Abstract
Nonviral nucleic acid delivery to cells and tissues is considered a standard tool in life science research. However, although an ideal delivery system should have high efficacy and minimal toxicity, existing materials fall short, most of them being either too toxic or little effective. We hypothesized that disulfide cross-linked low-molecular-weight (MW) linear poly(ethylene imine) (MW <4.6 kDa) would overcome this limitation. Investigations with these materials revealed that the extracellular high MW provided outstandingly high transfection efficacies (up to 69.62 ± 4.18% in HEK cells). We confirmed that the intracellular reductive degradation produced mainly nontoxic fragments (cell survival 98.69 ± 4.79%). When we compared the polymers in >1,400 individual experiments to seven commercial transfection reagents in seven different cell lines, we found highly superior transfection efficacies and substantially lower toxicities. This renders reductive degradation a highly promising tool for the design of new transfection materials.
Keywords: biodegradable, polyethylenimine, nucleic acid, disulfide bond, transfection
Gene transfer into cells has become a standard procedure in life science research. Nonviral carriers have gained in importance in recent years because of their safety in handling and ease of application compared with viral vectors. Browsing databases such as PubMed or Chemical Abstracts reveals that almost every publication related to genetics, biochemistry, molecular biology, developmental biology, or neurology incorporated lipid- or polymer-based transfection as a pivotal tool for the investigation of various cellular processes. Therefore, it appears that the delivery of nucleic acids is a well established method for the transfection of mammalian cells, and, given the amount of research data that has been produced, one would assume that the procedures for nonviral transfection would be well optimized. A closer look at the contemporary literature, however, reveals that doubts seem justified. Contemporary transfection reagents are almost universally toxic because of their cationic or amphiphilic character (1–4).
The dilemma that we face is exemplified by considering the polymeric transfection agent poly(ethylene imine) (PEI). Since its introduction in 1995, PEI has been considered the gold standard for polymer-based gene carriers because of the relatively high transfection efficacy of its polyplexes (complex of nucleic acid and polymer) (5, 6). However, it has been shown that both the efficacy and toxicity of PEI are strongly correlated with its molecular weight (MW) as well as its structure (branched or linear: bPEI or lPEI, respectively) (7–17). Efficacy and adverse reactions seem thereby to be strongly associated. PEI is not the only material that suffers from this “malignant” correlation. Nonviral transfection seems like choosing between Scylla and Charybdis: either a high transfection efficacy, which is associated with a devastating toxicity, or a low efficacy, which does not affect the cell viability at all.
Successful nonviral gene transfection currently requires compromise to achieve a useful level of transfection efficiency while minimizing the toxicity. It seems therefore obvious that excessive toxicity can easily turn nonviral nucleic acid delivery into a selection process that may render different sets of cell populations of transfected vs. nontransfected and live vs. dead cells depending on the reagent in use and may hence make the reagent an undesirably significant factor for the readout of an experiment. Surprisingly, little attention has been given in this context to the fact that toxicity and efficacy are cell type-dependent, because most of the studies describing nonviral transfection reagents were conducted with only one or two cell types.
One of the goals of our study was to close this gap in knowledge and to concomitantly show that a stringent analytical methodology is indispensable to successfully assess efficacies and toxicities. With our work, we further demonstrate an alternative that breaks up this correlation between efficacy and toxicity. We synthesized polymers that are reductively degradable inside cells and diminish their charge density to levels that minimize the risk of interactions with cellular components and, therefore, exert significantly less toxicity. In contrast to ester- or β-aminoester-linked polyamines, for which hydrolysis half-lives of hours to days (18–23) are a severe handicap, disulfides degrade rapidly not only inside endolysomes but also to a significant extent in the cell cytoplasm (24, 25), because they do not depend on acid catalysis. To demonstrate the potential of these materials, it was first necessary to assess the ability of reductively biodegradable polymers to condense plasmid DNA into polyplexes that are suitable for cellular uptake. Furthermore, to confirm the reductively cleavable principle of the polymers, we observed the intracellular trafficking of polyplexes and investigated their transfection efficacy in cells with only a low reductive potential. To substantiate our expectations and to give clear instructions for the transfection of various cell lines, we evaluated the transfection efficacy and toxicity of our polymeric transfection reagents in seven different cell lines and compared them with a number of commercially available transfection reagents.
