Abstract
The minimal set of genetic information necessary and sufficient to sustain a functioning cell contains not only trans-acting genes, but also cis-acting chromosomal regions that cannot be complemented by plasmids carrying these regions. In Escherichia coli (E. coli), only one chromosomal region, the origin of replication has been identified to be cis-acting. We constructed a series of mutants with long-range deletions, and the chromosomal regions containing trans-acting essential genes were deleted in the presence of plasmids complementing the deleted genes. The deleted regions cover all regions of the chromosome except for the origin and terminus of replication. The terminus affects cell growth, but is not essential. Our results indicate that the origin of DNA replication is the only vital, unique cis-acting DNA sequence in the E. coli chromosome necessary for survival.
Keywords: chromosome deletions, Escherichia coli, essential genes, FLP recombinase, red
Introduction
The experimental identification of essential genes has been carried out in some bacteria and yeast (Akerley et al, 2002; Forsyth et al, 2002; Giaever et al, 2002; Gerdes et al, 2003; Kobayashi et al, 2003; Baba et al, 2006; Glass et al, 2006). The methods often used to disrupt genes to determine whether they are essential or not included transposon mutagenesis and targeted disruption by homologous recombination. Using transposon mutagenesis, whole regions of chromosomes can be examined; however, the results are inconclusive, because not all regions are inactivated by random insertion. Targeted disruption, which can identify essential genes expressed in the diploid stage or expressed conditionally, is a suitable method to show whether annotated genes are essential. But intergenic regions have not been investigated. Although most essential trans-acting genes have been identified through gene disruption studies, the necessity of the intergenic regions has not been sufficiently clarified. For example, some genetic information in the intergenic regions is transcribed, whereas other genetic information is not. The former sites are trans-acting, and the latter sites are cis-acting.
In bacteria, chromosomes are generally uni-replicons; therefore, the origin of replication (oriC) is cis-acting and essential. In Escherichia coli, other cis-acting sites have also been reported. The dif is a cis-acting site, which is important for cell proliferation (Sherratt, 2003). This site was identified because a mutant with a very large deletion around the replication terminus (terC) grew slowly, and the chromosomal region responsible for this growth defect was identified and termed deletion-induced filamentation or dif (Kuempel et al, 1991). This dif site was eventually shown to be a site for recombination catalyzed by the XerC-XerD recombinase, which resolves chromosome multimers resulting from homologous recombination between replicated sister chromosomes. Although this site affects cell growth, its deletion leads to a relatively minor growth defect (Cui et al, 2007). Another cis-acting site, migS, was identified as being responsible for the polar movement of oriC, but this site was not essential for cell growth (Yamaichi and Niki, 2004).
To understand the essential genetic information of prokaryotic chromosomes, a genomic survey of cis-acting essential regions is necessary. An efficient way to identify essential factors, particularly cis-acting chromosome regions, is thought to be the systematic construction of large-scale chromosomal deletions. If unique and essential cis-acting regions are on a chromosome, the deletion mutants of these regions are no longer viable even in the presence of complementing plasmids. Previously, we constructed long-range deletions of the E. coli chromosome, which led to the reduction of the genome size (Hashimoto et al, 2005; Kato and Hashimoto (in press)). First, we constructed 75 deletions (medium-scale deletions (MD)) in regions lacking the essential genes, which were identified through a survey of the published literature, using the E. coli homologous recombination system. We then constructed a second series of deletions (large-scale deletions (LD)) and combined them to construct an engineered strain lacking 29.7% of the parental chromosome. In this study, we constructed deletion mutants for other chromosomal regions, particularly those containing essential genes, to identify additional essential cis-acting chromosome regions, while maintaining the viability of the mutants with complementing plasmids expressing the deleted genes.
