1999; Masutani et al. 1999), and steady-state kinetic studies have indicated that both these polymerases incorporate As opposite the two Ts of the TT dimer with the same efficiency and fidelity with which they incorporate As opposite undamaged Ts (Johnson et al. 2000c; Washington et al. 2000). Genetic studies in yeast have also implicated Polη in the error-free bypass of cyclobutane dimers formed at 5′-TC-3′ and 5′-CC-3′ sites (Yu et al. 2001). Polη can promote the efficient bypass of various other DNA lesions as well (Haracska et al. 2000a,b).
In addition to Polη, humans contain two other Y-family polymerases, Polι and Polκ. In contrast to Polη, which promotes replication through DNA lesions by both efficiently inserting the nucleotide opposite the lesion and by extending from the inserted nucleotide, Polι and Polκ promote TLS by promoting either the incorporation or the extension step, but not both (Prakash and Prakash 2002). For instance, Polι can incorporate nucleotides opposite the 3′T of a (6–4) TT photoproduct and opposite an abasic site, but it does not extend from the inserted nucleotide (Johnson et al. 2000b). Polκ, originally called Polθ by us (Johnson et al. 2000a), on the other hand, functions at the extension step; for example, it can extend from nucleotides inserted opposite the 3′T of a TT dimer and from nucleotides inserted opposite O6-methyl guanine, but it is highly inefficient at incorporating nucleotides opposite these DNA lesions (Haracska et al. 2002a; Washington et al. 2002).
Whereas Polη and Polι exist only in eukaryotes, homologs of Polκ are present in prokaryotes and eukaryotes, as well as in archaea. These Polκ-related polymerases include the dinB-encoded Pol IV of Escherichia coli, and the Dbh and Dpo4 polymerases of the thermophilic archaea, Sulfolobus solfataricus. One characteristic feature of all the Polκ homologs is their propensity to generate frameshift errors, particularly single-base deletions. Overexpression of Pol IV in E. coli results in a 1000-fold increase in spontaneous –1 frameshifts (Kim et al. 1997). In an in vitro DNA synthesis assay with Pol IV, single-base deletions and base substitutions were found to occur at a rate of 2 × 10–4 and 5 × 10–5, respectively (Kobayashi et al. 2002), and in similar assays with Dpo4, these changes occurred at a rate of 2.3 × 10–3 and 6.5 × 10–3, respectively (Kokoska et al. 2002). Genetic and biochemical studies have also indicated a role for Polκ in generating –1 frameshifts in addition to base substitutions. Transient overexpression of mouse DINB1-encoded Polκ in cultured mouse cells leads to enhanced spontaneous mutagenesis, with ∼70% of the mutations being base substitutions and ∼30% of the mutations being single-base deletions (Ogi et al. 1999). In an in vitro DNA synthesis reaction, human Polκ (hPolκ) generated single-base substitutions and deletions at a rate of ∼7 × 10–3 and ∼2 × 10–3, respectively (Ohashi et al. 2000a).
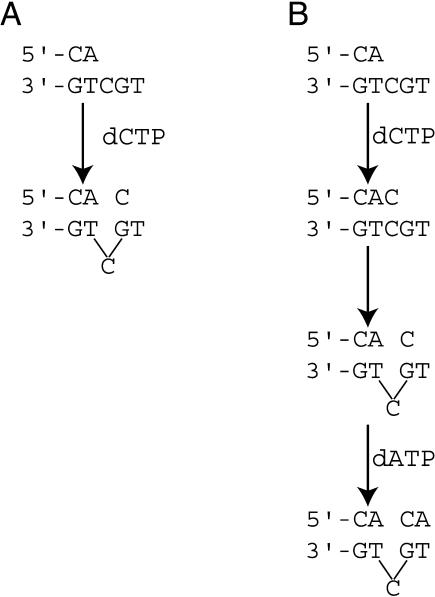
Deletions in homopolymeric runs could occur by the classical Streisinger model (Streisinger et al. 1966), resulting from the slippage of the primer and template strands relative to each other. This mechanism, however, does not explain the large fraction of deletions that occur in noniterated sequences in DinB-related polymerases. Two additional mechanisms, dNTP-stabilized misalignment (Efrati et al. 1997) and misinsertion misalignment (Kunkel and Soni 1988; Bebenek and Kunkel 1990), have been proposed to account for –1 deletion formation in noniterated sequences (Fig. 1). In dNTP-stabilized misalignment, the templating base becomes misaligned (“looped out”) in the polymerase active site, and the misalignment is stabilized by the pairing of the incoming dNTP with the complementary next template base (Fig. 1A). The evidence for the formation of –1 deletions by such a “dNTP-stabilized” misalignment mechanism is provided by the ternary complex of Dpo4, where, in type II crystals, the template base G becomes extrahelical and the incoming ddGTP bypasses this extrahelical base and pairs with the next template base C (Ling et al. 2001). Additionally, these structural data are supported by spectroscopic evidence showing that a template 2AP becomes extrahelical when Pol IV incorporates a dGMP opposite a template C located immediately downstream of the 2AP (Kobayashi et al. 2002). Misinsertion misalignment frameshifting occurs when nucleotide misincorporation is followed by template–primer slippage, and this results in the repositioning of the misincorporated nucleotide opposite the next complementary template base (Fig. 1B; Bebenek and Kunkel 1990). However, there has been no evidence that such a mechanism is used by any of the DinB-related polymerases.
Figure 1.