Results and Discussion
To obtain model polymers that allow for reductive degradation, we chose a simple synthesis approach: lPEI 2.6, 3.1, or 4.6 kDa were reversibly cross-linked by dithiodipropionic acid or cystine linkages to enable the polymer degradation in the presence of disulfide reducing agents (Fig. 1). Although intramolecular reactions within PEI molecules are possible, increases in molecular weight [see supporting information (SI) Fig. 7] suggests that it is not the dominating mechanism. We obtained a series of LRx-lPEIy and BCx-lPEIy polymers [LR and BC refer to the cross-linking reagents, Lomant's reagent and boc-cystine, respectively, x denotes the molar ratio cross-linker/lPEI, and y denotes the MW (kDa) of the lPEI component (SI Tables 2–4 and Fig. 1)]. We cross-linked single lPEI chains to produce a branched structure, which has been shown to more successfully form small polyplexes with narrow size distribution than the linear polymer (6). In the following, the results of the mechanistic studies are shown for two polymers, namely LR3-lPEI2.6 and BC8-lPEI2.6.
Fig. 1.
Schematic representation of the synthesis of biodegradable PEIs. LRx-lPEIy (A) and BCx-lPEIy (B) were prepared by cross-linking lPEI with 3,3′-dithiodipropionic acid di(N-succinimidyl ester) or a mixture of N, N′-bis-(tert-butoxycarbonyl)cystine and 4-(4,6-dimethoxy[1.3.5]triazin-2-yl)4-methyl-morpholiniumchlorid hydrate (DMT-MM), respectively, at various molar ratios.
As expected, the novel gene carriers complexed plasmid DNA to form nanoparticles in medium of low ionic strength (5% glucose) as well as at physiological salt conditions (150 mM sodium chloride). The polyplexes containing certain cross-linked lPEIs exhibited hydrodynamic diameters between 600 and 900 nm at NP ratios (ratio of nitrogens in polymer to phosphates in DNA) of 6–30 after exposure to serum-free cell culture medium as determined by laser light scattering (SI Fig. 8).
The capacity of the reductively biodegradable polymers to condense plasmid DNA into polyplexes that are appropriate for cellular uptake was additionally observed by confocal laser scanning microscopy (CLSM). The first polyplexes were detected inside CHO-K1 cells after 30 min of incubation (data not shown), and after 3 h, polyplexes were dispersed throughout the cytosol (Fig. 2A). Analysis of both whole cells and isolated nuclei with flow cytometry revealed the uptake of fluorescently labeled polyplexes by >95% of the cells and their corresponding nuclei after 6 h of incubation, irrespective of which cross-linked polymer was used for polyplex formation (Fig. 2B). The cell nuclei were isolated after incubation with polyplexes by a treatment of whole cells with hypotonic buffer after centrifugation at 600 × g. Control experiments with fluorescently labeled DNA only showed neither cellular nor nuclear uptake (data not shown). Although the percentage of cells showing polyplex uptake was consistent, the mean fluorescence intensity of whole cells that had incorporated DNA (and hence the absolute amount of plasmid DNA inside cells) depended on the polymer used for polyplex formation (Fig. 2C), but these subtle differences did not correspond to differences in the transfection efficiency. The mean fluorescence intensity of nuclei isolated after 6 h of incubation with polyplexes appeared steady at a value of 50 (Fig. 2C; dimensionless value), indicating that an equal amount of DNA had been transported to the nucleus, irrespective of the polymer used for polyplex formation. These results suggest that the nuclear transport may be the bottleneck in nonviral gene delivery, rather than the cellular uptake.
Fig. 2.
Uptake of polyplexes into CHO-K1 cells. (A) Observation of LR3-lPEI2.6-polyplexes formed at NP 18 in CHO-K1 cells 3 h after transfection by CLSM. The larger green dots (arrow) were polyplex aggregates that were not taken up by cells. The picture is an overlay of transmitted light and fluorescence image. (Scale bar, 10 μm.) (B and C) Uptake of polyplexes prepared with YOYO-1-labeled DNA and LR3-lPEI2.6 or BC8-LPEI2.6 in whole cells (■) or nuclei (□) after 6 h, as determined by flow cytometry. (B) Fluorescent cells/nuclei with polyplexes indicates the percentage of cells/nuclei that show a fluorescence because of intracellular/intranuclear YOYO-1-labeled DNA. (C) The mean fluorescence intensity is represented from those cells or nuclei that have incorporated YOYO-1-labeled DNA. Statistically significant differences of pairs are denoted by ★ (P < 0.01).