Results and discussion
First, for the chromosomal regions that were not deleted during the construction of the first MD series, the identified essential genes were cloned and the chromosomal regions containing these genes were deleted. We cloned the essential genes into mini-F plasmid vectors (Supplementary Figure 1 and Supplementary Information) either in vitro using restriction digestion and ligation methods or in vivo using the red recombination system (Supplementary Figure 2 and Supplementary Table I). Seven and 34 MDs were obtained in the absence and presence of the complementing plasmids, respectively. We also constructed one new LD. However, we did not succeed in constructing MD and LD deletions for the entire chromosome. Second, we investigated whether or not there were any essential cis-acting chromosome regions in the regions not deleted in the MD and LD. Therefore, we developed a system of moving chromosome regions into mini-F plasmids in vivo using the yeast FLP-FRT site-specific recombination system 1 (FLP-FRT1) (see Kato and Hashimoto, in press, for details). Figure 1 shows the improved system 2 (FLP-FRT2), which is essentially the same as FLP-FRT1. Using this system, 30 additional chromosome deletions were constructed, indicating that these regions have no essential cis-acting chromosome regions. Third, for these regions and the other regions that were not deleted, small-scale deletions (SD) were constructed using lambda (λ)-phage and the red recombination system (see Kato and Hashimoto, in press, for details.). Two hundred thirty-eight and 116 small-scale deletions (SD) were obtained using these methods in the absence and presence of complementing plasmids, respectively. Fourth, we tried to construct the deletions again for the other undeleted regions using improved systems. We obtained 5 and 15 deletions with system 2 (FLP-FRT2) and system 3 (FLP-FRT3), respectively. In total, 551 chromosome deletions mutations covering all of the E. coli genome, except for oriC and terC were constructed (Figures 2 and 3 and Supplementary Table I; see the profiling of E. coli chromosome (PEC) database (http://www.shigen.nig.ac.jp/ecoli/pec/index.jsp) for details).
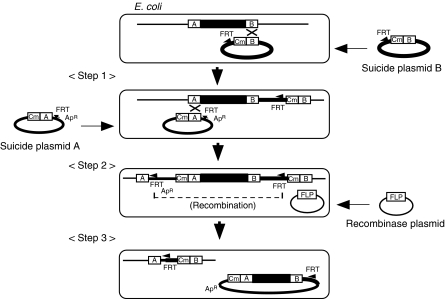
Figure 1.
The FRT2 system. A schematic drawing of the transfer of a chromosomal region to a mini-F plasmid with the FRT2 system. The chromosomal region to be deleted is represented by the bold line. The two chromosomal regions (A and B) flanking the region to be deleted are cloned into two kinds of suicide vectors. One is a mini-F plasmid, which has an FRT site and is replication defective at 42°C. The other is a R6K derivative, which has an FRT site and is replication defective due to the absence the pir gene necessary for replication. In step 1, the suicide plasmid carrying B is introduced into an E. coli strain. The chloramphenicol-resistant (CmR) colonies are isolated, representing step 1 recombinants in which the plasmid is integrated into the chromosome by homologous recombination. Next, the other suicide plasmid carrying A is introduced into step 1 recombinants, and the ampicillin-resistant (ApR) transformants are obtained at 42°C, representing step 2 recombinants. To inhibit homologous recombination beyond this stage, recA is disrupted by P1 transduction. In step 3, the FLP-plasmid, which is replication defective at 35°C, is introduced into step 2 recombinants, and the expression of the FLP recombinase is induced, resulting in simultaneous plasmid excision and chromosome deletion. To obtain a strain that did not carry the FLP-plasmid, cells were incubated at 35°C, at which point the FLP-plasmid does not replicate, but the miniF ts replicon remains functional.
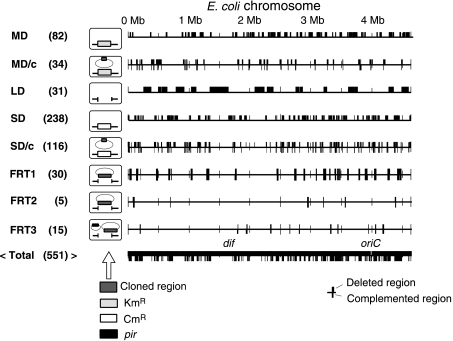
Figure 2.
