Abstract
Drosophila neuroblasts divide asymmetrically by aligning their mitotic spindle with cortical cell polarity to generate distinct sibling cell types. Neuroblasts asymmetrically localize Gαi, Pins, and Mud proteins; Pins/Gαi direct cortical polarity, whereas Mud is required for spindle orientation. It is currently unknown how Gαi–Pins–Mud binding is regulated to link cortical polarity with spindle orientation. Here, we show that Pins forms a “closed” state via intramolecular GoLoco–tetratricopeptide repeat (TPR) interactions, which regulate Mud binding. Biochemical, genetic, and live imaging experiments show that Gαi binds to the first of three Pins GoLoco motifs to recruit Pins to the apical cortex without “opening” Pins or recruiting Mud. However, Gαi and Mud bind cooperatively to the Pins GoLocos 2/3 and tetratricopeptide repeat domains, respectively, thereby restricting Pins–Mud interaction to the apical cortex and fixing spindle orientation. We conclude that Pins has multiple activity states that generate cortical polarity and link it with mitotic spindle orientation.
Keywords: cell polarity, cell signaling, differentiation, protein–protein interactions
In complex, multicellular organisms, differentiated cell types are needed to perform diverse functions. One common mechanism for cellular differentiation is asymmetric cell division, in which the mitotic spindle is aligned with the cell polarity axis to generate molecularly distinct sibling cells (1–4). Asymmetric divisions have been proposed to regulate stem cell pool size during development, adult tissue homeostasis, and the uncontrolled proliferation observed in cancer (5). Thus, understanding how the mitotic spindle is coupled to the cell polarity axis is relevant to stem cell and cancer biology. Here, we investigate this question in Drosophila neuroblasts, a model system for studying asymmetric cell division.
Drosophila neuroblasts are stem cell-like progenitors that divide asymmetrically to produce a larger self-renewing neuroblast and a smaller ganglion mother cell (GMC) that differentiates into neurons or glia (3). Mitotic neuroblasts segregate factors that promote neuroblast self-renewal to their apical cortex and differentiation factors to their basal cortex. Precise alignment of the mitotic spindle with the neuroblast apical/basal polarity is required for asymmetric cell division and proper brain development: spindle misalignment leads to symmetric cell divisions that expand the neuroblast population and brain size (6–8).
A key regulator of neuroblast cell polarity and spindle orientation is Partner of Inscuteable (Pins; LGN or mPins in mammals, GPR-1/2 in Caenorhabditis elegans). In metaphase neuroblasts, Pins is colocalized at the apical cortex with the heterotrimeric G protein subunit Gαi and the spindle-associated, coiled-coil mushroom body defect protein (8–18) (Mud; NuMA in mammals, Lin-5 in C. elegans). Pins and Gαi are interdependent for localization and for establishing cortical polarity (17, 18). Pins also binds directly to Mud and recruits it to the apical cortex; Mud is specifically required to align the mitotic spindle with Gαi/Pins but has no apparent role in establishing cortical polarity (8, 13, 14).
The mechanism underlying Pins regulation of cortical polarity and spindle–cortex coupling is unclear, and it is unknown how Gαi–Pins–Mud complex assembly is regulated. Pins has the potential to bind multiple Gαi·GDP molecules via three short GoLoco motifs (Fig. 1A), as do mammalian Pins homologs (19), but the role of these multiple binding sites is unknown. Moreover, via its tetratricopeptide repeats (TPRs), Pins can bind Mud (8, 13, 14), but the stoichiometry and regulation of this interaction has not been explored. Furthermore, we show below that, like its mammalian homolog LGN (19), the regions of Pins containing the TPRs and GoLocos interact, raising the possibility of cooperative “opening” of Pins by Gαi and Mud ligands. Here, we test the role of Pins intra- and intermolecular interactions in coupling cortical polarity with spindle orientation. We use biochemistry, genetics, and in vivo live imaging to test the role of Pins intramolecular interactions and whether Gαi and Mud bind Pins independently, cooperatively, or antagonistically. We conclude that Pins has multiple functional states—a form recruited by a single Gαi to the apical cortex that is unable to bind Mud but sufficient to induce cortical polarity, and a form saturated with Gαi that recruits Mud and links cortical polarity to the mitotic spindle. The multiple Pins states are due to cooperative binding of Mud and Gαi to Pins and result in a tight link between apical cortical polarity and mitotic spindle orientation.
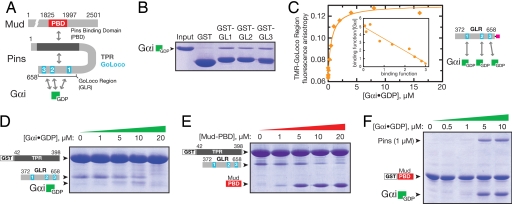
Fig. 1.
Pins intramolecular interaction regulates Gαi and Mud binding. (A) Domain structure of Pins and intra- and intermolecular interactions (PBD, region of Mud that binds Pins). (B) Each of the Pins GoLoco motifs can bind Gαi. Individual GST fusions of the three GoLocos bind Gαi·GDP at qualitatively similar levels. (C) The Pins GoLocos are intrinsically independent, equivalent Gαi binding sites. The extent of Gαi·GDP binding to the Pins GoLocos was monitored by the fluorescence anisotropy of a tetramethylrhodamine attached to a cysteine at its C terminus. The curve represents a model with three equivalent, independent binding sites of affinity Kd = 530 ± 80 nM. A Scatchard analysis is shown in Inset where the binding function is equal to the concentration of Pins-bound Gαi·GDP divided by the total concentration of Pins. (D) Gαi disrupts the Pins intramolecular interaction. In a qualitative “pull-down” assay, Gαi·GDP competes with the Pins GLR for binding to the Pins TPRs. Although Gαi ultimately disrupts the TPR–GLR interaction, at intermediate Gαi concentrations (5–10 μM), a Gαi–GLR–TPR complex can be formed, presumably those that result in occupation of GoLoco1 but not GoLoco2/3. At a higher concentration (20 μM), occupation of all three GoLoco motifs by Gαi interferes with the interaction of the GLR with the TPR region. Proteins are stained with Coomassie brilliant blue. (E) The Mud PBD disrupts the Pins intramolecular interaction. Binding of Mud to the Pins TPRs (as in B) competes with the Pins GoLocos. (F) Gαi increases the affinity of Pins for Mud. Full-length Pins binds weakly to the Mud PBD, but binding is enhanced by the presence of Gαi·GDP, indicating that Gαi and Mud bind cooperatively to Pins.