The disulfide cross-linked lPEIs were designed to exploit the differences in the degradation potential at different locations outside and inside cells, providing a clear opportunity to design vectors that are stable in the plasma but dissociate within the endolysosomal compartment or the cytoplasm. The concentration of extracellular glutathione (GSH) is usually 100–1,000 times less than intracellular GSH and hence favors the maintenance of disulfide bonds (24). In our initial experiments, we confirmed the destabilization of the cross-linked lPEI-derived polyplexes in the presence of disulfide reducing agents, indicating the degradability of the polymer via redox reactions (data not shown). Therefore, we were interested in the response of cross-linked lPEI-mediated gene transfer to changes in the reducing/oxidizing environment of cells. To this end, 1 h before transfection, CHO-K1 cells were treated with 50 μM duroquinone, an agent that oxidizes NADPH/H+, thereby lowering its reducing potential (26, 27) (see SI Fig. 9). Lower intracellular NADPH/H+ levels not only reduced the number of EGFP-positive cells significantly compared with untreated cells (Fig. 3A), but the amount of expressed EGFP also declined (Fig. 3B) after both 6 and 24 h. However, the intracellular NADPH/H+ concentration appears to recover with time, because after 24 h the transfection efficiency was reduced only 1.1- to 1.3-fold as compared with 1.7-fold after 6 h, depending on the polymer used for polyplex formation. When transfecting cells with low NADPH/H+ levels with lPEI polyplexes as a control, the efficacy was not affected at any time point (data not shown). Therefore, from the literature and our results, we conclude that the polyplexes may at least partially undergo an NADPH/H+-dependent, and therefore GSH-dependent, reduction inside the cell (28–30). The data in Fig. 3 also show that more than two-thirds of the transfected cells have already commenced EGFP production after 6 h, a very early time point that has not been reported for other polymers so far. Furthermore, the level of GSH in CHO-K1 cells appeared to be sufficient to activate the carriers at the tested NP ratios: A pretreatment of CHO-K1 cells with 1 and 5 mM GSH-monomethylester (GSH MME) to increase the cellular GSH pool (31) did not improve the gene expression of polyplexes that were built with biodegradable PEI (see SI Fig. 10).
Fig. 3.
Effect of reduced intracellular NADPH/H+ concentration, and hence reducing potential, on the transfection efficiency and mean fluorescence intensity in CHO-K1 cells. Polyplexes were prepared with either LR3-lPEI2.6 (■) or BC8-lPEI2.6 (□). Duroquinone at a concentration of 50 μM was added to the cells 1 hour before transfection. (A) Values represent the EGFP-positive cells as means ± SD as determined by flow cytometry 6 h and 24 h after transfection. (B) Values represent the corresponding mean fluorescence intensity of EGFP-positive cells as determined by flow cytometry 6 and 24 h after transfection. Statistically significant differences compared with untreated (without duroquinone) cells are denoted by  (P < 0.05) or ★ (P < 0.01).
(P < 0.05) or ★ (P < 0.01).
Observing the intracellular trafficking by CLSM revealed that many of the biodegradable PEI-derived polyplexes were colocalized with acidic organelles after 3 h of incubation in CHO-K1 cells (Fig. 4 A and B, arrows). One may expect that this sojourn in the lysosomes may result in damage to the transported plasmid DNA. However, by supplementing with 5 mM of sucrose, a lysosomotropic agent that accelerates and enhances the escape of polyplexes from the acidic vesicles (32, 33), we could disprove this hypothesis. The transfection efficiency with and without sucrose was not significantly different (Fig. 4C). Therefore, one could presume that disulfide bonds begin to degrade in the acidic environment of lysosomes, as is described for proteins (34), and that in the reductive environment of the cytosol, the degradation process proceeds.
Fig. 4.
Intracellular trafficking of polyplexes. (A and B) Tracking of Alexa Fluor 543-labeled plasmid DNA (shown in red) complexed with LR3-lPEI2.6 (A) or BC8-lPEI2.6 (B) cells by CLSM. The acidic organelles of CHO-K1 cells were stained with quinacrine (depicted in turquoise). Most polyplexes were colocalized with acidic vesicles, some examples are indicated by arrows. The larger red dots were polyplex aggregates that were not taken up by cells. Pictures are an overlay of transmitted light and fluorescence images. (Scale bars, 10 μm.) (C) The effect of the lysosomotropic agent sucrose at 5 mM (white bars) on EGFP expression of polyplexes prepared with LR3-lPEI2.6 or BC8-lPEI2.6 compared with transfection without sucrose (black bars) determined by flow cytometry. No statistically significant differences could be found with and without sucrose.