Model for generation of DNA polymerase-dependent single-base deletions. (A) In dNTP-stabilized misalignment, the templating base “loops out”, that is, is extrahelical, and the resulting misalignment is stabilized by the pairing of the incoming dNTP with the complementary next template base. (B) In misinsertion misalignment, nucleotide misincorporation is followed by template–primer slippage and the repositioning of the misincorporated nucleotide opposite the next complementary template base.
Most DNA polymerases, including the replicative polymerases, yeast and human Polη (Mendelman et al. 1990; Goodman et al. 1993; Washington et al. 2001a), as well as human Polι (Vaisman et al. 2001), extend a given mispair with nearly the same kinetic efficiency as the efficiency of incorporating the incorrect nucleotide to form that mispair. hPolκ, however, is more adept at mispair extension than at mispair formation (Washington et al. 2002). In this regard, hPolκ differs strikingly from E. coli Pol IV, as Pol IV extends mismatches far less efficiently than hPolκ (Kobayashi et al. 2002).
Here we show that mispaired primer termini can be extended by hPolκ with nearly equivalent efficiencies, either by direct extension or by the realignment of the mispaired primer-terminal base with the next complementary template base. Hence, hPolκ could contribute to rescuing the stalled replication fork when primer-terminal mismatches are not removed by the proofreading activity of Polδ. Such a mismatched primer end could then be extended by hPolκ either directly, by incorporating the next correct nucleotide, resulting in base substitution mutagenesis, or, by misalignment of the template base and the pairing of the primer-terminal base with the next complementary template base, resulting in –1 deletion formation (see Fig. 1B). The ability of hPolκ to efficiently use such a template–primer misalignment mechanism provides for a novel means of extending mispaired termini and for generating single-base deletions.
Results
Extension of mispaired primer termini by hPolκ by template–primer misalignment and by the incorporation of the next correct nucleotide
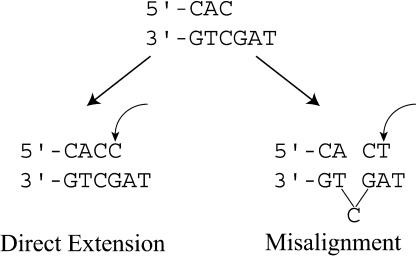
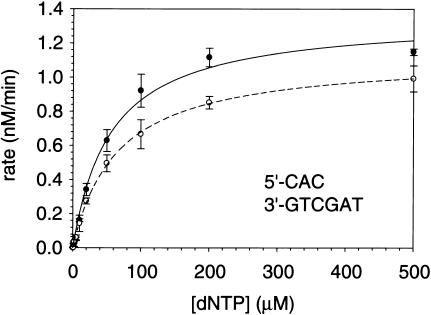
To compare the ability of hPolκ to extend mispaired primer termini by incorporating the correct next nucleotide (direct extension) or by template–primer misalignment, we used steady-state kinetics to measure the rate of nucleotide incorporation following a mismatched template–primer terminus. Appropriate DNA substrates were constructed to distinguish between these two modes of mispair extension (Table 1). DNA substrate 1, for example, contains a primer-terminal C · C mispair followed by a G and an A in the consecutive downstream template positions (Table 2). In this substrate, the C · C mispair can be extended by the direct incorporation of a C opposite the next 5′G; alternatively, this mispair can be extended by template–primer misalignment, in which case the primer-terminal C pairs with the next 5′G, and the misalignment is followed by the incorporation of a T opposite the next template base A (Fig. 2). hPolκ was incubated with different concentrations of dATP, dTTP, dCTP, or dGTP, and the rate of nucleotide incorporation plotted as a function of nucleotide concentration (Fig. 3). The kcat and Km values for nucleotide incorporation were then determined from the best fit to the Michaelis-Menten equation by using nonlinear regression. As shown in Figure 4, with this DNA substrate (#1), only the C and T nucleotides were incorporated, and the efficiencies (kcat/Km) of their incorporation were quite similar (Fig. 3; Table 2), indicating that, in this sequence context, the C · C mispair can be extended nearly equally well by direct extension or by misalignment. With DNA substrates 2 and 3, on the other hand, in which the C · C mispair is followed by a C or a T, respectively, in the next 5′ position in the template, the primer-terminal C is extended only by the incorporation of the next correct nucleotide, G or A, respectively (Table 2). Thus, in the absence of a complementary base in the next 5′ position in the template, the template–primer misalignment seen with substrate 1 does not occur.
Table 1.
DNA substrates used in this study
| DNA substrate | Sequencea |
|---|---|
| 1 | 5′-...GCAC |
| 3′-...CGTCGATCTTGAGAAGCACGTCCGTA | |
| 2 | 5′-...GCAC |
| 3′-...CGTCCATCTTGAGAAGCACGTCCGTA | |
| 3 | 5′-...GCAC |
| 3′-...CGTCTATCTTGAGAAGCACGTCCGTA | |
| 4 | 5′-...GCAC |
| 3′-...CGTCGTTCTTGAGAAGCACGTCCGTA | |
| 5 | 5′-...GCAC |
| 3′-...CGTCGCTCTTGAGAAGCACGTCCGTA | |
| 6 | 5′-...GCAC |
| 3′-...CGTCGGTCTTGAGAAGCACGTCCGTA | |
| 7 | 5′-...GCAGT |
| 3′-...CGTCGACGTTGAGAAGCACGTCCGTA | |
| 8 | 5′-...GCAGT |
| 3′-...CGTCGAGCTTGAGAAGCACGTCCGTA | |
| 9 | 5′-...GCAGT |
| 3′-...CGTCGATCTTGAGAAGCACGTCCGTA | |
| 10 | 5′-...GCAGT |
| 3′-...CGTCGAGGTTGAGAAGCACGTCCGTA | |
| 11 | 5′-...GCA |
| 3′-...CGTXATTCTTGAGAAGCACGTCCGTA | |
| 12 | 5′-...GCA |
| 3′-...CGTXGTTCTTGAGAAGCACGTCCGTA | |
| 13 | 5′-...GCA |
| 3′-...CGTXCTTCTTGAGAAGCACGTCCGTA | |
| 14 | 5′-...GCAC |
| 3′-...CGTXGTTCTTGAGAAGCACGTCCGTA | |
| 15 | 5′-...GCAG |
| 3′-...CGTXCTTCTTGAGAAGCACGTCCGTA | |
| 16 | 5′-...GCAT |
| 3′-...CGTXATTCTTGAGAAGCACGTCCGTA |
X corresponds to an abasic site.