Results
Gαi and Mud Bind Cooperatively to Pins.
The NH2-terminal half of Pins contains seven TPRs, and the COOH-terminal half contains three GoLoco motifs, which we term the GoLoco region, or GLR (Fig. 1A). Each of the three GoLocos has the potential to bind GDP-bound Gαi (20), whereas the TPRs bind the Mud protein (8, 13, 14). Before testing whether the Pins intramolecular interaction regulates Pins–Gαi–Mud complex assembly, we first characterized each of the relevant individual domain interactions: TPR–Mud, GLR–Gαi, and TPR–GLR. First, the Pins TPRs bind Mud with a 1:1 stoichiometry as judged by the elution profile of the TPR–Mud complex on a calibrated gel-filtration column [shown in supporting information (SI) Fig. 5A], indicating that Pins contains a single Mud binding site. Second, each of the three Pins GoLoco domains binds Gαi·GDP (hereafter Gαi) equally well in a qualitative pull-down assay (Fig. 1B) as well as in a more quantitative assay measuring Gαi binding by using the fluorescence anisotropy of tetramethylrhodamine attached to the COOH terminus of the Pins GLR (Fig. 1C). A binding isotherm describing three equivalent, independent sites with submicromolar Gαi affinities (Kd = 530 ± 80 nM) fits the data well and yields a linear Scatchard relationship (Fig. 1C Inset). We conclude that each GoLoco in the Pins GLR binds Gαi with a similar affinity and without cooperativity in the absence of the TPRs, similar to a three-GoLoco region of the protein AGS3 (21). Finally, the interaction between the TPRs and GLR has an affinity of Kd = ≈2 μM in trans (SI Fig. 5B), which may be enhanced in intact Pins because of the increase in effective concentration.
To test whether the Pins intramolecular interaction regulates Pins–Gαi–Mud complex assembly, we first determined whether Gαi or Mud binding disrupts TPR–GLR. Using a qualitative assay in which the TPRs and GLR are expressed as separate fragments, we find that increasing concentrations of Gαi completely disrupt the trans TPR–GLR complex (Fig. 1D). The region of Mud that binds to Pins (Pins binding domain or PBD; contained within Mud residues 1825–1997) also disrupts the TPR–GLR complex, although not as efficiently as Gαi (Fig. 1E). Thus, Pins contains an intramolecular interaction that competes against both Gαi and Mud binding.
Because Gαi and Mud are both coupled to the Pins intramolecular interaction, we tested whether the two proteins bind cooperatively to Pins by determining whether Gαi could enhance the affinity of Pins for Mud. As shown in Fig. 1F, 1 μM Pins binds weakly to a GST fusion of the Mud PBD. However, addition of Gαi induces a large increase in Pins binding and the formation of a Mud–Pins–Gαi ternary complex. We conclude that Gαi increases the affinity of Pins for Mud (i.e., Gαi and Mud bind cooperatively to Pins).
Differential GoLoco Regulation by the Pins Intramolecular Interaction.
Because Pins contains three GoLoco motifs and the Pins intramolecular interaction competes against Gαi binding, we next tested whether these Gαi binding sites are repressed equally in intact Pins. We used gel-filtration chromatography of full-length Pins and Gαi to determine how Gαi–GoLoco binding is affected by the intramolecular interaction. Pins elutes as a single peak with an elution volume consistent with the molecular weight for a monomer (Fig. 2A). (The protein composition of the peaks, as determined by SDS/PAGE, is shown in SI Fig. 5C.) Addition of low Gαi concentrations leads to formation of a 1:1 Gαi:Pins complex peak (we assigned peaks to 1:1 or 2:1 complexes using Pins mutants with one or two GoLocos inactivated). Higher Gαi concentrations lead to the formation of a 3:1 Gαi:Pins complex with a very broad peak, suggestive of a lower affinity interaction. We conclude that full-length Pins contains a single high-affinity Gαi-binding GoLoco and two low-affinity GoLocos.
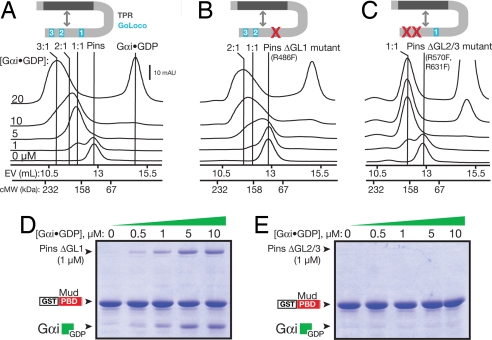
Fig. 2.