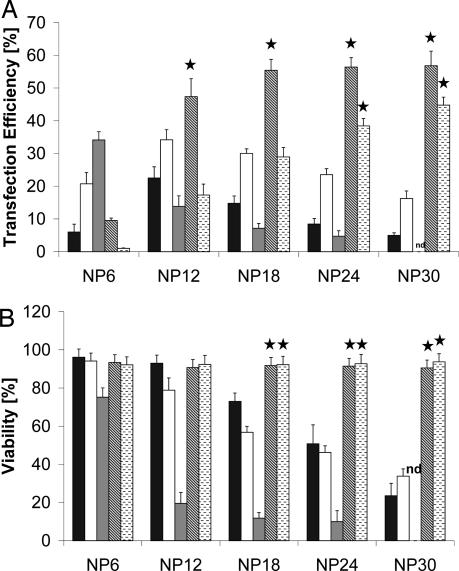
After demonstrating that our cross-linked polymers can be used to form polyplexes, that they are activated in the reductive environment of the cell, are transported to the nucleus, and have the power to efficiently transfect CHO-K1 cells, we compared LR3-lPEI2.6 and BC8-lPEI2.6 with the standard polymer-based transfection reagents, namely bPEI 25 kDa, lPEI 25 kDa, and lPEI 22 kDa (ExGen) in CHO-K1 cells (Fig. 5). We achieved an outstanding maximum efficacy of ≈60% in CHO-K1 cells using LR3-lPEI2.6 as the gene transfer vehicle, which was by nearly 2-fold higher than the best of the polymer standards (ExGen at NP 6 or lPEI 22 kDa at NP 12) (Fig. 5A). Compared with the other polymers, LR3-lPEI2.6 and BC8-lPEI2.6 reached their maximum efficacy at relatively high NP ratios (NP 18 and NP 30, respectively). But despite the high polymer amount applied for polyplex formation, >90% of cells survived the transfection process at all NP ratios (Fig. 5B).
Fig. 5.
Comparison with standard polymer-based transfection reagents. Transfection efficiency (A) and corresponding cell viability (B) of bPEI 25-kDa (black bars), lPEI 22 kDa (white bars), ExGen (gray bars), LR3-lPEI2.6 (diagonally hatched bars), and BC8-lPEI2.6 (horizontally hatched bars) complexed with pEGFP-N1 in CHO-K1 cells as determined by flow cytometry. NP ratios at which biodegradable PEIs were statistically significant different compared with all other polymers are denoted by ★ (P < 0.01).
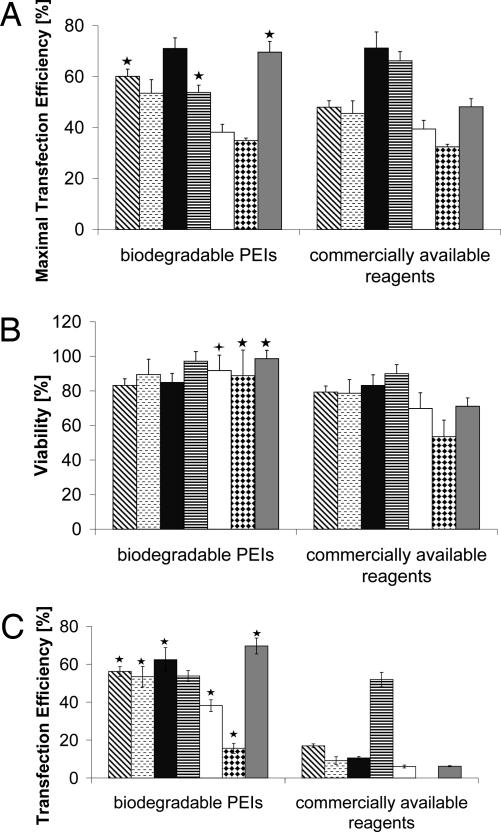
These very promising results prompted us to expand our transfection study. We chose various lipid- and polymer-based, commercially available transfection reagents (Table 1) and compared them with seven of our best cross-linked lPEIs in CHO-K1, COS-7, NIH/3T3, HepG2, HCT116, HeLa, and HEK-293 cells. For each reagent, we evaluated the optimal DNA/reagent ratio in each cell line (for the commercially available reagents according to the supplier's protocol), in terms of the transfection efficiency and cell viability. Altogether, we tested ≈340 various transfection conditions in the above-mentioned seven cell lines and obtained a detailed knowledge of the transfection efficacy and corresponding cell viability of the most commonly used transfection reagents in comparison with our cross-linked polymers. The maximal transfection efficiency when cross-linked lPEIs were used as gene delivery vehicles was as high or significantly higher than the commercially available reagents in six of seven cell lines, reaching efficiencies between 40% and 70% (Fig. 6A). In addition, the corresponding cell viability was as high or even significantly higher in three cell lines, giving our polymers a clear superiority (Fig. 6B). Only in HepG2 cells was the transfection efficiency ≈13% higher when a commercially available transfection reagent was used, in this case, Lipofectamine (66.23 ± 2.23% vs. 53.79 ± 2.86%). But this higher transfection efficacy was also accompanied by a lower cell viability (89.93 ± 6.16% vs. 97.21 ± 5.56%). The most important point in our study is that if one requires a cell viability >90% after the transfection process, our biodegradable PEIs were greatly superior to the other lipid- or polymer-based transfection reagents (Fig. 6C) (for detailed information on which transfection reagent reached the maximal efficiency, please refer to SI Table 5). In all cell lines tested, the efficacy was 5- to 7-fold higher than that of the commercially available reagents when transfection was conducted under conditions that maintained a cell viability >90%. Moreover, in contrast to the biodegradable PEIs, no commercially available transfection reagent was able to maintain a cell viability of >90% in HeLa cells after the transfection process.