Summary of the E. coli chromosome deletions. Deleted and complemented regions are drawn using a linear chromosome map. Horizontal lines represent chromosomes, and upper boxes and lower boxes indicate deleted and complemented regions, respectively. MD, LD, SD, and FRT represent the systems used to construct the deletions and c indicates the presence of a complementing plasmid. Numbers in parentheses indicate the number of deletion mutations constructed using each system. Depictions of cells containing deletion mutations are provided to show markers inserted into the deleted chromosome regions and plasmids. For details and methods, see Supplementary information; Hashimoto et al, 2005, Kato and Hashimoto, in press.
Figure 3.
E. coli chromosome deletions. Deleted and complemented regions of each deletion mutant are shown using a linear chromosome map. Thick black horizontal lines indicate the chromosomes. Thin colored lines show the deleted and complemented regions. These colors are different due to the types of the deletion mutants and are indicated at the bottom of the Figure. The names of MD and LD deletions are shown below the thin lines. The gene names are shown above the line as a reference for the map locations. The black and red gene names indicate genes that are deleted and complemented, respectively. Details of deletions are shown in the profiling of E. coli chromosome (PEC) database (http://www.shigen.nig.ac.jp/ecoli/pec/index.jsp).
The terC region does not contain a site that affects cell growth, other than dif (Kuempel et al, 1991); therefore, the results of this work indicate that there are no unique, cis-acting, and essential regions other than oriC. Eukaryotic chromosomes are multireplicons, and thus each origin of replication is not necessarily essential. Apart from the origin of replication, other cis-acting chromosome regions in eukaryotic cells include telomeric sequences, which are necessary for chromosome maintenance and centromeric regions, which are required for stable segregation of eukaryotic chromosomes. The centromere is a unique region in each chromosome: in theory, two centromeres on one chromosome can pull apart the chromosomal DNA between two daughter cells during mitosis (Mann and Davis, 1983). In prokaryotic cells, the mechanism underlying bacterial chromosome segregation is not understood. So far, a prokaryotic centromere has not been identified and it is not known if one exists. Low-copy number bacterial plasmids have their own partition systems, in which a cis-acting DNA region plays an essential role (Hayes and Barilla, 2006). But unlike eukaryotic centromeres, a plasmid carrying two copies of the cis-acting sequence is structurally stable. It is not known whether a eukaryote-like centromere functions in chromosome segregation in prokaryotes. Here, we did not identify any cis-acting and essential sites other than oriC. Because oriC and dif regions are not thought to contain a site for chromosome stability (Kogoma and Meyenburg, 1983; Ogura and Hiraga, 1983; Tecklenburg et al, 1995), our results suggest that a potential cis-acting site for chromosome segregation may be dispensable or redundant. Alternatively, prokaryotic cell sequences equivalent to the eukaryotic centromere may not exist. Global reorganization of chromosomes triggered by a loss of this cohesion resembles eukaryotic prometaphase (Sunako et al, 2001; Bates and Kleckner, 2005). It is suggested that the bacterial mechanism of chromosome segregation is a primordial one to which microtubule-based processes were added later.
Our results also show that all of the trans-acting essential genes were cloned on the complementing plasmids; however, the cloned genes (501 genes) are not necessarily essential. Baba et al (2006) reported 303 trans-acting essential genes by targeted disruption, but 35 of them were not cloned on our complementing plasmids, indicating that these genes are nonessential (‘Class A' in the Supplementary Table II). The discrepancy between the two studies may be due to a difference in the strains and culture conditions used. For example, the culture media Antibiotic medium 3 (this work) contains a glucose, but LB (Baba et al, 2006) does not. Alternatively, it may be ascribed to the difference between a single gene knockout (Baba et al, 2006) and a large-scale deletion of genes (this work). For example, the anti-toxin genes yefM and chpR, which were identified as essential genes of the toxin-antitoxin system (Baba et al, 2006), were deleted with the toxin genes yoeB and mazF, respectively, in our study. In addition, the dicA gene encoding a repressor of a cell division inhibitor was deleted in our study with the dicB, the inhibitor gene, whereas the dicA gene disruptant was not obtained in a previous study (Baba et al, 2006). Furthermore, we identified 25 genes (‘Class B') and 15 genes encoding small RNA (‘Class C') as essential genes and 2 genes as nonessential genes, which were determined from the results of our gene disruption experiments (J Kato, unpublished data, 2006) and other reports (We listed the relevant PMID number in the Supplementary Table II). In total, we identified 303 essential genes (Supplementary Table II). Sequence comparison of these essential genes with those of Bacillus subtilis (271 genes) revealed that 177 were conserved between these two genera (Supplementary Table II; Kobayashi et al, 2003). When functionally classified, genes involved in translation, protein translocation, and lipid synthesis were well conserved, whereas those involved in cell wall and membrane synthesis were not (Supplementary Table II), which may reflect structural differences in the cell wall and membrane.
