Abstract
We study theoretically the feasibility of using transverse electronic transport within a nanopore for rapid DNA sequencing. Specifically, we examine the effects of the environment and detection probes on the distinguishability of the DNA bases. We find that the intrinsic measurement bandwidth of the electrodes helps the detection of single bases by averaging over the current distributions of each base. We also find that although the overall magnitude of the current may change dramatically with different detection conditions, the intrinsic distinguishability of the bases is not significantly affected by pore size and transverse field strength. The latter is the result of very effective stabilization of the DNA by the transverse field induced by the probes, so long as that field is much larger than the field that drives DNA through the pore. In addition, the ions and water together effectively screen the charge on the nucleotides, so that the electron states participating in the transport properties of the latter ones resemble those of the uncharged species. Finally, water in the environment has negligible direct influence on the transverse electrical current.
INTRODUCTION
Now that the first full human genome has been sequenced (1,2), new uses of sequencing in medicine seem to be on the horizon. One of the most ambitious goals is to be able to sequence an entire human genome in less than an hour for ∼US$1000, allowing for every-day sequencing in medicine (3).
Several intriguing sequencing methods (4–8) have been proposed that would lead us closer to achieving this goal. Many of these methods are based on the idea of translocating DNA through a nanopore (4,9–26). In their pioneering work Kasianowicz et al. demonstrated that DNA can be pulled through a biological nanopore roughly the size of the DNA itself (4). The translocation of the DNA can be detected by measuring a blockade current when ions are partially prevented from entering the pore. More recent experiments have been based on solid state pores made of silicon-based materials (22–29). The advantage of solid state pores is that it may be possible to embed single molecule sensors in the pore to measure various physical properties of the DNA during translocation, allowing the DNA to be directly sequenced by detecting specific signatures of individual bases.
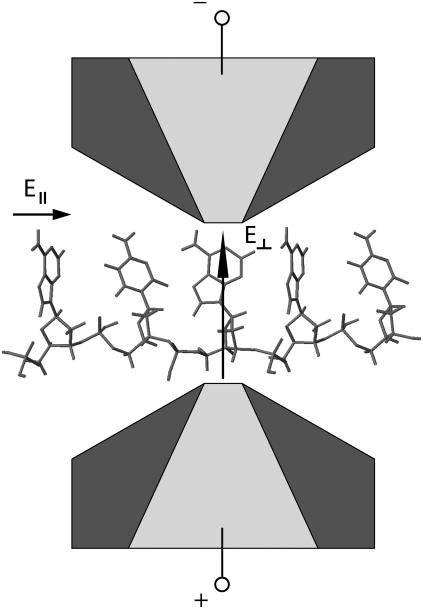
Previous work has shown the potential for sequencing DNA by measuring a transverse electronic current (6) as single-stranded DNA (ss-DNA) translocates through a nanopore (8). The concept envisions a nanopore device with embedded nanoscale gold electrodes, as schematically shown in Fig. 1. In these proof-of-concept calculations, we assume gold electrodes. One can also envision electrodes made out of other materials, such as carbon nanotubes. Change of material will, for example, change the coupling in between the DNA and electrodes. The main conclusions drawn in this and previous work (8) will, however, not change because the calibration of the device (as discussed later) will take into account the microscopic details of the nanopore and electrodes. Operating such a device with a transverse field, E⊥, (the field that drives the electronic current) greater than the longitudinal pulling field, E‖, (i.e., the field that drives DNA translocation) stabilizes the motion of the nucleotide between the electrodes (8). This creates a very desirable situation where structural fluctuations (the most important source of intrinsic noise (8)) are reduced to such a level that distributions of currents for each base, while still overlapping, are different enough to allow for high statistical distinguishability between the different bases. In previous work, however, it was assumed that each measurement could be performed almost instantaneously. This is just a theoretical assumption, and a more realistic treatment of the measurement probes needs to be taken into account.