Differential repression of the three GoLocos by the Pins intramolecular interaction. (A) Analysis of Pins binding to Gαi·GDP by gel-filtration chromatography. Pins and mixtures of Pins and Gαi·GDP were separated by gel filtration. Marks indicating the elution volumes of 2:1 and 1:1 Gαi:Pins complexes were determined by using single and double Pins GoLoco mutants, respectively. The column elution volumes (EV) of standard proteins to give a calibrated molecular weight (cMW) are shown on the x axis. The protein composition of the 20 μM Gαi eluate for this and B and C are shown in SI Fig. 5C. (B) Analysis of Pins with an inactive GoLoco1 (Pins ΔGL1) binding to Gαi·GDP by gel-filtration chromatography. Loss of GoLoco1 causes loss of the high-affinity peak that occurs at low Gαi concentrations. (C) Analysis of Pins with an inactive GoLoco 2 and 3 (Pins ΔGL2/3) binding to Gαi·GDP by gel-filtration chromatography. Only the high-affinity interaction remains after loss of GoLocos 2 and 3. (D) Cooperative binding of Gαi and Mud to Pins does not require GoLoco1. In the absence of GoLoco1, Gαi·GDP enhances the affinity of Pins for Mud. (E) GoLocos 2 and 3 are required for cooperative binding of Gαi and Mud to Pins. Although Gαi·GDP can bind to Pins in which GoLocos 2 and 3 are inactivated (C), binding does not lead to cooperative Mud binding.
Because the three GoLocos are intrinsically equivalent, independent Gαi binding sites (Fig. 1D), the distinct Gαi binding behavior in full-length Pins suggests that Pins contains one GoLoco domain that is unregulated or only partially regulated by the intramolecular interaction and two GoLoco domains that are cooperatively repressed. To further explore this model, we inactivated one or more GoLocos by mutating a single critical arginine residue (20) to phenylalanine in the context of full-length Pins. These mutations do not inhibit the ability of the TPRs and GoLocos to interact (SI Fig. 5D). Inactivation of GoLoco1 (Pins ΔGL1; R486F) specifically abolishes the high-affinity 1:1 complex (Fig. 2B), whereas inactivation of either GoLoco 2 or 3 has no effect on the high-affinity complex (unpublished observations). We therefore classify GoLoco1 as a high-affinity GoLoco in the context of full-length Pins. Disruption of GoLocos 2 and 3 (Pins ΔGL2/3; R570F, R631F) leads to the formation of a 1:1 complex at low concentrations of Gαi, further confirming that GoLoco1 is not repressed by the TPRs (Fig. 2C). We conclude that the three GoLoco motifs are differentially regulated by the Pins intramolecular interaction: Gαi shows unregulated high-affinity binding to GoLoco1 and low-affinity, cooperative binding to GoLocos 2 and 3.
We next asked how Gαi binding to the different Pins GoLoco domains affects cooperative Gαi–Pins–Mud complex assembly. When GoLoco1 is inactivated (Pins ΔGL1), Gαi can still enhance Mud binding (Fig. 2D), in a manner similar to the WT Pins (Fig. 1D). The activation is more efficient, however, presumably because of the lack of Gαi “buffering” by GoLoco1. In contrast, in the Pins ΔGL2/3 mutant, Gαi does not enhance Mud binding (Fig. 2E) even though it binds GoLoco1 with high affinity (Fig. 2C). Thus, Pins differentially regulates the ability of Gαi to promote Pins–Mud binding: Gαi binding to GoLoco1 has no effect on Pins–Mud binding, whereas Gαi binding to GoLocos 2 and 3 strongly enhances Pins–Mud association.
The Conformational Transition to “Open” Pins Requires both Mud and Gαi.
Our results suggest that Gαi binding to GoLocos 2 and 3 “opens” Pins to allow Mud binding to the TPRs. To directly monitor the Pins conformational transition between “closed” and “open” states, we constructed a Pins fluorescence resonance energy transfer (FRET) sensor with YFP and CFP at the NH2 and COOH termini, respectively (Fig. 3A). This type of sensor has been used successfully to monitor the conformational transition of a mammalian Pins homolog, LGN (19). Surprisingly, addition of Gαi or Mud alone did not cause a significant change in the YFP–Pins–CFP FRET signal, even at high concentrations (Fig. 3B), suggesting that Gαi or Mud alone is insufficient to “open” Pins. The addition of both ligands together, however, leads to a large change in the FRET signal (nearly complete loss of energy transfer), indicating that Mud and Gαi are both required to induce the “open” Pins conformation (Fig. 3B). To test the model that Gαi binding to GoLoco1 cannot open Pins, we analyzed a Pins ΔGL2/3 FRET sensor. As shown in Fig. 3C, Mud and Gαi fail to induce the conformational change seen with the WT FRET sensor, consistent with Gαi binding at GoLoco1 not being coupled to the intramolecular interaction.
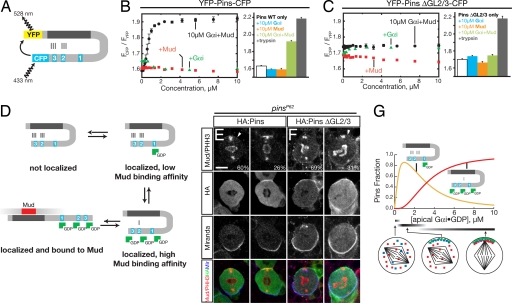
Fig. 3.