In our study, 50 chromosome regions were moved to a mini-F plasmid using the FLP-FRT systems. Forty-six of them were found to contain essential gene(s), whereas 2 regions had no essential genes and were deleted without complementing plasmids. The other two regions (OCL30 and OCL34) did not contain any essential genes, but these regions were essential and therefore were not deleted without the complementing plasmids. In these regions, there may be the functionally redundant genes; one of which may be at least essential. Thirty chromosome regions were first moved to a mini-F plasmid using system 1 (FLP-FRT1). When we tried to move the other 20 regions to the plasmids using the improved system 2, (FLP-FRT2), 5 regions were moved, but the other 15 regions were not. For the regions affected by these deletions, the chromosomes and plasmids of the strains obtained at each step of system 2 (FLP-FRT2) were analyzed. The results indicated that the chromosomal regions flanked by two FRT sites had been excised after induction of the FLP recombinase, but the resultant plasmids were not stably maintained even in the wild-type strain using the mini-F temperature-sensitive replicon. Analyses of some of the excised plasmids suggested a rearrangement of the plasmid structure (data not shown). Unexpected recombination between the cloned chromosomal regions and a mini-F plasmid may cause instability of the excised plasmids. Therefore, we developed an improved system, termed system 3 (FLP-FRT3), in which the excised chromosome region was maintained by an R6K replicon. This finally allowed us to construct the remaining 15 deletion mutations.
Developments in synthetic biology have made it possible to reduce the size of the genome of E. coli K-12 (Kolisnychenko et al, 2002; Yu et al, 2002; Hashimoto et al, 2005; Posfai et al, 2006), and recent work indicates that genome reduction can have unanticipated benefits (Posfai et al, 2006). To further engineer E. coli and to make useful improvements for industry and therapeutics, such as facilitating the production of metabolites and proteins, it is important to understand both the cis- and trans-acting essential genetic information. Further analyses are necessary to experimentally clarify the minimal set of genetic information necessary and sufficient to sustain a functioning cell.
Materials and methods
Strains and media
All E. coli strains used are derivatives of MG1655. The MD series was constructed in MG1655 rpsL polA12. The LD and SD deletion series were built using MG1655 rsh3 (red:tet (Δ(recC ptr recB recD)∷Plac-red) rpsL hsdR:Ap), which was constructed using KM22 (Murphy, 1998). MG1655 rpsL was used to combine LD deletion units. Antibiotic Medium 3 (Becton Dickinson, USA) was used for all experiments except for those involving sacB selection, for which LB (-N) Suc was used (LB broth with 10% sucrose and lacking NaCl). The approximate formula per liter of the Antibiotic Medium 3 is beef extract 1.5 g, yeast extract 1.5 g, peptone 5.0 g, dextrose 1.0 g, sodium chloride 3.5 g, dipotassium phosphate 3.68 g, and monopotassium phosphate 1.32 g.
Construction of the complementing plasmids
Complementing plasmids were constructed in vitro or in vivo. In vitro, chromosome regions were amplified by PCR using primers flanked by restriction sites, digested with restriction enzymes, and ligated into mini-F vectors (Supplementary Figure 1 and Supplementary Information) with T4 DNA ligase. In vivo (Supplementary Figure 2), DNA fragments to be cloned were prepared and flanked by two DNA fragments, ‘KmN' and ‘mF', by two successive PCR reactions and introduced into the E. coli strain with the red gene of λ-phage and a mini-F vector, mFCm4-2. Introduced fragments were cloned into the mini-F vector by red recombination, resulting in kanamycin-resistant (KmR) and chloramphenicol-sensitive (CmS) complementing plasmids.