For all of the DNA substrates, the upstream template—primer sequence is:
5′-CGACGATGCTCCGGTACTCCAGTGTAG...;
3′-GCTGCTACGAGGCCATGAGGTCACATC....
Table 2.
Steady-state kinetic parameters for C · C mispair extension by Polκ on undamaged DNA
| DNA substrate | Sequence | dNTP | kcat (min-1) | Km (μM) | kcat/Km (μM-1 min-1) | frel |
|---|---|---|---|---|---|---|
| 1 | CAC | dATP | n.d. | n.d. | <0.0001 | |
| GTCGAT | dTTP | 1.1 ± 0.02 | 65 ± 3 | 0.017 | 0.74 | |
| dCTP | 1.3 ± 0.06 | 56 ± 7 | 0.023 | |||
| dGTP | n.d. | n.d. | <0.0001 | |||
| 2 | CAC | dGTP | 0.40 ± 0.02 | 9.1 ± 2 | 0.04 | |
| GTCCAT | dTTP | n.d. | n.d. | <0.0001 | ||
| 3 | CAC | dATP | 0.34 ± 0.02 | 2.2 ± 0.5 | 0.15 | |
| GTCTAT | dTTP | n.d. | n.d. | <0.0001 | ||
| 4 | CAC | dATP | 0.89 ± 0.03 | 30 ± 4 | 0.030 | |
| GTCGTT | dTTP | n.d. | n.d. | <0.0001 | 0.3 | |
| dCTP | 1.0 ± 0.06 | 10 ± 3 | 0.1 | |||
| dGTP | n.d. | n.d. | <0.0001 | |||
| 5 | CAC | dATP | n.d. | n.d. | <0.0001 | |
| GTCGCT | dTTP | n.d. | n.d. | <0.0001 | 1.0 | |
| dCTP | 0.61 ± 0.05 | 18 ± 6 | 0.034 | |||
| dGTP | 0.34 ± 0.09 | 9.9 ± 1.2 | 0.034 | |||
| 6 | CAC | dATP | n.d. | n.d. | <0.0001 | |
| GTCGGT | dTTP | n.d. | n.d. | <0.0001 | N/A | |
| dCTP | 0.57 ± 0.042 | 19 ± 5 | 0.03 | |||
| dGTP | n.d. | n.d. | <0.0001 |
(frel) The relative efficiency (kcat/Km) of misalignment to direct extension; (n.d.) no nucleotide incorporation was detected.
Figure 2.
Two modes of mismatch extension by hPolκ. (Left) In direct mismatch extension, following a primer-terminal mispair, the incoming dCTP (indicated by the curved arrow) base pairs with the correct templating base G. (Right) In extension by misalignment, the C · C mispair realigns so that the template C becomes extrahelical and the primer-terminal C pairs with the next template base G. This is followed by the pairing of the incoming dTTP (indicated by the curved arrow) with the subsequent complementary template base.
Figure 3.
Steady-state kinetics of nucleotide incorporation by hPolκ on DNA substrate 1. The rate of dCTP (•) and dTTP (○) incorporation following a C · C primer-terminal mispair was graphed as a function of [dNTP]. The solid and broken lines represent the respective best fit to the Michaelis-Menten equation. The error bars represent the standard error of three experimental replicates. The steady-state parameters, kcat and Km, are listed in Table 2.
Figure 4.
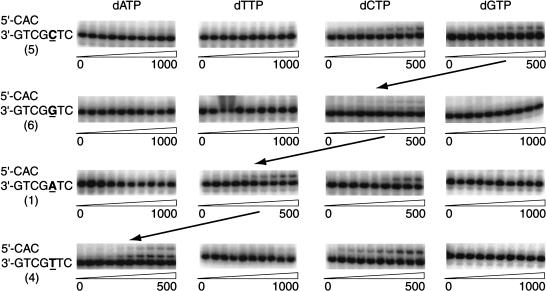
hPolκ-catalyzed extension of a primer-terminal mispair by direct extension and by misalignment. The extension of a primer-terminal mispair by misalignment is inferred from the sequence specificity of nucleotides incorporated with various DNA substrates in which the base 5′ to the templating residue (bold and underlined) is varied. Incorporation of dCTP with each DNA substrate reflects direct extension of the mispair, and incorporation of the dNTP complementary to the base 5′ of the templating residue (indicated by diagonal arrows) reflects misalignment. dNTP concentration, in micromoles per liter, is indicated below each panel. The numbers below each DNA sequence refer to the substrates listed in Table 1. The steady-state kinetic parameters kcat and Km are listed in Table 2.