Table 1.
Commercially available transfection reagents used in our study
| Transfection reagent | Supplier | Description |
|---|---|---|
| PolyFect | Qiagen | Polycationic dendrimer, branched structure |
| SuperFect | Qiagen | Polycationic dendrimer, branched structure |
| Lipofectamine | Invitrogen | Liposome formulation |
| TransIT-LT1 | Mirus Bio | Liposome formulation |
| Effectene | Qiagen | Nonliposomal lipid formulation with an enhancer |
| FuGENE 6 | Roche | Blend of lipids in 80% ethanol |
| JetPEI | Polyplus | Linear PEI 22 kDa |
Fig. 6.
Comparison of biodegradable PEIs with commercially available transfection reagents. (A and B) EGFP-positive cells expressed as maximal transfection efficiency (A) and corresponding cell viability (B) of various biodegradable PEIs and commercially available transfection reagents complexed with pEGFP-N1 in (from left to right) CHO-K1 (diagonally hatched bars), COS-7 (horizontally hatched bars), NIH/3T3 (black bars), HepG2 (horizontally striped bars), HCT116 (white bars), HeLa (diamond-hatched bars), and HEK-293 (gray bars) cells as determined by flow cytometry. (C) Transfection efficiency under conditions where cell viability is >90%. Statistically significant differences of biodegradable PEIs compared with other transfection reagents are denoted by  (P < 0.05) or by ★ (P < 0.01).
(P < 0.05) or by ★ (P < 0.01).
We have synthesized potentially biodegradable polymers. We adjusted the degradability to the time scale of cellular events such as endocytosis. This was achieved by taking advantage of disulfide bonds that are cleaved by redox reactions inside cells. By a transfection screening in different cell lines and a comparison with commercially available transfection reagents, we identified promising candidates for gene delivery. We conclude that use of the reductive principle in combination with a low-MW polycationic starting material seems to be a promising strategy for the delivery of macromolecular, negatively charged substances into cells. In contrast to other studies that investigated a cross-linking of either low-MW branched (35) or linear (20) polyamines via disulfide bonds, our choice of lPEI 2.6, 3.1 or 4.6 kDa as linear starting material not only entailed a much higher cell viability compared with the commercially available standard transfection reagents but also superior gene transfer efficacy. We could show that it is possible to achieve a high transfection efficiency (≈60%) while maintaining a high cell viability. Nonviral transfection no longer requires choosing between Scylla and Charybdis, and we can hope that polycationic polymers will indeed find their entry into clinics.
Materials and Methods
Synthesis.
lPEI.
lPEIs with Mn of 2.6, 3.1, and 4.6 kDa were synthesized by ring-opening polymerization of 2-ethyl-2-oxazoline (Sigma–Aldrich, Deisenhofen, Germany) and acid-catalyzed hydrolysis of the corresponding poly(2-ethyl-2-oxazoline) as described (36).
LRx-lPEIy.
Anhydrous lPEI was dissolved in 12 ml of dichlormethane (DCM) and heated to 50°C. The 3,3′-dithiopropionic acid di-(N-succinimidyl ester) (Lomant's reagent, LR) (Fluka, Buchs, Switzerland) was dissolved in 5 ml of DCM (specific amounts listed in SI Table 2). The clear solution was added drop-wise to the lPEI solution under vigorous stirring, and the mixture was kept at 50°C overnight.
BCx-lPEIy.
Anhydrous lPEI, 4-(4,6-dimethoxy [1.3.5] triazin-2-yl) 4-methylmorpholiniumchlorid hydrate (DMT-MM) (Acros Organics, Geel, Belgium), and N, N′-bis-(tert-butoxycarbonyl) cystine (boc-cystine, BC) (Advanced Chem Tech, Louisville, KY) were each dissolved in 10 ml, 6 ml, and 4 ml of ethanol, respectively (specific amounts are listed in SI Table 3). The lPEI and BC solutions were transferred into the glass tube of a parallel synthesis block and mixed by vigorous shaking. The DMT-MM solution was then added to the clear mixture, and the mixture was vigorously shaken at room temperature overnight.