Pins undergoes a conformational change into a high Mud binding affinity state in response to Gαi binding to GoLocos 2 and 3. (A) Architecture of the Pins FRET sensor. (B) Full transition to the Pins open state requires both Gαi·GDP and Mud. Shown in the bar graph are the FRET ratios for the WT Pins FRET sensor ([YFP–Pins–CFP] = 200 nM) with various combinations of Gαi·GDP and Mud ligands present at 10 μM concentration and after trypsin digestion. Error bars equal one SD (n = 3). (C) Pins with inactivated GoLocos 2 and 3 is unable to transition to open state with Gαi·GDP and Mud present. Shown in the bar graph are the FRET ratios for the Pins ΔGL2/3 FRET sensor ([YFP–Pins–CFP] = 200 nM) with various combinations of Gαi·GDP and Mud ligands present at 10 μM concentration. Error bars equal one SD (n = 3). (D) Model for coupled Gαi·GDP and Mud binding and relationship to Pins conformational states. (E) HA:Pins can rescue Mud localization in pins larval neuroblasts. Pins localization is detected by anti-HA antibody, Mud is detected by anti-Mud antibody, and the mitotic stage is determined by anti-phospho histone H3 (α-PHH3) antibody. Moderate (60%) or strong (26%) Mud apical crescents were observed in metaphase neuroblasts. The localization of Mud to the spindle poles is independent of Pins activity (13). Miranda was used as a basal marker. (F) Pins with inactivated GoLocos 2 and 3 fails to rescue Mud apical enrichment in pins larval neuroblasts. Staining is as in E. Although HA:Pins ΔGL2/3 is correctly localized, only weak or moderate Mud apical staining was observed in 31% of metaphase neuroblasts. (Scale bar: E and F, 5 μm.) (G) Pins–Gαi response profile. The concentration of Pins with Gαi bound at GoLoco1 and Gαi-saturated Pins is shown as a function of Gαi concentration based on the model shown in D. This model uses the intrinsic affinity of Gαi for the GoLocos (Kd = 530 nM) and assumes that the inactive form of Pins is favored 1,000:1 over active Pins in the absence of Gαi or Mud. Simulations were performed with Berkeley Madonna. Predicted neuroblast phenotypes with the state of cortical polarity (red, Mud; blue, Pins; green, Gαi) and spindle positioning are shown at low, medium, and high Gαi concentration ranges.
Because Mud or Gαi alone are not able to “open” Pins, we can exclude a simple model in which Mud and Gαi directly compete in a mutually exclusive fashion (e.g., sterically) with the intramolecular interaction. Although we observe disruption of the Pins TPR–GLR interaction in trans (Fig. 1 D and E), this is likely to result from effective concentration effects in which the interaction is weaker when the two domains are not in the same polypeptide. We conclude that Mud and Gαi allosterically modulate the TPRs and GoLocos, respectively, in a manner that leaves the intramolecular interaction intact but in a weakened state, poised to open upon binding of the second ligand. Thus, Pins can exist in a “closed” state (no Gαi or Mud bound), a “potentiated” closed state (with Gαi or Mud bound), and an “open” state (with both Gαi and Mud bound) (Fig. 3D).
Neuroblasts Expressing Pins with Inactive GoLocos 2 and 3 Fail to Induce Pins–Mud Apical Coupling.
Based on the network of interactions present in Pins, Gαi binding to GoLoco1 should recruit Pins to the neuroblast apical cortex but not lead to Mud recruitment. To test this model, we expressed either HA:Pins WT or HA:Pins ΔGL2/3 in pins mutant neuroblasts and examined both Pins and Mud localization. In third-instar larval central brain neuroblasts, both WT and ΔGL2/3 Pins localized to the apical cortex at metaphase (Fig. 3 E and F). However, Mud was correctly recruited to the apical cortex in neuroblasts expressing WT Pins (Fig. 3E, 86%; n = 15), and Mud recruitment in ΔGL2/3 neuroblasts was significantly reduced (Fig. 3F, 31%; n = 13). Thus, Gαi binding to GoLoco1 is sufficient for Pins localization but not for efficient Mud targeting.
To understand how cortically localized and Mud-recruiting Pins states are populated as Gαi accumulates at the apical cortex, we simulated Pins–Gαi binding based on the parameters described earlier (Fig. 3G). At low Gαi concentration, Pins with Gαi bound to GoLoco1 predominates because of its higher affinity relative to the other two GoLocos (which are repressed by the TPRs). Although this Pins form does not bind to Mud with high affinity, we hypothesize that it is sufficient to induce aspects of cortical polarity (e.g., Insc polarization). At higher Gαi concentrations, GoLoco1 becomes saturated and binding can occur at GoLocos 2 and 3, allowing for Mud recruitment to the apical cortex (Fig. 3D). Thus, we predict that as Gαi accumulates at the apical cortex, it first recruits Pins in a form that is competent for cortical polarization but not spindle positioning. As Gαi levels further increase, however, GoLocos 2 and 3 become populated, weakening the intramolecular interaction and freeing the TPRs to recruit Mud to the apical cortex.
Reduced Gαi Impairs Pins–Mud Interaction and Spindle Orientation in Larval Neuroblasts.
We tested the model that the population of Pins activation states is very sensitive to Gαi concentration by examining Pins localization, Mud localization, and spindle orientation in larval neuroblasts with different levels of Gαi protein. Our model strongly predicts that normal Gαi and Mud levels should “open” Pins to form a ternary complex at the apical cortex that is functional for spindle alignment, low Gαi levels would bind Pins GoLoco1 and recruit Pins to the apical cortex without allowing Mud binding or spindle orientation, and no Gαi protein would result in a failure to recruit Pins or Mud to the cortex. To test this model, we examined larval neuroblasts with normal, low, or no Gαi protein (WT zygotic mutants and maternal zygotic mutants, respectively). As expected, neuroblasts with WT levels of Gαi invariably colocalize Gαi, Pins, and Mud to an apical cortical crescent that is tightly coupled with the mitotic spindle (Fig. 4A, C, E, F, and K; quantified in Fig. 4J), consistent with the activity of both Gαi and Mud “opening” Pins to form a ternary complex that is functional for spindle orientation. In contrast, neuroblasts with reduced Gαi levels formed robust Pins and Insc crescents (Fig. 4 B, D, and G–I; quantified in Fig. 4J) but typically failed to localize Mud to the apical cortex (Fig. 4 G–I; quantified in Fig. 4J) and showed defects in spindle orientation (Fig. 4 H, I, and K). Neuroblasts lacking all Gαi protein fail to recruit Pins to the cortex and have spindle orientation defects (17, 18). These results strongly support our model: low Gαi levels can recruit “closed” Pins to the cortex without recruiting Mud or promoting spindle orientation, whereas higher Gαi levels function together with Mud to “open” Pins and promote spindle orientation.