Next, we examined the efficiency of hPolκ in extending C · C primer termini by template–primer misalignment and by direct extension using DNA substrates 4, 5, and 6, in which the base 5′ to the templating G was modified to a T, a C, or a G, respectively. As shown in Figure 4, with DNA substrate 4, hPolκ incorporates a C, which would occur by direct mispair extension, and also an A, incorporation of which would occur by template–primer misalignment, with efficiencies of 0.1 and 0.03, respectively (Table 2). Thus, in this case, direct mispair extension is approximately threefold more efficient than misalignment. On substrate 5, the nucleotides C and G are incorporated with the same relative efficiency (Fig. 4; Table 2), indicating that, in this sequence context, mispair extension by these two means occurs equally well. With substrate 6, both modes of mispair extension would cause the insertion of a C, as the two template bases 5′ to the mispair are Gs, and that is what is observed (Fig. 4; Table 2). In summary, for all four C · C mispair substrates 1, 4, 5, and 6, mismatch extension is accomplished either directly through the incorporation of dCTP, or by incorporation of the nucleotide complementary to the base 5′ to the templating base via template–primer misalignment, and the two modes of extension occur with nearly equal efficiencies.
We also examined the extension of a G · T mispair in DNA substrates with different sequence contexts (Table 3). With DNA substrate 7, only the incorporation of a T or a G nucleotide, which would occur by direct extension or by template–primer misalignment, respectively, was observed, and the efficiencies of extension by these two modes were nearly equivalent. On substrates 8 and 9, however, the extension of the G · T mispair by the direct incorporation of the next correct nucleotide was favored by ∼5- to 15-fold over template–primer misalignment. On substrate 10, direct extension was approximately twofold more efficient than misalignment.
Table 3.
Steady-state kinetic parameters for G-T mispair extension by Polκ on undamaged DNA
| DNA substrate | Sequence | dNTP | kcat (min-1) | Km (μM) | kcat/Km (μM-1 min-1) | frel |
|---|---|---|---|---|---|---|
| 7 | AGT | dTTP | 0.54 ± 0.009 | 190 ± 9 | 0.0028 | 0.96 |
| TCGACG | dGTP | 0.47 ± 0.005 | 177 ± 8 | 0.0027 | ||
| 8 | AGT | dTTP | 2.2 ± 0.05 | 57 ± 5 | 0.039 | 0.2 |
| TCGAGC | dCTP | 1.5 ± 0.09 | 193 ± 33 | 0.0077 | ||
| 9 | AGT | dTTP | 2.0 ± 0.04 | 43 ± 4 | 0.047 | 0.057 |
| TCGATC | dATP | 0.85 ± 0.05 | 314 ± 50 | 0.0027 | ||
| 10 | AGT | dTTP | 0.28 ± 0.02 | 17.9 ± 5 | 0.016 | 0.54 |
| TCGAGG | dCTP | 0.74 ± 0.02 | 85.3 ± 10 | 0.0087 |
(frel) The relative efficiency (kcat/Km) of misalignment to direct extension. No incorporation of the other two nucleotides was observed.
hPolκ extends from the nucleotide opposite an abasic site only by template–primer misalignment
hPolκ is unable to replicate through DNA lesions such as a cis–syn TT dimer, a (6–4) TT photoproduct, or an abasic site (Johnson et al. 2000a; Ohashi et al. 2000b). The inability of hPolκ to replicate through these DNA lesions is primarily due to its highly inefficient incorporation of nucleotides opposite them. hPolκ, however, is able to extend from a nucleotide inserted opposite the 3′T of the TT dimer by another DNA polymerase, and it does so by the incorporation of an A opposite the 5′T of the lesion (Washington et al. 2002). hPolκ is unable to carry out the nucleotide incorporation or the extension step opposite the (6–4) TT lesion (Washington et al. 2002). DNA synthesis by hPolκ stalls one nucleotide before an abasic site, indicating that this lesion, too, presents a block to nucleotide incorporation by this enzyme (Johnson et al. 2000a). hPolκ, however, can weakly incorporate an A opposite an abasic site, but it is more than 250-fold less efficient at it than it is at incorporating an A opposite an undamaged T (Haracska et al. 2002b).
Kinetic studies with DNA polymerase β have indicated that it skips an abasic site and incorporates the nucleotide that is complementary to the next 5′ template base (Efrati et al. 1997). In this dNTP-stabilized mode of frameshifting, the looped-out template abasic moiety is stabilized by the hydrogen bonding of the incoming dNTP bound in the polymerase active site to the complementary downstream base. Structural studies with S. solfataricus Dpo4 (Ling et al. 2001) and biochemical studies with E. coli Pol IV (Kobayashi et al. 2002) have also provided support for this means of frameshifting. To examine if hPolκ uses such a frameshifting mechanism in bypassing an abasic site, we used DNA substrates 11–13, wherein a looping out of the template abasic site followed by the incorporation of the nucleotide complementary to the next 5′ template base would result in the incorporation of a T, C, or G nucleotide, respectively (Table 4). With all these DNA substrates, hPolκ incorporated the nucleotides opposite the abasic site quite inefficiently; the A nucleotide, however, was incorporated preferentially over the others (Table 4). From these observations, we conclude that hPolκ does not use a dNTP-stabilized frameshifting mechanism at template abasic sites.
Table 4.