FIGURE 1.
Schematic of a nanopore (dark gray) with embedded electrodes (light gray) attached to the edges of the pore. The electrodes are used to inject a current through the nucleotides in the direction transverse to the backbone. The electronic signature can then be used to sequence the DNA.
In this article we will examine the fact that, contrary to naive expectation, the measurement bandwidth of the electrical probes reduces these overlapping distributions into sharply peaked and disjoint distributions, rather than just limiting the sampling rate. Therefore, assuming that no external sources of noise are present other than shot, thermal, and structural fluctuation noise, a single current measurement may be sufficient to distinguish each individual base. Thus by measuring the current as the nucleotides translocate through the pore, the DNA may be accurately sequenced in extremely short timescales.
In Lagerqvist et al. (8) it was estimated that the raw sequencing throughput of a single 12.5-Å pore, operating with a 1-V transverse bias, could be as high as 3,000,000,000 bases in 7 h. In a real device, however, the pore diameter will not be easy to control and it may not be possible to operate the device at high transverse biases. Also, ions and water in the pore are additional sources of noise on top of the structural fluctuations of the ss-DNA. In this article we will examine in detail these effects on the distinguishability of the bases. To do this, we use all-atom molecular dynamics simulations coupled with quantum mechanical current calculations. We find that although the overall magnitude of current can change dramatically, the intrinsic distinguishability of the bases is not significantly affected by pore size and transverse field strength. The latter is the result of very effective stabilization of the DNA by the transverse field, E⊥, so long as that field is much larger than the pulling field, E‖. In addition, the ions and water together effectively screen the charge on the nucleotides, so that the electron states participating in the transport properties of nucleotides in solution resemble those of uncharged species. Finally, water in the environment has a negligible direct influence on the electrical current through the DNA.
SETUP AND METHODS
To calculate the current, we use a scattering approach with a tight-binding Hamiltonian to represent the electronic structure of the system. For each carbon, nitrogen, oxygen, and phosphorus atom s−, px−, py−, and pz− orbitals are used, while s-orbitals are used for hydrogen and gold. We take the Fermi level to be that of bulk gold, which is identical to that of the extended molecule. However, for the biases we consider, the current calculations are relatively insensitive to the exact position of the Fermi level, as it falls within the highest occupied molecular orbital (HOMO)-lowest unoccupied molecular orbital (LUMO) gap. (8) The retarded Green's function, 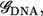 can be written as
can be written as
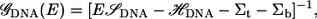 |
(1) |
where 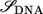 and
and 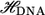 are the overlap and the Hamiltonian matrices and Σt(b) are the self-energy terms that describe the interaction in between the leads and the DNA. For a given
are the overlap and the Hamiltonian matrices and Σt(b) are the self-energy terms that describe the interaction in between the leads and the DNA. For a given 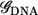 the transmission coefficient can then be calculated as
the transmission coefficient can then be calculated as
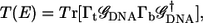 |
(2) |
where Γt(b) = i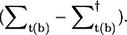 Finally the current in between two electrodes is given by
Finally the current in between two electrodes is given by
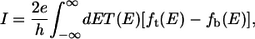 |
(3) |
where ft(b) is the Fermi-Dirac function of top (bottom) electrode (6). Room temperature is assumed throughout this article and we have assumed that the voltage drops uniformly in the space between the DNA molecule and electrodes. Unless otherwise stated, water and ions are not directly included in the Hamiltonian for transport. Below we do, however, discuss the effect of water and ions on the HOMO-LUMO gap. We will consider transverse biases small enough that we do not expect chemical reactions to occur, and definitely smaller than the electrolysis threshold for water of ∼1.2 V.