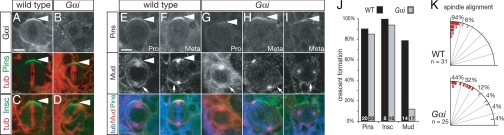
Fig. 4.
Neuroblasts with reduced Gαi levels can recruit Pins/Insc but not Mud to the apical cortex and have spindle alignment defects. (A and B Top) Gαi apical cortical protein levels are reduced in Gαi zygotic mutant larval neuroblasts (B) compared with WT (A). (A and B Lower) Same neuroblast labeled for α-tubulin (tub) and Pins. Strong Pins crescents were observed in both WT (Left) and zygotic Gαi mutant (Right) metaphase neuroblasts. (C and D) Strong Insc crescents were observed in both WT (C) and zygotic Gαi mutant (D) metaphase neuroblasts (cortical fluorescence intensities Insc 84% of WT; see Methods for details). (E–I) Gαi zygotic mutant larval neuroblasts have robust Pins apical crescents but a loss of Mud apical protein crescents (cortical fluorescence intensities of Pins 90% of WT; see Methods for details). (E and F) WT larval neuroblasts have apical crescents of Pins and Mud at prophase (pro) and metaphase (meta) (arrowheads), as well as strong Mud staining on centrosomes and at the basal cortex (arrows). (G–I) Gαi mutant larval neuroblasts show Pins apical crescents but have defects in forming Mud apical protein crescents at prophase and metaphase (arrowheads), although Mud at the basal cortex is unaffected (arrows). (Scale bar: E–I, 5 μm.) (J) Quantification of apical crescent formation at metaphase in WT and Gαi mutant neuroblasts (see Methods for details). The number in each bar represents the number of neuroblasts examined. (K) Gαi zygotic mutant neuroblasts show spindle alignment defects. Quantification of apical spindle pole alignment (red ticks) relative to the center of the Pins cortical crescent (vertical line). WT spindles are tightly aligned, but Gαi mutant spindles are frequently misaligned.
To further test our model, we used time-lapse video microscopy to examine the dynamics of spindle behavior using a GFP-tagged microtubule-associated protein (see Methods). In WT neuroblasts, the apical centrosome/spindle pole is anchored at the center of the Gαi/Pins/Mud crescent from prometaphase through telophase (13), although slight spindle rocking can be observed (SI Fig. 6 and SI Movie 1). In neuroblasts with reduced Gαi levels, where Gαi/Pins but not Mud are present at the apical cortex, we find that the centrosome/spindle pole is not stably attached to the apical cortex and often shows excessive rotation (SI Fig. 6 and SI Movie 2). These data provide further support for our model that low levels of Gαi are sufficient to recruit Pins to the cortex via GoLoco1 binding but are insufficient to allow Pins to bind Mud and capture the apical spindle pole.
Discussion
Through interactions with Gαi and Mud, Pins regulates two fundamental aspects of asymmetric cell division: cortical polarity and alignment of the spindle with the resulting polarity axis. In this study, we have investigated the mechanism by which Gαi regulates Pins interactions with the spindle orientation protein Mud. We have found that, although the three Pins GoLocos are intrinsically equivalent, independent Gαi binding sites, an intramolecular interaction with the Pins TPRs leads to differential Gαi binding. Gαi binding to GoLoco1 is not coupled to the Pins intramolecular interaction and therefore does not influence Mud binding but is sufficient to localize Pins to the cortex for Mud-independent functions (e.g., recruitment of Insc to the apical cortex). Gαi binding to GoLocos 2 and 3 destabilizes the Pins intramolecular interaction leading to cooperative Mud binding, and together the ligands induce an “open” Pins conformational state. This leads to a model in which Gαi induces multiple Pins activation states: one that localizes cortically but is not competent for Mud binding, and one that binds Mud linking localized Gαi to the mitotic spindle (Fig. 3D).
The Pins Intramolecular Interaction as a Mechanism for Localizing Mud Activity to the Cortex.
Intramolecular interactions are common features of signaling proteins that typically act through “autoinhibition” of an enzymatic or ligand binding activity (22). Such interactions allow for coupling of regulatory molecule binding to an increase or decrease in downstream function, a critical aspect of information flow in signaling pathways (22–24). Pins is involved in the regulation of multiple downstream functions, and our results support the notion that the multiple Gαi binding sites present in Pins allow for the signal to branch into two pathways, one controlling cortical polarity and the other spindle positioning. A notable exception to the multiple GoLocos present in Pins-like proteins is the C. elegans Pins homologue GPR-1/2, which contains a single GoLoco domain. The lack of multiple GoLocos in GPR-1/2 may be consistent with their more limited role in C. elegans asymmetric cell division, where they regulate spindle positioning but not cortical polarity (25–27).