Steady-state kinetic parameters for nucleotide incorporation opposite an abasic site by Polκ
| DNA substrate | Sequence | dNTP | kcat (min-1) | Km (μM) | kcat/Km (μM-1 min-1) |
|---|---|---|---|---|---|
| 11 | GCA | dATP | 1.6 ± 0.057 | 235 ± 27 | 0.007 |
| CGTXAT | dTTP | 0.090 ± 0.0056 | 212 ± 55 | 0.0004 | |
| dCTP | 0.19 ± 0.024 | 409 ± 180 | 0.0005 | ||
| dGTP | 0.10 ± 0.0068 | 281 ± 62 | 0.0004 | ||
| 12 | GCA | dATP | 0.25 ± 0.017 | 269 ± 47 | 0.0009 |
| CGTXGT | dTTP | n.d. | n.d. | <0.0001 | |
| dCTP | n.d. | n.d. | <0.0001 | ||
| dGTP | 0.074 ± 0.005 | 321 ± 88 | 0.0002 | ||
| 13 | GCA | dATP | 0.44 ± 0.013 | 216 ± 17 | 0.002 |
| CGTXCT | dTTP | n.d. | n.d. | <0.0001 | |
| dCTP | 0.069 ± 0.0064 | 295 ± 96 | 0.0002 | ||
| dGTP | 0.046 ± 0.0040 | 296 ± 82 | 0.0002 |
(n.d.) No nucleotide incorporation was detected; (X) abasic site.
We have also examined the ability of hPolκ to carry out dNTP-stabilized misalignment on nondamaged DNA templates, using the DNA substrate shown in Figure 1A. This substrate has an A · T primer-terminal base pair, a C as the templating base, and a G as the next 5′ template base. In this case, normal nucleotide incorporation would result in a dGTP being inserted opposite the template C, and dNTP-stabilized misalignment would result in the enzyme skipping over the template C residue and incorporating a dCTP opposite the next 5′ template G residue. We found that only dGTP incorporation was detectible with this DNA substrate, and, given the detection limit of the assay, incorporation of the other dNTPs were at least 5000-fold lower than the efficiency (kcat/Km) of dGTP incorporation. We conclude from this that hPolκ does not use a dNTP-stabilized mechanism of misalignment on nondamaged DNA.
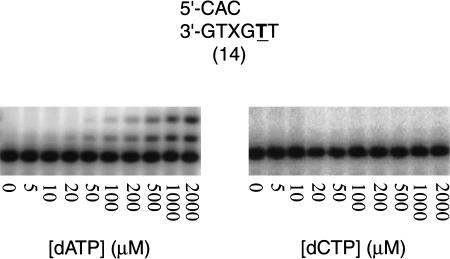
Next, we used DNA substrates 14–16 to examine the ability of hPolκ to extend from nucleotides placed opposite an abasic site (Table 5). With all these substrates, hPolκ extends by template–primer misalignment and not by direct extension. For instance, with DNA substrate 14, we observed only the incorporation of the A nucleotide (Fig. 5; Table 5), which would occur by the looping out of the abasic moiety followed by the pairing of primer-terminal C with the next 5′ template base G and the subsequent incorporation of an A opposite the next template base T. In contrast, the lack of any C incorporation indicates the absence of direct extension (Fig. 5; Table 5). With substrates 15 and 16 also, only the incorporation of A, which would occur by template–primer misalignment, was observed (Table 5). Because of the inability to distinguish between direct extension and misalignment in the sequence context of DNA substrates 14–16, we did not examine the kinetics of extension from an A placed at the primer terminus directly opposite the template abasic site. However, we have confirmed the lack of direct extension from an A opposite an abasic site by hPolκ using another sequence context.
Table 5.
Steady-state kinetic parameters for extension from nucleotides opposite an abasic site by Polκ
| DNA substrate | Sequence | dNTP | kcat (min-1) | Km (μM) | kcat/Km (μM-1 min-1) |
|---|---|---|---|---|---|
| 14 | CAC | dATP | 2.4 ± 0.052 | 313 ± 20 | 0.008 |
| GTXGTT | dCTP | n.d. | n.d. | <0.0001 | |
| 15 | CAG | dATP | 2.1 ± 0.044 | 168 ± 14 | 0.01 |
| GTXCTT | dGTP | n.d. | n.d. | <0.0001 | |
| 16 | CAT | dATP | 1.6 ± 0.057 | 235 ± 27 | 0.007 |
| GTXATT | dTTP | n.d. | n.d. | <0.0001 |
(n.d.) No nucleotide incorporation was detected. Also, no incorporation of the other two nucleotides was observed; (X) abasic site.
Figure 5.
hPolκ-catalyzed extension from nucleotides opposite an abasic site by misalignment. The dATP and dCTP concentrations were varied from 0 to 2000 μM. (Left) On DNA substrate 14, the incorporation of dATP opposite the downstream template T (bold and underlined) occurs via “looping out” of the abasic site and the subsequent pairing of the primer-terminal C with the next 5′ template G. (Right) The lack of direct extension is illustrated by the absence of any dCTP incorporation. The steady-state kinetic parameters, kcat and Km, are listed in Table 5.
Discussion
On undamaged DNA, hPolκ is quite adept at extending mispaired termini by the incorporation of the next correct nucleotide (Washington et al. 2002). In copying undamaged DNA, however, Polκ generates both base substitution and single-base deletion mutations (Ogi et al. 1999; Ohashi et al. 2000a). In this study, we have examined if hPolκ could extend mispaired primer termini by using a template–primer misalignment mechanism, wherein the template base at the primer terminus “loops out,” followed by the pairing of the primer-terminal base with the complementary 5′-template base and the incorporation of the nucleotide complementary to the next 5′-template base (see Fig. 2). Such a mechanism would then result in single-base deletions. Using C · C and G · T primer-terminal mispairs, we show here that such a template–primer misalignment mechanism, in fact, is operative during DNA synthesis by hPolκ, and in many of the sequence contexts, mispair extension by such a misalignment process is as frequent as that by the incorporation of the next correct nucleotide (direct extension).