The justification for using a tight-binding approach is twofold. First, the current through the junction is strongly dependent upon the coupling of the nucleotide (in the junction region) to the electrodes. Thus, the important quantity to reproduce is the molecular orbitals of the bases and backbone relative to the electrodes to get the DNA-electrode coupling. We find that the tight-binding calculations reproduce very well the molecular orbitals of charge-neutral nucleotides when compared to density functional theory (DFT) calculations. We have performed DFT calculations with DZVP basis set and GGA functional B88-PW91. The HOMO, LUMO, and nearby electronic levels have similar molecular wavefunctions in both the DFT and tight-binding calculations. Second, the presence of a nearby counterion (see below) in addition to the nearby water effectively brings the HOMO-LUMO gap of the nucleotide close to the one of the charge neutral case (although the positioning of the gold Fermi level may change with respect to these states), whereas the molecular orbitals of the bases remain nearly identical regardless of the presence of the counterion. However, the position of the counterion is not fixed, but it fluctuates around the charge on the backbone at an average distance of 6.5 Å. This will cause additional fluctuations in the current across the junction. Nonetheless, since the current value is mainly controlled by the DNA-electrode coupling, these fluctuations are less likely to contribute than those due to structural motion.
To analyze the dynamics of the DNA strand as it propagates through the pore, classical molecular dynamics simulations have been performed with the package NAMD2 (30). For the interaction in between the DNA, water, and ions the CHARMM27 force field (31,32) was used, while UFF parameters (33) were used for interactions between the Si3N4 membrane and other atoms. The location of the Si3N4 atoms was assumed to be frozen throughout the simulation. This also prevents the system from drifting due to the external electric field. When integrating over time, a time step of 1 fs was used and the temperature of the system was kept constant at 300 K throughout the whole simulation by coupling all but the hydrogen atoms to a thermal bath with a Langevin damping constant of 0.2 ps−1. The van der Waals interactions were gradually cut off starting at 10 Å from the atom until reaching zero interaction 12 Å away. The DNA strand was driven through the pore with a large electric field of 6 kcal/(mol Å e) to achieve feasible simulation times. This field is larger than the ones used experimentally and results in a negligible stabilizing electric field, E⊥. Thus, for sampling current distributions of the nucleotides, we turn off E‖ when a base is aligned in between the electrodes. This gives an adequate representation of the structural fluctuations when 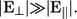 One of our main conclusions is that the device has to be operated in this regime for sequencing to be possible.
One of our main conclusions is that the device has to be operated in this regime for sequencing to be possible.
The pore analyzed in this article is made up of a 24-Å thick silicon nitride membrane in the β-phase (34). The experimental etching of the pore is mimicked by removing the atoms inside a double conical shape with a minimum diameter of 1.4 nm located at the center of the membrane and with an outer diameter of 2.5 nm (see Fig. 1 for a schematic representation of the pore). This corresponds to a cone angle of 20° for each of the cones. In actual experimental realizations of nanopores such geometry may not always be realized. For the conclusions of this article this would not matter as the calibration of the device would correct for geometrical imperfections in the pore, as discussed later in the article. A sphere, with a radius of 6 nm, of TIP3 water (35) is placed around the pore and 1 M of potassium and chlorine ions are added. Spherical boundary conditions are used under NVT conditions and the size of the membrane is chosen slightly smaller than the water sphere, such that water molecules can just pass through on the sides of the membrane. Finally, a single-stranded poly(X) (where X is A, T, C, or G) molecule is generated by removing one strand from a helical double stranded polynucleotide. At the initial time of the simulation, this molecule is placed parallel to the pore such that the tip of the single strand is just inside the pore.
RESULTS AND DISCUSSIONS
We take a two-step computational approach to examine the issues described above: 1), molecular dynamics simulations are used to sample real time atomistic coordinates of the DNA, water, and ions in a prototypical Si3N4 nanopore, and 2), these coordinates are used in quantum mechanical calculations to find the current across the nanostructure (see also the section Setup and Methods).
We discuss our results in three subsections: In “Setup”, we examine a larger pore diameter and weaker transverse field than in Lagerqvist et al. (8); in “Nucleotide Stabilization”, we look at the current distribution with varying transverse field strength; in “Influence of the Environment”, we look at the role of the environment on the electronic conductance.