For both polymer types, the volatiles were then removed under reduced pressure, the yellow waxen residue was dissolved in 2 M HCl, and the cross-linked lPEI was precipitated with concentrated aqueous sodium hydroxide. The white gel-like residue was washed with water until the supernatant became neutral. For analysis, the purified amine bases were dried at 70°C under reduced pressure. The amine base was transformed into the corresponding hydrochloride with 2 M HCl to facilitate water solubility.
Formation of Polyplexes.
Polyplexes were prepared at NP ratios of 6, 12, 18, 24, and 30. Polyplexes were formed by mixing 2 μg of plasmid DNA (pEGFP-N1) (Clontech, Germany) with the appropriate amount of polymer solution, each diluted in 150 mM NaCl. The resulting polyplexes were incubated for 20 min at room temperature before use. The complexes with commercially available transfection reagents were prepared according to the supplier's protocol.
Functionality of Polymers in Vitro: Transfection and Cytotoxicity Experiments.
For gene transfer studies, various cell lines (CHO-K1, COS-7, NIH/3T3, HepG2, HCT116, HeLa, and HEK-293) were grown in 24-well plates at an initial density of 38,000–50,000 cells per well. Experiments were performed as described previously and evaluated by flow cytometry using a FACSCalibur (Becton–Dickinson, Heidelberg, Germany) (37). The cell population referred to the number of whole cells (dead or live) after the transfection process, in contrast to the cellular debris. EGFP-positive cells were detected by using a 515- to 545-nm band-pass filter, whereas the propidium iodide (Sigma–Aldrich) emission was measured with a 670-nm long-pass filter. The mean fluorescence intensity was determined by using the EGFP-positive cells. The transfection efficiency and cell viability were calculated as follows: First, the cell population of the samples was normalized to the cell population of untreated cells as well as the cell viability. In a next step, the number of EGFP-positive or propidium iodide-negative cells were referred to the calculated cell population, expressing the transfection efficiency or cell viability, respectively.
Cellular and Nuclear Uptake of Polyplexes.
YOYO-1 (Molecular Probes, Eugene, OR) -labeled nucleic acid was used to monitor polyplex delivery as described (37, 38). Cells were incubated with polyplexes for 6 h, followed by flow-cytometry analysis of whole cells and nuclei. Briefly, the nuclei were isolated as follows: cells were incubated for 5 min in a buffer with low salt concentration (Tris·HCl 10 mmol/liter, KCl 60 mmol/liter, and EDTA 1 mmol/liter). Thereafter, cells were treated with the same buffer containing a nonionic detergent (Tergitol Type Nonidet P-40, 0.5%) (Sigma–Aldrich) and protease inhibitors (Roche, Mannheim, Germany). Nuclei were pelleted at 600 × g, rinsed three times with nuclei isolation buffer without Tergitol Type Nonidet P-40, and then resuspended in PBS. The percentage of cells and nuclei that have taken up polyplexes and their mean fluorescence intensity were determined by flow cytometry after excitation with a 488-nm argon laser and detection with a 515- to 545-nm band-pass filter.
Intracellular Trafficking: Confocal Laser Scanning Microscopy (CLSM).
A Zeiss Axiovert 200 M microscope coupled to a Zeiss LSM 510 scanning device (Zeiss, Oberkochen, Germany) was used for CLSM experiments. The inverted microscope was equipped with a Plan-Apochromat ×63 objective. Cells were plated in an eight-well Lab-Tek chambered coverglass (Nunc, Wiesbaden, Germany), and measurements were directly performed in each well at 37°C. The thickness of the optical sections was between 0.7 and 1.2 μm.
For the investigation of the uptake of polyplexes, YOYO-1-labeled plasmid DNA was used and depicted in green. The fluorescent dye was excited with a 488-nm argon laser, and images were taken by using a band-pass filter of 505–530 nm in the single-track mode at the indicated time after the addition of the polyplexes.
For the detection of polyplexes in acidic compartments of the cell, Alexa Fluor 546-labeled plasmid DNA was applied. Polyplexes and quinacrine mustard (Sigma–Aldrich) at a concentration 10−6 M were added to the cells at the same time. Quinacrine mustard was excited at 458 nm, the fluorescence was imaged by using a band-pass filter of 475–525 nm, and is depicted in turquoise. Alexa Fluor (Molecular Probes) -labeled DNA was excited at 543 nm, and the fluorescence was recorded with a 560-nm long-pass filter and is shown in red. A mixture of both colors indicates a close proximity and therefore an interaction. Images were taken in the multitracking modus.