Although most mismatches generated during normal DNA replication by Polδ would be removed by its proofreading 3′→5′ exonuclease, some of the mispaired termini could be refractory to this activity. In that case, the proficient ability of hPolκ to extend mispaired primer termini by direct extension or by misalignment could contribute to the continued and efficient progression of the replication fork. However, we expect that, in this role, Polκ would compete with Polζ in humans, as Polζ, too, is an avid extender of mispaired primer termini (Johnson et al. 2000b). Genetic studies in yeast have indicated a role for Polζ in the generation of base substitution and deletion mutations, as inactivation of REV3, which encodes the catalytic subunit of Polζ, decreases the rate of spontaneous single-base substitutions as well as single-base deletions by ∼60% (Roche et al. 1994; Kunz et al. 1998). These observations raise the possibility that Polζ, too, can extend mispaired primer termini by using the template–primer misalignment mechanism described here for Polκ, generating single-base deletions in the process. In that case, a mispaired primer terminus formed during normal DNA replication would be extended by Polκ or by Polζ, either directly or by misalignment.
Although hPolκ can extend a mispaired primer terminus on undamaged DNA either directly or by misalignment, it extends from a nucleotide placed opposite an abasic site only by misalignment, wherein the base opposite the abasic site pairs with the complementary 5′-template base, and this misalignment is followed by the incorporation of a nucleotide complementary to the next 5′-template base (see Table 5; Fig. 5). Although hPolκ can incorporate an A opposite an abasic site, it is quite inefficient in doing so, and therefore, we do not expect it to make a significant contribution to AP bypass by a process in which it first incorporates the nucleotide opposite the abasic site and then extends from the incorporated nucleotide either directly or by misalignment (Ohashi et al. 2000b). However, in sequence contexts in which template–primer misalignment is possible, hPolκ might contribute to AP bypass by extending from the nucleotide incorporated opposite the lesion site by another DNA polymerase, such as Polδ, Rev1, or Polη (Haracska et al. 2001). Nevertheless, because of the highly proficient ability of Polζ to directly extend from nucleotides opposite an abasic site (Haracska et al. 2001), we expect this polymerase to make a much greater contribution to its bypass than hPolκ.
The lack of direct extension from nucleotides opposite an abasic site by hPolκ contrasts with its proficient ability to extend directly from nucleotides placed opposite DNA lesions such as a cis–syn TT dimer, an O6-methylguanine, or an 8-oxoguanine lesion (Haracska et al. 2002a; Washington et al. 2002). Moreover, this extension is not predicated by the presence of a correct nucleotide opposite the lesion site. For example, hPolκ efficiently extends from a G nucleotide opposite the 3′T of the TT dimer, from a T opposite an O6-methylguanine lesion, or from an Aopposite an 8-oxoguanine lesion (Haracska et al. 2002a; Washington et al. 2002). This suggests that hPolκ can directly extend from a mispaired primer terminus irrespective of whether the template base at the primer terminus is damaged. From these observations, we infer that direct mispair extension by hPolκ requires the presence of a base opposite the primer-terminal residue, whereas, in the absence of the said template base, only the misalignment mechanism is possible.
Although hPolκ belongs to the DinB family, it differs from the other members of this family in its proficient ability to extend mispaired primer termini. For example, E. coli Pol IV misincorporates nucleotides with a frequency of ∼10–3 to 10–5, and it extends mispaired termini with about the same frequency (Kobayashi et al. 2002). We have found that the archaean Dpo4, too, is not an efficient extender of mismatched primer termini (W.T. Wolfle, R.E. Johnson, L. Prakash, and S. Prakash, unpubl.). hPolκ differs also from these other polymerases in its virtual inability to carry out the dNTP-stabilized mode of misalignment, and, as we show here, hPolκ can proficiently extend mispaired primer termini by using a different sort of template–primer misalignment mechanism.
The proficient ability of hPolκ to extend mispaired primer termini directly or by template–primer misalignment suggests that its active site is particularly less constrained at the template–primer junction so that it can tolerate the geometric distortions conferred by mismatched base pairs or those resulting from template–primer misalignment. In contrast to its proficient ability to extend mispaired primer termini, foext 10–1 to 10–2, hPolκ misincorporates nucleotides with a frequency of ∼10–3 to 10–4. hPolκ is also much less efficient at incorporating nucleotides opposite DNA lesions than at extending from nucleotides incorporated opposite the lesion site by another DNA polymerase (Haracska et al. 2002a; Washington et al. 2002). These properties of hPolκ would suggest that its active site is much more constrained at the site of the templating base and the incoming dNTP than at the primer terminus.
In summary, in its proficient ability to extend mispaired primer termini either directly or by misalignment, hPolκ has diverged significantly from its E. coli and archaean counterparts. Furthermore, it uses these two modes of mispair extension to different degrees, depending on whether the template base is present or absent at the primer terminus. These properties of hPolκ distinguish it also from the other two human Y family polymerases, Polη and Polι, as neither of them is a proficient extender of mispaired primer termini, and additionally, these polymerases differ in their damage bypass abilities. These Y-family polymerases have thus become highly specialized, able to perform different tasks in replication and in damage bypass.