Setup
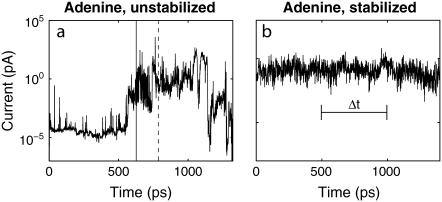
Fig. 2 a shows the current as a function of time as a ss-DNA strand made up of 15 adenine bases propagates through a 14-Å diameter pore with embedded electrodes. The distance is measured from the atomic coordinates of the outermost atoms. A pore with a diameter <10 Å does not allow the translocation of ss-DNA, whereas a pore with a diameter >∼15 Å causes a much smaller conductance. Fig. 2 b shows the current as a function of time when E‖ has been turned off. The starting condition was taken from the simulation used for Fig. 2 a at the time indicated by the dashed line in Fig. 2 a, when a base was aligned in between the electrodes. We can see that as time progresses the nucleotide stays aligned in between the electrodes due to the interaction of the nucleotide with the transverse electric field. This is discussed in further detail later on in the article. Similar curves have been found for all other DNA bases. The tunneling current of bare electrodes can be estimated as 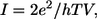 where V is the bias,
where V is the bias, 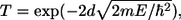 where e is the electron charge, d the electrode spacing, m the electron mass, and E the work function of gold. For d = 14 Å and E = 5 eV, we find that the current with vacuum in between the electrodes is ∼0.1 aA at a bias of 0.1 V, i.e., orders of magnitude lower than the currents obtained with DNA in between the electrodes.
where e is the electron charge, d the electrode spacing, m the electron mass, and E the work function of gold. For d = 14 Å and E = 5 eV, we find that the current with vacuum in between the electrodes is ∼0.1 aA at a bias of 0.1 V, i.e., orders of magnitude lower than the currents obtained with DNA in between the electrodes.
FIGURE 2.
Current, at a bias of 0.1 V, as a function of time for poly(A)15, as (a) it is propagating through a pore with two electrodes without a stabilizing field, and (b) when the driving field is turned off at a time (indicated by the dashed line in a) while a base is aligned in between the electrode pair. Solid vertical line in panel a indicates the time at which the DNA starts propagating through the pore. The transverse electric field is included in the simulations for panel b. Δt represents a finite inverse bandwidth.
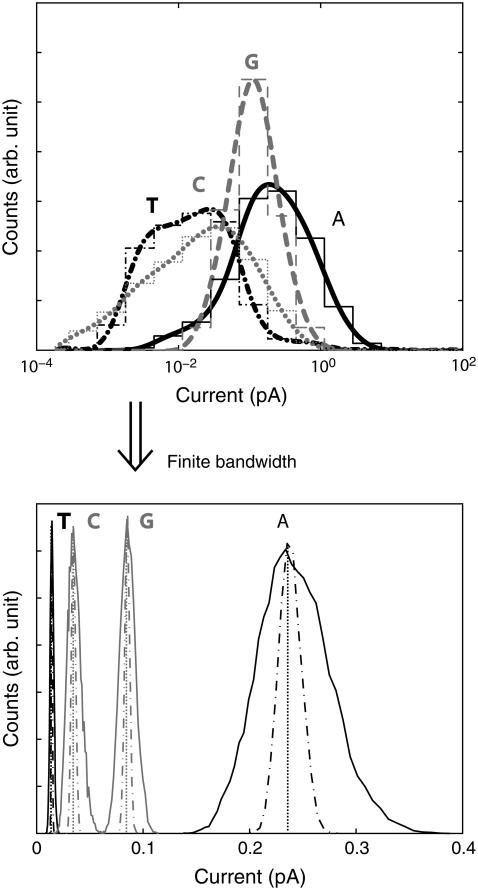
Since we envision operating the nanopore/electrode device in a regime where the transverse field is much stronger than the driving field, we can examine the real time structural fluctuations by sampling the current with the driving field off. Note that it could be possible to operate a device in this regime as a thermal ratchet, e.g., the thermal fluctuations overcome the barrier to moving each nucleotide away from the junction region. The distributions of these currents, with this particular pore geometry, for all four bases are shown in the top section of Fig. 3, assuming each current is measured instantaneously. From the molecular dynamics simulations we observe that ions fluctuate inside the pore at timescales of the order of 5 ps due to thermal fluctuations. As the total current sampling time per base is 1.2 ns, we effectively sample over multiple ionic configurations. We can see that these distributions are unique, but overlapping. This means that a handful of measurements of a base to be sequenced would not be enough to distinguish it from the other bases. However, in a real experiment each measurement would take a finite amount of time to be performed (finite inverse bandwidth indicated with Δt in Fig. 2 b). In other words, the electric probes have a finite bandwidth. Hence, a real measurement averages over a time interval, which is determined by the sampling frequency of the electrodes/probes, causing the distributions to narrow around their average current.