Statistical Analysis.
All measurements were collected (n = 3–6) and expressed as means ± SD. Single ANOVA was used in conjunction with a multiple comparison test (Tukey test) to assess the statistical significance.
Supplementary Material
Acknowledgments
This work was supported, in part, by Deutsche Forschungsgemeinschaft (DFG) Grant BR 3566/1-1. We thank Allison Dennis (Georgia Institute of Technology, Atlanta, GA) for the careful revision of this manuscript.
Abbreviations
- PEI
poly(ethylene imine)
- lPEI
linear PEI
- bPEI
branched PEI
- LRx-lPEIy
lPEI cross-linked with Lomant's reagent
- BCx-lPEIy
lPEI cross-linked with boc-cystine.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0703882104/DC1.
References
- 1.Lv H, Zhang S, Wang B, Cui S, Yan J. J Control Release. 2006;114:100–109. doi: 10.1016/j.jconrel.2006.04.014. [DOI] [PubMed] [Google Scholar]
- 2.Wagner E, Kloeckner J. Adv Polym Sci. 2006;192:135–173. [Google Scholar]
- 3.Park TG, Jeong JH, Kim SW. Adv Drug Deliv Rev. 2006;58:467–486. doi: 10.1016/j.addr.2006.03.007. [DOI] [PubMed] [Google Scholar]
- 4.Hunter AC. Adv Drug Deliv Rev. 2006;58:1523–1531. doi: 10.1016/j.addr.2006.09.008. [DOI] [PubMed] [Google Scholar]
- 5.Lungwitz U, Breunig M, Blunk T, Gopferich A. Eur J Pharm Biopharm. 2005;60:247–266. doi: 10.1016/j.ejpb.2004.11.011. [DOI] [PubMed] [Google Scholar]
- 6.Neu M, Fischer D, Kissel T. J Gene Med. 2005;7:992–1009. doi: 10.1002/jgm.773. [DOI] [PubMed] [Google Scholar]
- 7.Godbey WT, Wu KK, Mikos AG. J Biomed Mater Res. 1999;45:268–275. doi: 10.1002/(sici)1097-4636(19990605)45:3<268::aid-jbm15>3.0.co;2-q. [DOI] [PubMed] [Google Scholar]
- 8.Wightman L, Kircheis R, Rossler V, Carotta S, Ruzicka R, Kursa M, Wagner E. J Gene Med. 2001;3:362–372. doi: 10.1002/jgm.187. [DOI] [PubMed] [Google Scholar]
- 9.Schaffer DV, Fidelman NA, Dan N, Lauffenburger DA. Biotechnol Bioeng. 2000;67:598–606. doi: 10.1002/(sici)1097-0290(20000305)67:5<598::aid-bit10>3.0.co;2-g. [DOI] [PubMed] [Google Scholar]
- 10.Bettinger T, Carlisle RC, Read ML, Ogris M, Seymour LW. Nucleic Acids Res. 2001;29:3882–3891. doi: 10.1093/nar/29.18.3882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Reschel T, Konak C, Oupicky D, Seymour LW, Ulbrich K. J Control Release. 2002;81:201–217. doi: 10.1016/s0168-3659(02)00045-7. [DOI] [PubMed] [Google Scholar]
- 12.Thomas M, Klibanov AM. Proc Natl Acad Sci USA. 2002;99:14640–14645. doi: 10.1073/pnas.192581499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kunath K, Von Harpe A, Fischer D, Petersen H, Bickel U, Voigt K, Kissel T. J Control Release. 2003;89:113–125. doi: 10.1016/s0168-3659(03)00076-2. [DOI] [PubMed] [Google Scholar]
- 14.Bieber T, Meissner W, Kostin S, Niemann A, Elsasser HP. J Control Release. 2002;82:441–454. doi: 10.1016/s0168-3659(02)00129-3. [DOI] [PubMed] [Google Scholar]
- 15.Plank C, Mechtler K, Szoka FCJ, Wagner E. Hum Gene Ther. 1996;7:1437–1446. doi: 10.1089/hum.1996.7.12-1437. [DOI] [PubMed] [Google Scholar]
- 16.Ahn CH, Chae SY, Bae YH, Kim SW. J Control Release. 2004;97:567–574. doi: 10.1016/j.jconrel.2004.04.002. [DOI] [PubMed] [Google Scholar]