In the model presented here, the Pins intramolecular interaction serves to regulate Mud binding. This may occur for several reasons. First, localization of Mud activity to the apical cortex appears to be important for aligning the spindle with the axis of cortical polarity (8, 13, 14). In this context, the Pins intramolecular interaction may be important for restricting Mud activity to the apical cortex. Mutant pins or Gαi neuroblasts may have low ectopic Mud activity at the basal or lateral cortex that leads to the observed misdirected spindle rotation seen in our live neuroblast imaging (SI Movie 2). This observation is consistent with our previous observations that too little Mud (in mud mutant neuroblasts) results in spindle position defects without any rotation (13). Second, Mud activity may be affected by its interaction with Pins. For example, LGN binds to a region of NuMA near its microtubule binding site such that LGN binding to NuMA competes with microtubule binding (28).
Unequal Regulation of the GoLocos by the TPRs Leads to a Complex Gαi Response Profile.
A unique feature of the Pins intramolecular interaction is that autoinhibition is incomplete. Binding of GoLocos 2 and 3 to Gαi is repressed by the TPRs, but binding to GoLoco1 is not. This has two important consequences. First, whereas the three GoLocos are intrinsically equivalent and independent Gαi binding sites, TPR repression of GoLocos 2 and 3 significantly lowers the affinity of these GoLocos relative to GoLoco1. This leads to preferential population of GoLoco1, which may be important for temporal regulation of asymmetric cell division by ensuring that cortical polarity is established before the spindle is positioned. Second, the TPRs appear to repress GoLocos 2 and 3 cooperatively (Gαi binding to 2 or 3 increases the affinity at the other site). Cooperativity is a common property of signaling pathways that is used generate complex input–output profiles (29). Pins exhibits both homotropic (Gαi) and heterotropic (Gαi and Mud) binding cooperativity. In both cases, cooperativity is not an inherent property of the binding sites but is generated through the competition that results from the intramolecular interaction between the TPRs and GoLocos. Such “cooperative repression” of inherently equivalent binding sites through intramolecular interactions may be a general mechanism for generating cooperativity in signaling proteins.
Methods
Protein Expression and Purification.
DNA encoding full-length Drosophila Pins was amplified from an embryonic cDNA library. Drosophila Gαi was completely insoluble, and therefore mouse Gαi3 25–354 (which is 76% identical to the Drosophila protein) cloned from a macrophage cDNA library was used for these studies. A plasmid containing Mud residues 1825–1997 was generated as described in ref. 13.
All proteins were expressed by using the Escherichia coli strain BL21(DE3) as a host strain with pGEX 4T-1-based vectors for GST fusions and pBH-based vectors for hexahistidine fusions, which were isolated and purified as described in ref. 30.
Gαi was either used directly after purification or loaded with GDP or GMPPNP subsequent to purification (GDP-loaded and unloaded behaved identically). Nucleotide was added at a 5-fold molar excess in 10 mM Hepes/100 mM NaCl/1 mM DTT/1 mM EDTA (pH 7.5) and incubated at room temperature for 30 min. The final buffer conditions contained 10 mM MgCl2.
In Vitro Binding Assays.
GST pull-down assays were performed as described in ref. 31. Briefly, ligands were added to glutathione agarose with adsorbed GST fusion proteins in binding buffer (10 mM Hepes/100 mM NaCl/1 mM DTT) at the indicated concentrations to a final reaction volume of 50 μl and incubated at room temperature for 15 min before washing, elution, and analysis by gel electrophoresis.
Fluorescence anisotropy binding assays were performed as described in ref. 31. For labeling of the Pins GoLocos, a cysteine was added at the COOH terminus of residues 372–658, and the two naturally occurring cysteines, residues 487 and 561, were mutated to serines. For binding experiments, solutions were prepared with increasing amount of ligand and constant dye-labeled component (100 nM) in binding buffer with the temperature maintained at 20°C by using a circulating water bath. Data series were fit to an equation describing binding to three independent, equivalent sites using nonlinear regression.
Gel-filtration studies were carried out on a Superdex 200 molecular sizing column calibrated with a series of molecular weight standards (GE Healthcare) equilibrated in binding buffer. Proteins at the indicated concentrations in a volume of 100 μl were incubated at 4°C for 15 min before being loaded on the column at a flow rate of 0.5 ml/min with 300-μl fractions collected for analysis. Protein elution was detected by absorbance at 280 nm.
Construction and Analysis of the Pins FRET Sensor.
Full-length Pins, with yellow fluorescent protein (EYFP 1–239) and cyan fluorescent protein (ECFP 1–239) coding sequences at the NH2 and COOH termini, respectively, was expressed and purified as described above except that gel-filtration chromatography was added as a final purification step. The proteins at 200 nM in binding buffer were excited with a wavelength of 433 nm (to minimize direct YFP excitation), and the amount of energy transfer was measured by taking the ratio of CFP (475 nm) and YFP (525 nm) emission. For proteolysis experiments, proteins were incubated with 0.9 nM trypsin (Sigma) at 18°C for 15 min, followed by FRET measurement.
Fly Strains.
The Oregon R strain was used as WT control for the analysis of cell polarity and spindle orientation in larval neuroblasts of fixed specimens. The gene trap line P{PTT-GA}JupiterG00147 [also known as G147-GFP (31)], which expresses a GFP-tagged microtubule-associated protein, was used to visualize microtubules in live larval neuroblast preparations. The GαiP8 allele is described in ref. 17. We used zygotic GαiP8 mutants that contained maternally contributed Gαi that allowed for survival into late larval stages unlike maternal zygotic GαiP8 mutants. The GαiP8 allele and GαiP8, P{PTT-GA}JupiterG00147 recombinant chromosome were rebalanced over TM3 actin-GFP Ser and TM6B Tb Hu chromosomes, respectively. Newly hatched mutant larvae were identified based on the absence of GFP expression in the gut or the absence of the dominant Tb marker.