FIGURE 3.
Top graph shows the probability distributions, assuming instantaneous measurements, of currents at a bias of 0.1 V for poly(X)15, where X is A/T/C/G for the solid black, dash-dotted black, dotted gray, dashed gray curves, respectively. The thin lines show the actual current intervals used for the count, whereas the thick lines are an interpolation. After the driving electric field is turned off, the system is left to equilibrate for 200 ps before samples are taken. Each distribution is made up of 1200 samples, each taken with a 1-ps interval. The bottom graph again shows the probability distributions, but now with the assumption that each measurement is time averaged in between each sample. The solid/dashed-dotted line assumes that the distributions in the top graphs are sampled 100/1000/107 times for each new data point.
To accurately determine the new shape of the distributions when each measurement is time averaged, one would ideally need multiple ensembles, where, for example, one would be the current shown in Fig. 2 b. This is however difficult to realize numerically. On the other hand, one can assume, as a reasonable starting point, that the interpolated distributions shown in the top half of Fig. 3 are exact. That this is indeed a good approximation is justified by the following (see also below). Starting a nucleotide out with the base parallel to the electrode surfaces, we find that it takes ∼100 ps for the transverse field |E⊥| to align it perpendicular to the electrode surfaces. If one thus waits for a time longer than 100 ps to sample the distributions, the latter ones must be weakly dependent on initial conditions. We can then assume that the average current for another ensemble can be generated by sampling these interpolated distributions. By repeating this process multiple times, one can then calculate the new distributions, where each measurement is time averaged. These new distributions are shown in the bottom part of Fig. 3. Clearly, the new distributions become sharper the more times one samples the original distributions. The number of samples that should be taken is determined by the ratio between the period of physical measurement and the time interval for two measurements to be considered independent of each other. While the sampling frequency can be determined exactly, it is hard to give an exact value of the time needed in between two instantaneous measurements for them to be considered independent. However, we can take the timescale for atomic movements in the simulation, which is ∼1 ps, as a rough estimate.