Materials and methods
Nucleotides and DNA substrates
Solutions of each dNTP (100 mM) were purchased from Roche Diagnostics and stored in aliquots at –80°C. Synthetic oligode-oxynucleotides were used to prepare the DNA substrates listed in Table 1. Substrates 1–6 contain a primer-terminal C · C mispair, substrates 7–10 contain a primer-terminal G · T mispair, and substrates 11–16, contain a template strand abasic site. 32P-5′-end labeled primers (1 μM) were annealed to templates (1.5 μM) in 50 mM Tris · HCl (pH 7.5) and 100 mM NaCl by heating to 95°C for 2 min, followed by slow cooling to room temperature.
Protein expression and purification
Yeast strain BJ5464 was transformed with plasmid pPOL42, which carries the gene encoding wild-type hPolκ fused in frame with glutathione S-transferase. The protein was overexpressed and purified as described previously for Polη (Washington et al. 2001b). Cleavage by PreScission protease resulted in an additional 7 amino acid leader sequence attached to the full-length hPolκ protein. Purified hPolκ was stored in 5-μL aliquots at –80°C.
Steady-state kinetic measurements
The DNA polymerase assay contained 25 mM Tris HCl (pH 7.5), 5 mM dithiothreitol, 0.1 mg/mL bovine serum albumin, 5 mM MgCl2, 50 nM DNA, 1 nM Polκ, and various concentrations of dNTP in a range appropriate for Km determination. Values of the steady-state kinetic parameters kcat and Km for nucleotide incorporation for all substrates, except 14–16, were determined as follows: The reactions were carried out for 10 min at 24°C and were quenched with the addition of four volumes of ice-cold 95% formamide loading dye and placed on ice. Product formation was monitored using 10% polyacrylamide gel electrophoresis (8 M urea) and the respective gel band intensities were quantified using the PhosphorImager (Molecular Dynamics). The mean and standard error values for the rate of nucleotide incorporation at each nucleotide concentration were obtained from a set of three independent experiments and were used to determine the kcat and Km parameters from the best fit of the data to the Michaelis-Menten equation (Sigma Plot 7.0). For substrates 14–16, deoxynucleotide incorporation was measured at multiple time points for each nucleotide concentration, and the observed rate of nucleotide incorporation was determined by linear regression and used to determine kcat and Km. The efficiency of nucleotide incorporation (kcat/Km) was determined and used to calculate the efficiency of misalignment relative to direct extension (frel).
Acknowledgments
This work was supported by National Institutes of Health Grant GM19261.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.1108603.
References
- Bebenek K. and Kusnkel, T.A. 1990. Frameshift errors initiated by nucleotide misincorporation. Proc. Nat. Acad. Sci. 87: 4946–4950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Efrati E., Tucco, G., Eritja, R., Wilson, S.H., and Goodman, M.F. 1997. Abasic translesion synthesis by DNA polymerase β violates the “A-rule”: Novel types of nucleotide incorporation by human DNA polymerase β at an abasic lesion in different sequence contexts. J. Biol. Chem. 272: 2559–2569. [DOI] [PubMed] [Google Scholar]
- Goodman M.F., Creighton, S., Bloom, L.B., and Petruska, J. 1993. Biochemical basis of DNA replication fidelity. Crit. Rev. Biochem. Mol. Biol. 28: 83–126. [DOI] [PubMed] [Google Scholar]
- Haracska L., Prakash, S., and Prakash, L. 2000a. Replication past O6-methylguanine by yeast and human DNA polymerase η. Mol. Cell. Biol. 20: 8001–8007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haracska L., Yu, S.-L., Johnson, R.E., Prakash, L., and Prakash, S. 2000b. Efficient and accurate replication in the presence of 7,8-dihydro-8-oxoguanine by DNA polymerase η. Nat. Genet. 25: 458–461. [DOI] [PubMed] [Google Scholar]
- Haracska L., Unk, I., Johnson, R.E., Johansson, E., Burgers, P.M.J., Prakash, S., and Prakash, L. 2001. Roles of yeast DNA polymerases δ and ζ and of Rev1 in the bypass of abasic sites. Genes & Dev. 15: 945–954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haracska L., Prakash, L., and Prakash, S. 2002a. Role of human DNA polymerase κ as an extender in translesion synthesis. Proc. Nat. Acad. Sci. 99: 16000–16005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haracska L., Unk, I., Johnson, R.E., Phillips, B.B., Hurwitz, J., Prakash, L., and Prakash, S. 2002b. Stimulation of DNA synthesis activity of human DNA polymerase κ by PCNA. Mol. Cell. Biol. 22: 784–791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson R.E., Prakash, S., and Prakash, L. 1999. Efficient bypass of a thymine–thymine dimer by yeast DNA polymerase, Polη. Science 283: 1001–1004. [DOI] [PubMed] [Google Scholar]