For transgenic analysis, the full-length WT or ΔGL2/3 Pins were subcloned into pUAST containing an N-terminal hemagglutinin (HA) epitope. Transgenic flies carrying P{UAS-HA:Pins} or P{UAS-HA:PinsΔGL2/3} on the second chromosome were crossed with the pinsP62 allele to form P{UAS-HA:Pins}; pinsP62/TM3-Sb or P{UAS-HA:PinsΔGL2/3}; pinsP62/TM3-Sb lines. These flies were crossed at 18°C to the worniu-GAL4; pinsP62/TM3-actin-GFP Ser driver line, and mutant larvae in the progeny were analyzed. Mutant larvae were identified based on the absence of GFP expression and the presence of smaller central brain neuroblasts.
Immunocytochemistry.
Freshly hatched WT pinsP62 and GαiP8 zygotic mutant larvae were aged for 96–120 h at 25°C and prepared for immunofluorescent antibody labeling as described previously, with the modification that 5% normal goat serum was added to the larval blocking and primary antibody solutions (32). Primary antibodies were rat anti-Pins [#2, 1:500 (9)], rabbit anti-Gαi (1:500; raised against peptide, amino acids 327–355), rabbit anti-Insc (1:1,000; W. Chia), rabbit anti-Mud (raised against amino acids 375–549; 1:2,000; H. Nash), mouse anti-α-tubulin (1:2,000; DM1A, Sigma), and mouse-anti-HA (1:1,000; Covance). Fluorescently conjugated secondary antibodies were obtained from Jackson ImmunoResearch and Molecular Probes. For DNA labeling, fixed specimens were incubated in PBS 0.1% Triton X-100 containing 1 mg/ml RNase A for 1 h at room temperature and counterstained with 4 g/ml propidium iodide. Confocal images were acquired on a Leica TCS SP2 microscope equipped with a ×63 1.4-N.A. oil-immersion objective. Panels were arranged by using ImageJ (National Institutes of Health), Photoshop (Adobe Systems), and Illustrator (Adobe Systems).
To determine the frequency of cortical Pins, Insc, and Mud crescents, the ratio of background-corrected cortical and cytoplasmic fluorescence intensity (FI) was determined. A crescent was scored to be present if the ratio of background-corrected cortical and cytoplasmic FI exceeded 1.75. To calculate background corrected FIs, mean FIs were determined for three 2 × 2-pixel areas at the cortex and in the cytoplasm of each metaphase neuroblasts. From these measured cortical and cytoplasmic FIs, the mean background fluorescence (measured in three different areas of interphase nuclei, which do not express Pins, Insc, or Mud proteins) was subtracted to obtain the background-corrected cortical and cytoplasmic FIs.
Analysis of Spindle Orientation in Fixed and Live Larval Neuroblasts.
In fixed specimens, spindle orientation was measured at metaphase as the angle between the spindle axis (defined by position of the two spindle poles) and the cell polarity axis (defined as a line through the cell center and the center of the Pins or Insc crescents).
For the analysis of spindle orientation in live neuroblasts, freshly hatched larvae expressing a GFP-tagged MT-associated protein (G147-GFP) were aged at 25°C for 96–120 h, and whole brains were mounted and imaged as described in ref. 32. Brains were imaged by using a Bio-Rad Radiance 2100 laser scanning confocal microscope equipped with a ×60 1.4-N.A. oil-immersion objective. Image stacks were processed by using ImageJ and converted into movies by using QuickTime. The beginning of prometaphase (0:00 [min:sec]) was determined as the time point at which microtubules emanating from centrosomes penetrated the cell center to form the spindle. Anaphase onset was determined by the apparent shortening of kinetochore fibers. Orientation of centrosome pairs/spindles over time was measured as the angular position relative to an axis defined by the position of the centrosome pair at the beginning of prometaphase (0:00, 0°).
Supplementary Material
Acknowledgments
We thank V. DeRose, K. Guillemin, R. Newman, S. Siegrist, and T. Stevens for critical reading of the manuscript; members of the K.E.P. laboratory for helpful comments and suggestions; W. Breyer for her work at the inception of this project; W. Chia (Temasek Lifesciences Laboratory, National University of Singapore), X. Morin (Developmental Biology Institute of Marseilles, France), H.A.N., and J. Knoblich (Molecular Biotechnology of the Austrian Academy of Sciences, Vienna, Austria) for sharing reagents; and S. Siegrist and S. Shrimpton for generation of the GαiP8, P{PTT-GA}JupiterG00147 recombinant line. This work was supported by an American Heart Association predoctoral fellowship (to R.W.N.), the Howard Hughes Medical Institute (for which C.Q.D. is an Investigator), a Damon Runyon Scholar Award (to K.E.P.), and National Institutes of Health Grant GM068032 (to K.E.P.).
Abbreviations
- GLR
GoLoco region
- PBD
Pins binding domain
- TPR
tetratricopeptide repeat.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0701812104/DC1.