Assuming no external noise, the distributions in the lower half of Fig. 3 show that as long as one samples the instantaneous distributions at least 100 times (solid lines), the new distributions will be completely separated from each other, and if sampled 1000 times the new distributions assume much sharper shapes. Assuming that the timescale for independent sample measurements is of the order of 1 ps, one would need an apparatus sampling at a rate no faster than ∼1/(1000 × 1 ps) = 1 GHz to obtain disjoint distributions. Since sampling frequencies of the order of gigahertz or less are relatively easy to obtain, we conclude that a single current measurement may be sufficient to distinguish the different bases. We would like to stress again that, in this analysis, we have assumed no other external source of noise is present (like, for instance, telegraph noise or 1/f noise). If thermal noise is of concern, one may reduce the sampling frequency, which both reduces the thermal noise and sharpens the intrinsic current distributions. For example, at a sampling frequency of 100 kHz, the root mean square current thermal noise for Guanine is of the order of 40 fA. The root mean-square current thermal noise is given by 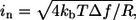 where T is the temperature, Δf is the sampling frequency, and R is the resistance of the nucleotide in the junction. The distributions due to structural fluctuations assume δ-function-like shapes. A larger average current can be obtained by either increasing the bias or by slightly reducing the pore size. If the distributions of the four bases were to begin overlapping due to external noise, multiple measurements per base would be needed, and a statistical analysis similar to the one presented in Lagerqvist et al. (8) could be performed. In the rest of the article we will keep on discussing the case where each measurement is assumed to be instantaneous because this case contains more information; one can always transform any distribution given below into a “finite-bandwidth” one as described above.
where T is the temperature, Δf is the sampling frequency, and R is the resistance of the nucleotide in the junction. The distributions due to structural fluctuations assume δ-function-like shapes. A larger average current can be obtained by either increasing the bias or by slightly reducing the pore size. If the distributions of the four bases were to begin overlapping due to external noise, multiple measurements per base would be needed, and a statistical analysis similar to the one presented in Lagerqvist et al. (8) could be performed. In the rest of the article we will keep on discussing the case where each measurement is assumed to be instantaneous because this case contains more information; one can always transform any distribution given below into a “finite-bandwidth” one as described above.
Current distributions for a different pore diameter and transverse field were examined previously in Lagerqvist et al. (8), assuming instantaneous measurements. Although the electrode spacing is larger in this article, 14 Å compared to 12.5 Å, and the electrode bias is smaller, 0.1 V compared to 1.0 V, the distributions show remarkable similarities. Adenine shows the largest mode in both cases and Guanine the second largest. Notice, however, that in the previous article Thymine had a slightly larger mode current than Cytosine, whereas the results presented here are reversed. This can be attributed to the change in electrode spacing and highlights the importance of calibrating the device (see discussion below). It also demonstrates the importance of the DNA-electrode coupling: in the upright configuration for which the nucleotides are held, the Thymine nucleotide has the largest base-electrode distance, and therefore its coupling to the second electrode is most sensitive to the electrode spacing. For larger electrode spacing, the nucleotide conductance is strongly dependent on the base-electrode distance. As the electrode spacing gets smaller, the current depends more on the spatial character and energies of the molecular orbitals. On the other hand the currents are orders of magnitude larger for the smaller pore/larger bias case. For example, the mode conductance for Adenine is roughly 1000 times larger. This difference can be attributed to the change in the electrode spacing; a smaller pore radius will result in a larger current.
Following the discussion in the previous paragraph, we would like to emphasize that it is highly unlikely that the exact geometry of two pores would be identical. Since in nanoscale systems a single atom change in the contact geometry or local environment may lead to a substantial change in the current (36,37), the first step to sequence a DNA strand would be to calibrate the device by creating these distributions for the pore at hand. Then a strand can be sequenced by comparing its current, at each base location, to these target distributions.
One final comment before we go on to discuss stabilization. When E‖ is turned off, we observe a mild drift of the Thymine (T) nucleotide, which causes a slightly different form for its current distribution when assuming that each measurement is instantaneous. In our previous work we found nearly Gaussian distributions for A and G nucleotides but not C and T. Since C and T are smaller bases, we believe their homogeneous strands have more freedom to move within the pore. When the strand moves enough, a nearest neighbor base can come in close vicinity of the electrodes. For the same reasons, the nearest neighbor base makes a larger difference in the conductance of C and T bases. In a stretched configuration, the nearest neighbor bases do not affect the conductance very much so long as the electrode width is on the order of a single nucleotide (6). However, the molecular dynamics simulation allows many configurations to be explored, including ones where the nucleotide is near the edge of the electrode and thus the neighboring nucleotide can make a substantial contribution to the conductance of the junction. Thus, a potential future direction of research may be to examine the role of base sequence on DNA motion within the nanopore.