- ____. 2000a. The human DINB1 gene encodes the DNA polymerase Polθ. Proc. Nat. Acad. Sci. 97: 3838–3843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson R.E., Washington, M.T., Haracska, L., Prakash, S., and Prakash, L. 2000b. Eukaryotic polymerases ι and ζ act sequentially to bypass DNA lesions. Nature 406: 1015–1019. [DOI] [PubMed] [Google Scholar]
- Johnson R.E., Washington, M.T., Prakash, S., and Prakash, L. 2000c. Fidelity of human DNA polymerase η. J. Biol. Chem. 275: 7447–7450. [DOI] [PubMed] [Google Scholar]
- Kim S.-R., Maenhaut-Michel, G., Yamada, M., Yamamoto, Y., Matsui, K., Sofuni, T., Nohmi, T., and Ohmori, H. 1997. Multiple pathways for SOS-induced mutagenesis in Escherichia coli: An overexpression of dinB/dinP results in strongly enhancing mutagenesis in the absence of any exogenous treatment to damage DNA. Proc. Nat. Acad. Sci. 94: 13792–13797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi S., Valentine, M.R., Pham, P., O'Donnell, M., and Goodman, M.F. 2002. Fidelity of Escherichia coli DNA polymerase IV: Preferential generation of small deletion mutations by dNTP-stabilized misalignment. J. Biol. Chem. 277: 34198–34207. [DOI] [PubMed] [Google Scholar]
- Kokoska R.J., Bebeneck, K., Boudsocq, F., Woodgate, R., and Kunkel, T.A. 2002. Low fidelity DNA synthesis by a Y family DNA polymerase due to misalignment in the active site. J. Biol. Chem. 277: 19633–19638. [DOI] [PubMed] [Google Scholar]
- Kunkel T.A. and Soni, A. 1988. Mutagenesis by transient misalignment. J. Biol. Chem. 263: 14784–14789. [PubMed] [Google Scholar]
- Kunz B.A., Ramachandran, K., and Vonarx, E.J. 1998. DNA sequence analysis of spontaneous mutagenesis in Saccharomyces cerevisiae. Genetics 148: 1491–1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling H., Boudsocq, F., Woodgate, R., and Yang, W. 2001. Crystal structure of a Y-family DNA polymerase in action: A mechanism for error-prone and lesion-bypass replication. Cell 107: 91–102. [DOI] [PubMed] [Google Scholar]
- Masutani C., Kusumoto, R., Yamada, A., Dohmae, N., Yokoi, M., Yuasa, M., Araki, M., Iwai, S., Takio, K., and Hanaoka, F. 1999. The XPV (xeroderma pigmentosum variant) gene encodes human DNA polymerase η. Nature 399: 700–704. [DOI] [PubMed] [Google Scholar]
- Mendelman L.V., Petruska, J., and Goodman, M.F. 1990. Base mispair extension kinetics. Comparison of DNA polymerase α and reverse transcriptase. J. Biol. Chem. 265: 2338–2346. [PubMed] [Google Scholar]
- Ogi T., Kato Jr., T., Kato, T., and Ohmori, H. 1999. Mutation enhancement by DINB1, a mammalian homologue of the Escherichia coli mutagenesis protein DinB. Genes Cells 4: 607–618. [DOI] [PubMed] [Google Scholar]
- Ohashi E., Bebenek, K., Matsuda, T., Feaver, W.J., Gerlach, V.L., Friedberg, E.C., Ohmori, H., and Kunkel, T.A. 2000a. Fidelity and processivity of DNA synthesis by DNA polymerase κ, the product of the human DINB1 gene. J. Biol. Chem. 275: 39678–39684. [DOI] [PubMed] [Google Scholar]
- Ohashi E., Ogi, T., Kusumoto, R., Iwai, S., Masutani, C., Hanaoka, F., and Ohmori, H. 2000b. Error-prone bypass of certain DNA lesions by the human DNA polymerase κ. Genes & Dev. 14: 1589–1594. [PMC free article] [PubMed] [Google Scholar]
- Prakash S. and Prakash, L. 2002. Translesion DNA synthesis in eukaryotes: A one- or two-polymerase affair. Genes & Dev. 16: 1872–1883. [DOI] [PubMed] [Google Scholar]
- Roche H., Gietz, R.D., and Kunz, B.A. 1994. Specificity of the yeast rev3Δ antimutator and REV3 dependency of the mutator resulting from a defect (rad1Δ) in nucleotide excision repair. Genetics 137: 637–646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Streisinger G., Okada, Y., Emrich, J., Newton, J., Tsugita, A., Terzaghi, E., and Inouye, M. 1966. Frameshift mutations and the genetic code. Cold Spring Harbor Symp. Quant. Biol. 31: 77–84. [DOI] [PubMed] [Google Scholar]
- Vaisman A., Tissier, A., Frank, E.G., Goodman, M.F., and Woodgate, R. 2001. Human DNA polymerase ι promiscuous mismatch extension. J. Biol. Chem. 276: 30615–30622. [DOI] [PubMed] [Google Scholar]
- Washington M.T., Johnson, R.E., Prakash, S., and Prakash, L. 2000. Accuracy of thymine–thymine dimer bypass by Saccharomyces cerevisiae DNA polymerase η. Proc. Nat. Acad. Sci. 97: 3094–3099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ____. 2001a. Mismatch extension ability of yeast and human DNA polymerase η. J. Biol. Chem. 276: 2263–2266. [DOI] [PubMed] [Google Scholar]
- Washington M.T., Prakash, L., and Prakash, S. 2001b. Yeast DNA polymerase η utilizes an induced fit mechanism of nucleotide incorporation. Cell 107: 917–927. [DOI] [PubMed] [Google Scholar]
- Washington M.T., Johnson, R.E., Prakash, L., and Prakash, S. 2002. Human DINB1-encoded DNA polymerase κ is a promiscuous extender of mispaired primer termini. Proc. Nat. Acad. Sci. 99: 1910–1914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu S.-L., Johnson, R.E., Prakash, S., and Prakash, L. 2001. Requirement of DNA polymerase η for error-free bypass of UV-induced CC and TC photoproducts. Mol. Cell. Biol. 21: 185–188. [DOI] [PMC free article] [PubMed] [Google Scholar]