References
- 1.Betschinger J, Knoblich JA. Curr Biol. 2004;14:R674–R685. doi: 10.1016/j.cub.2004.08.017. [DOI] [PubMed] [Google Scholar]
- 2.Cowan CR, Hyman AA. Annu Rev Cell Dev Biol. 2004;20:427–453. doi: 10.1146/annurev.cellbio.19.111301.113823. [DOI] [PubMed] [Google Scholar]
- 3.Yu F, Kuo CT, Jan YN. Neuron. 2006;51:13–20. doi: 10.1016/j.neuron.2006.06.016. [DOI] [PubMed] [Google Scholar]
- 4.Wodarz A. Curr Opin Cell Biol. 2005;17:475–481. doi: 10.1016/j.ceb.2005.08.005. [DOI] [PubMed] [Google Scholar]
- 5.Morrison SJ, Kimble J. Nature. 2006;441:1068–1074. doi: 10.1038/nature04956. [DOI] [PubMed] [Google Scholar]
- 6.Lee CY, Andersen RO, Cabernard C, Manning L, Tran KD, Lanskey MJ, Bashirullah A, Doe CQ. Genes Dev. 2006;20:3464–3474. doi: 10.1101/gad.1489406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang H, Somers GW, Bashirullah A, Heberlein U, Yu F, Chia W. Genes Dev. 2006;20:3453–3463. doi: 10.1101/gad.1487506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bowman SK, Neumuller RA, Novatchkova M, Du Q, Knoblich JA. Dev Cell. 2006;10:731–742. doi: 10.1016/j.devcel.2006.05.005. [DOI] [PubMed] [Google Scholar]
- 9.Yu F, Morin X, Cai Y, Yang X, Chia W. Cell. 2000;100:399–409. doi: 10.1016/s0092-8674(00)80676-5. [DOI] [PubMed] [Google Scholar]
- 10.Schaefer M, Shevchenko A, Knoblich JA. Curr Biol. 2000;10:353–362. doi: 10.1016/s0960-9822(00)00401-2. [DOI] [PubMed] [Google Scholar]
- 11.Parmentier ML, Woods D, Greig S, Phan PG, Radovic A, Bryant P, O'Kane CJ. J Neurosci. 2000;20:RC84. doi: 10.1523/JNEUROSCI.20-14-j0003.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Siegrist SE, Doe CQ. Cell. 2005;123:1323–1335. doi: 10.1016/j.cell.2005.09.043. [DOI] [PubMed] [Google Scholar]
- 13.Siller KH, Cabernard C, Doe CQ. Nat Cell Biol. 2006;8:595–600. doi: 10.1038/ncb1412. [DOI] [PubMed] [Google Scholar]
- 14.Izumi Y, Ohta N, Hisata K, Raabe T, Matsuzaki F. Nat Cell Biol. 2006;8:586–593. doi: 10.1038/ncb1409. [DOI] [PubMed] [Google Scholar]
- 15.Guan Z, Prado A, Melzig J, Heisenberg M, Nash HA, Raabe T. Proc Natl Acad Sci USA. 2000;97:8122–8127. doi: 10.1073/pnas.97.14.8122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Prokop A, Technau GM. Dev Biol. 1994;161:321–337. doi: 10.1006/dbio.1994.1034. [DOI] [PubMed] [Google Scholar]
- 17.Yu F, Cai Y, Kaushik R, Yang X, Chia W. J Cell Biol. 2003;162:623–633. doi: 10.1083/jcb.200303174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schaefer M, Petronczki M, Dorner D, Forte M, Knoblich JA. Cell. 2001;107:183–194. doi: 10.1016/s0092-8674(01)00521-9. [DOI] [PubMed] [Google Scholar]
- 19.Du Q, Macara IG. Cell. 2004;119:503–516. doi: 10.1016/j.cell.2004.10.028. [DOI] [PubMed] [Google Scholar]
- 20.Willard FS, Kimple RJ, Siderovski DP. Annu Rev Biochem. 2004;73:925–951. doi: 10.1146/annurev.biochem.73.011303.073756. [DOI] [PubMed] [Google Scholar]
- 21.Adhikari A, Sprang SR. J Biol Chem. 2003;278:51825–51832. doi: 10.1074/jbc.M306300200. [DOI] [PubMed] [Google Scholar]
- 22.Lim WA. Curr Opin Struct Biol. 2002;12:61–68. doi: 10.1016/s0959-440x(02)00290-7. [DOI] [PubMed] [Google Scholar]
- 23.Rohatgi R, Ma L, Miki H, Lopez M, Kirchhausen T, Takenawa T, Kirschner MW. Cell. 1999;97:221–231. doi: 10.1016/s0092-8674(00)80732-1. [DOI] [PubMed] [Google Scholar]
- 24.Prehoda KE, Scott JA, Mullins RD, Lim WA. Science. 2000;290:801–806. doi: 10.1126/science.290.5492.801. [DOI] [PubMed] [Google Scholar]
- 25.Srinivasan DG, Fisk RM, Xu H, van den Heuvel S. Genes Dev. 2003;17:1225–1239. doi: 10.1101/gad.1081203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Colombo K, Grill SW, Kimple RJ, Willard FS, Siderovski DP, Gonczy P. Science. 2003;300:1957–1961. doi: 10.1126/science.1084146. [DOI] [PubMed] [Google Scholar]
- 27.Gotta M, Dong Y, Peterson YK, Lanier SM, Ahringer J. Curr Biol. 2003;13:1029–1037. doi: 10.1016/s0960-9822(03)00371-3. [DOI] [PubMed] [Google Scholar]
- 28.Du Q, Taylor L, Compton DA, Macara IG. Curr Biol. 2002;12:1928–1933. doi: 10.1016/s0960-9822(02)01298-8. [DOI] [PubMed] [Google Scholar]
- 29.Huang CY, Ferrell JE., Jr Proc Natl Acad Sci USA. 1996;93:10078–10083. doi: 10.1073/pnas.93.19.10078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Morin X, Daneman R, Zavortink M, Chia W. Proc Natl Acad Sci USA. 2001;98:15050–15055. doi: 10.1073/pnas.261408198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Peterson FC, Penkert RR, Volkman BF, Prehoda KE. Mol Cell. 2004;13:665–676. doi: 10.1016/s1097-2765(04)00086-3. [DOI] [PubMed] [Google Scholar]
- 32.Siller KH, Serr M, Steward R, Hays TS, Doe CQ. Mol Biol Cell. 2005;16:5127–5140. doi: 10.1091/mbc.E05-04-0338. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.