MD mutant construction
The MD series was constructed with the E. coli homologous recombination system using ColE1-related plasmids and the polA mutant (Hashimoto et al, 2005; Kato and Hashimoto, in press). The vector 664BSCK2-4 has two positive selection markers (CmR and KmR), two negative selection markers (rpsL+ streptomycin-sensitive (SmS) and sacB+), and multicloning sites flanking the KmR marker. Both chromosomal regions flanking the targeted region were cloned into 664BSCK2-4, and the resulting plasmid introduced into MG1655 rpsL polA12. A CmR transformant, in which the plasmid was integrated by homologous recombination between the cloned region and the same region on the chromosome, was selected at 42°C. After incubation at 30°C, a SmR KmR CmS colony, in which the plasmid was excised by another homologous recombination between the other cloned chromosomal region and the same region on the chromosome, was isolated and deletions were confirmed by PCR.
SD mutant construction
The SD system has been described previously (Kato and Hashimoto, in press). Briefly, a linear DNA fragment encoding the CmR gene was generated by PCR using oligonucleotide primers with a 40-base-pair region of homology to regions flanking the targeted deletion. The frequency of recombination was low using primers containing a 40-base-pair region of homology, but improved upon attachment of an approximately 1-kb region of homology to either end of the CmR gene. Fragments were introduced into the E. coli strain MG1655 red by electroporation and CmR recombinants were isolated. Deletions were confirmed by PCR analysis.
LD mutant construction
The LD series was constructed by the ‘CRS cassette method' using the red gene-mediated λ-phage homologous recombination system and linear DNA fragments (Hashimoto et al, 2005; Kato and Hashimoto, in press). The CRS cassette is approximately 5 kb and bears one positive selection marker, CmR, and two negative selection markers, rpsL+ (SmS) and sacB+. A DNA fragment in which chromosomal regions flanking the region to be deleted were joined to the ends of the CRS cassette was introduced into MG1655 rsh3. CmR colonies were selected and deletions confirmed by PCR. To remove the CRS cassette, a DNA fragment in which the same flanking chromosomal regions were directly joined to each other was introduced into CmR colonies. SmR and sucrose-resistant colonies were selected and deletions confirmed by PCR.
FLP-FRT mutant construction
The FLP-FRT2 system is shown in Figure 1 (for details, see Kato and Hashimoto, in press). In our FLP-FRT 1 prototype system, A-Km and B-Km DNA fragments, which contain the KmR gene joined to two (A and B) chromosomal regions flanking the region to be deleted, were prepared by PCR using 664BSCK2-4 derivative plasmids used to construct MD deletions described above. The A-Km DNA fragment was inserted into a mini-F plasmid, mini-FtsFA (suicide plasmid A with a CmR marker), which is replication-defective at 42°C, and the B-Km DNA fragment was inserted into a R6K-related plasmid, pSG76SA (suicide plasmid B with an ApR marker), which lacks pir necessary for replication (Posfai et al, 1994; Kato and Hashimoto, in press). First, pSG76SA carrying B-Km was introduced into the wild-type strain MG1655 and the KmR recombinants in which the plasmid was integrated by homologous recombination were isolated. Second, the plasmid mini-FtsFA carrying A-Km was introduced into KmR colonies obtained as described, and CmR colonies were obtained. Third, recA was disrupted by P1 transduction to inhibit homologous recombination beyond this stage. The FLP-containing plasmid (recombinase plasmid) was introduced and recombinase expression was induced, resulting in plasmid excision and chromosome deletion. To obtain a strain lacking the FLP-plasmid, cells were incubated at 35°C, at which the FLP-plasmid does not replicate but the miniF ts replicon remains functional.