- 17.Godbey WT, Wu KK, Mikos AG. Biomaterials. 2000;22:471–480. doi: 10.1016/s0142-9612(00)00203-9. [DOI] [PubMed] [Google Scholar]
- 18.Kim YH, Park JH, Lee M, Kim YH, Park TG, Kim SW. J Control Release. 2005;103:209–219. doi: 10.1016/j.jconrel.2004.11.008. [DOI] [PubMed] [Google Scholar]
- 19.Forrest ML, Koerber JT, Pack DW. Bioconjug Chem. 2003;14:934–940. doi: 10.1021/bc034014g. [DOI] [PubMed] [Google Scholar]
- 20.Kloeckner J, Wagner E, Ogris M. Eur J Pharm Sci. 2006;29:414–425. doi: 10.1016/j.ejps.2006.08.002. [DOI] [PubMed] [Google Scholar]
- 21.Kloeckner J, Bruzzano S, Ogris M, Wagner E. Bioconjug Chem. 2006;17:1339–1345. doi: 10.1021/bc060133v. [DOI] [PubMed] [Google Scholar]
- 22.Thomas M, Lu JL, Zhang C, Chen J, Klibanov AM. Pharm Res. 2007;24:1564–1571. doi: 10.1007/s11095-007-9279-3. [DOI] [PubMed] [Google Scholar]
- 23.Thomas M, Ge Q, Lu JJ, Chen J, Klibanov AM. Pharm Res. 2005;22:373–380. doi: 10.1007/s11095-004-1874-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schafer FQ, Buettner GR. Free Radical Biol Med. 2001;30:1191–1212. doi: 10.1016/s0891-5849(01)00480-4. [DOI] [PubMed] [Google Scholar]
- 25.Saito G, Swanson JA, Lee KD. Adv Drug Deliv Rev. 2003;55:199–215. doi: 10.1016/s0169-409x(02)00179-5. [DOI] [PubMed] [Google Scholar]
- 26.Chernyak BV, Dedov VN, Chernyak VY. FEBS Lett. 1995;365:75–78. doi: 10.1016/0014-5793(95)00411-2. [DOI] [PubMed] [Google Scholar]
- 27.Balakirev MY, Zimmer G. Arch Biochem Biophys. 1998;356:46–54. doi: 10.1006/abbi.1998.0738. [DOI] [PubMed] [Google Scholar]
- 28.Read ML, Bremner KH, Oupicky D, Green NK, Searle PF, Seymour LW. J Gene Med. 2003;5:232–245. doi: 10.1002/jgm.331. [DOI] [PubMed] [Google Scholar]
- 29.Balakirev M, Schoehn G, Chroboczek J. Chem Biol. 2000;7:813–819. doi: 10.1016/s1074-5521(00)00030-2. [DOI] [PubMed] [Google Scholar]
- 30.Carlisle RC, Etrych T, Briggs SS, Preece JA, Ulbrich K, Seymour LW. J Gene Med. 2004;6:337–344. doi: 10.1002/jgm.525. [DOI] [PubMed] [Google Scholar]
- 31.Levy EJ, Anderson ME, Meister A. Proc Natl Acad Sci USA. 1993;90:9171–9175. doi: 10.1073/pnas.90.19.9171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ciftci K, Levy RJ. Int J Pharm. 2001;218:81–92. doi: 10.1016/s0378-5173(01)00623-8. [DOI] [PubMed] [Google Scholar]
- 33.Wattiaux R, Wattiaux-De Coninck S, Rutgeerts MJ, Tulkens P. Nature. 1964;203:757–758. doi: 10.1038/203757a0. [DOI] [PubMed] [Google Scholar]
- 34.Collins DS, Unanue ER, Harding CV. J Immunol. 1991;147:4054–4059. [PubMed] [Google Scholar]
- 35.Gosselin MA, Guo W, Lee RJ. Bioconjug Chem. 2001;12:989–994. doi: 10.1021/bc0100455. [DOI] [PubMed] [Google Scholar]
- 36.Lungwitz U. Regensburg, Germany: Univ of Regensburg; 2006. pp. 85–112. PhD thesis. [Google Scholar]
- 37.Breunig M, Lungwitz U, Liebl R, Fontanari C, Klar J, Kurtz A, Blunk T, Goepferich A. J Gene Med. 2005;7:1287–1298. doi: 10.1002/jgm.775. [DOI] [PubMed] [Google Scholar]
- 38.Breunig M, Lungwitz U, Liebl R, Klar J, Obermayer B, Blunk T, Goepferich A. Biochim Biophys Acta. 2007;1770:196–205. doi: 10.1016/j.bbagen.2006.10.005. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.