The FRT2 system introduces improvements to the FRT1 system (Kato and Hashimoto, in press). Briefly, the two chromosomal regions (A and B) flanking the targeted region were joined to the CmR gene to create A-Cm and B-Cm by PCR. A-Cm was inserted into mini-FtsFAK, and B-Cm into pSG76SA. pSG76SA carrying B-Cm was introduced into the wild-type strain MG1655. CmR colonies in which the plasmid was integrated were isolated. Next, mini-FtsFAK carrying A-Cm was introduced into the CmR recombinants, and the ampicillin-resistant (ApR) recombinants were isolated at 42°C. To inhibit homologous recombination beyond this stage, recA was disrupted by P1 transduction. The FLP-plasmid was introduced into CmR ApR recombinants and plasmid excision and chromosome deletion was induced. In the FRT3 system, the plasmid 184 Km pir, encoding a functional copy of pir, was co-introduced with the FLP-plasmid, and the excised plasmid was maintained with the R6K replicon in addition to the miniF ts replicon (Kato and Hashimoto, in press). In all other aspects, the FRT3 system was the same as the FRT2 system.
Supplementary Material
Supplementary Figures
Supplementary Table 1
Supplementary Table 2
Supplementary Information
Acknowledgments
We thank Dr Yukiko Yamazaki for PEC. We are also grateful to A Takahashi, H China, S Fukuda, S Ito, S Koiwai, M Tanimoto and K Kin for technical assistance. This work was supported by KAKENHI (Grant-in-Aid for Scientific Research) on Priority Areas ‘Systems Genomics' from the Ministry of Education, Culture, Sports, Science and Technology of Japan.
References
- Akerley BJ, Rubin EJ, Novick VL, Amaya K, Judson N, Mekalanos JJ (2002) A genome-scale analysis for identification of genes required for growth or survival of Haemophilus influenzae. Proc Natl Acad Sci USA 99: 966–971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baba T, Ara T, Hasegawa M, Takai Y, Okumura Y, Baba M, Datsenko KA, Tomita M, Wanner BL, Mori H (2006) Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol Syst Biol 2: 2006.0008 10.1038/msb4100050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bates D, Kleckner N (2005) Chromosome and replisome dynamics in E.coli: loss of sister cohesion triggers global chromosome movement and mediates chromosome segregation. Cell 121: 899–911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui T, Moro-oka N, Ohsumi K, Kodama K, Ohshima T, Ogasawara N, Mori H, Wanner B, Niki H, Horiuchi T (2007) Escherichia coli with a linear genome. EMBO Rep 8: 181–187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forsyth RA, Haselbeck RJ, Ohlsen KL, Yamamoto RT, Xu H, Trawick JD, Wall D, Wang L, Brown-Driver V, Froelich JM, C KG, King P, McCarthy M, Malone C, Misiner B, Robbins D, Tan Z, Zhu Zy ZY, Carr G, Mosca DA et al. (2002) A genome-wide strategy for the identification of essential genes in Staphylococcus aureus. Mol Microbiol 43: 1387–1400 [DOI] [PubMed] [Google Scholar]
- Gerdes SY, Scholle MD, Campbell JW, Balázsi G, Ravasz E, Daugherty MD, Somera AL, Kyrpides NC, Anderson I, Gelfand MS, Bhattacharya A, Kapatral V, D'Souza M, Baev MV, Grechkin Y, Mseeh F, Fonstein MY, Overbeek R, Barabási AL, Oltvai ZN et al. (2003) Experimental determination and system level analysis of essential genes in Escherichia coli MG1655. J Bacteriol 185: 5673–5684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giaever G, Chu AM, Ni L, Connelly C, Riles L, Véronneau S, Dow S, Lucau-Danila A, Anderson K, André B, Arkin AP, Astromoff A, El-Bakkoury M, Bangham R, Benito R, Brachat S, Campanaro S, Curtiss M, Davis K, Deutschbauer A et al. (2002) Functional profiling of the Saccharomyces cerevisiae genome. Nature 418: 387–391 [DOI] [PubMed] [Google Scholar]
- Glass JI, Assad-Garcia N, Alperovich N, Yooseph S, Lewis MR, Maruf M, Hutchison CA III, Smith HO, Craig V (2006) Essential genes of a minimal bacterium. Proc Natl Acad Sci USA 103: 425–430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto M, Ichimura T, Mizoguchi H, Tanaka K, Fujimitsu K, Keyamura K, Ote T, Yamakawa T, Yamazaki Y, Mori H, Katayama T, Kato J (2005) Cell size and nucleoid organization of engineered Escherichia coli cells with a reduced genome. Mol Microbiol 55: 137–149 [DOI] [PubMed] [Google Scholar]
- Hayes F, Barilla D (2006) Assembling the bacterial segrosome. Trends Biochem Sci 31: 247–250 [DOI] [PubMed] [Google Scholar]
- Kato J, Hashimoto M. Construction of long chromosomal deletion mutants of Escherichia coli and minimization of the genome. In: Methods in Molecular Biology (Gene Essentiality at Genome Scale: Protocols and Bioinformatics), Osterman A (ed) Humana Press: NJ, USA in press [DOI] [PubMed] [Google Scholar]
- Kobayashi K, Ehrlich SD, Albertini A, Amati G, Andersen KK, Arnaud M, Asai K, Ashikaga S, Aymerich S, Bessieres P, Boland F, Brignell SC, Bron S, Bunai K, Chapuis J, Christiansen LC, Danchin A, Débarbouille M, Dervyn E, Deuerling E et al. (2003) Essential Bacillus subtilis genes. Proc Natl Acad Sci USA 100: 4678–4683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kogoma T, Meyenburg K (1983) The origin of replication, oriC, and the dnaA protein are dispensable in stable DNA replication (sdrA) mutants of Escherichia coli K-12. EMBO J 2: 463–468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolisnychenko V, Plunkett G III, Herring CD, Feher T, Posfai J, Blattner FR, Posfai G (2002) Engineering a reduced Escherichia coli genome. Genome Res 12: 640–647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuempel PL, Henson JM, Dircks L, Tecklenburg M, Lim DF (1991) dif, a recA-independent recombination site in the terminus region of the chromosome of Escherichia coli. New Biol 3: 799–811 [PubMed] [Google Scholar]
- Mann C, Davis RW (1983) Instability of dicentric plasmids in yeast. Proc Natl Acad Sci USA 80: 228–232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy KC (1998) Use of bacteriophage λ recombination functions to promote gene replacement in Escherichia coli. J Bacteriol 180: 2063–2071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogura T, Hiraga S (1983) Partition mechanism of F plasmid: two plasmid gene-encoded products and a cis-acting region are involved in partition. Cell 32: 351–360 [DOI] [PubMed] [Google Scholar]
- Posfai G, Koob M, Hradecna Z, Hasan N, Filutowicz M, Szybalski W (1994) In vivo excision and amplification of large segments of the Escherichia coli genome. Nucleic Acids Res 22: 2392–2398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Posfai G, Plunkett G III, Feher T, Frisch D, Keil GM, Umenhoffer K, Kolisnychenko V, Stahl B, Sharma SS, de Arruda M, Burland V, Harcum SW, Blattner FR (2006) Emergent properties of reduced-genome Escherichia coli. Science 312: 1044–1046 [DOI] [PubMed] [Google Scholar]
- Sherratt DJ (2003) Bacterial chromosome dynamics. Science 301: 780–785 [DOI] [PubMed] [Google Scholar]
- Sunako Y, Onogi T, Hiraga S (2001) Sister chromosome cohesion of Escherichia coli. Mol Microbiol 42: 1233–1241 [DOI] [PubMed] [Google Scholar]
- Tecklenburg M, Naumer A, Nagappan O, Kuempel PL (1995) The dif resolvase locus of the Escherichia coli chromosome can be replaced by a 33-bp sequence, but function depends on location. Proc Natl Acad Sci USA 92: 1352–1356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaichi Y, Niki H (2004) migS, a cis-acting site that affects bipolar positioning of oriC on the Escherichia coli chromosome. EMBO J 23: 221–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu BJ, Sung BH, Koob MD, Lee CH, Lee JH, Lee WS, Kim MS, Kim SC (2002) Minimization of the Escherichia coli genome using a Tn5-targeted Cre/loxP excision system. Nat Biotechnol 20: 1018–1023 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figures
Supplementary Table 1
Supplementary Table 2
Supplementary Information