Nucleotide stabilization
Each phosphate group carries a negative charge in solution. In addition, as we have anticipated, we find that there is always a counterion fluctuating about the backbone charge, at an average distance of 6.5 Å. This helps neutralizing the DNA charge inside the pore as confirmed by other molecular dynamics simulations (38) and also experiment (39,40). However, our results show that the transverse field can still act as a very effective stabilizer on the resulting nucleotide-counterion dipole.
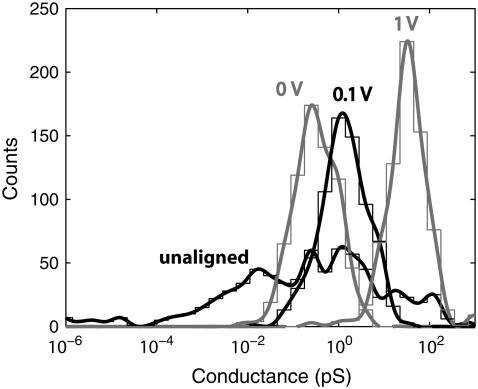
To better understand the effect of the stabilizing field on the current, one can compare the conductance distributions for varying transverse electric fields. In Fig. 4 the conductance distributions for four different cases are shown. The black curve shows the conductance distribution for the completely unstabilized and unaligned case, generated by transforming the current in Fig. 2 a into a distribution starting from the time when the strand starts to translocate. The blue curve is the conductance distribution generated by turning E‖ off at a time when a base is aligned in between the electrodes, as indicated by the dashed line in Fig. 2 a, while not including any stabilizing field. This corresponds to the limiting case in which both E‖ and E⊥ are allowed to approach zero with 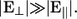 Red and green distributions are generated in the same manner as the blue curve, but with both a bias and stabilizing field of 0.1 and 1.0 V, respectively. As expected, the distribution for the unaligned case is much broader, compared to the other cases, as the bases are oriented in all possible directions while the strand propagates through the pore with a driving field much larger than the stabilizing field. When starting with a base aligned in between the two electrodes the distributions are clearly much sharper. One also notices an increase of conductance of almost an order of magnitude for a stabilizing field of 0.1 V compared to the one with no stabilizing field. Furthermore, the conductance increases by almost two orders of magnitude when increasing the stabilizing field from 0.1 to 1.0 V. This confirms the effect of the stabilizing field: as the field increases in strength it pulls the backbone closer to one electrode and aligns the base toward the other electrode, which increases the conductance. The alignment of the base with the field is also favored by the steric effect of the alignment of the backbone with one of the electrodes.
Red and green distributions are generated in the same manner as the blue curve, but with both a bias and stabilizing field of 0.1 and 1.0 V, respectively. As expected, the distribution for the unaligned case is much broader, compared to the other cases, as the bases are oriented in all possible directions while the strand propagates through the pore with a driving field much larger than the stabilizing field. When starting with a base aligned in between the two electrodes the distributions are clearly much sharper. One also notices an increase of conductance of almost an order of magnitude for a stabilizing field of 0.1 V compared to the one with no stabilizing field. Furthermore, the conductance increases by almost two orders of magnitude when increasing the stabilizing field from 0.1 to 1.0 V. This confirms the effect of the stabilizing field: as the field increases in strength it pulls the backbone closer to one electrode and aligns the base toward the other electrode, which increases the conductance. The alignment of the base with the field is also favored by the steric effect of the alignment of the backbone with one of the electrodes.
FIGURE 4.
Probability distributions of the conductance for varying stabilizing fields for poly(A)15. Leftmost curve labeled “unaligned” corresponds to the completely unstabilized case as shown in Fig. 2 a, whereas other curves correspond to various stabilizing fields when the driving field is turned off. The symbol 0 V corresponds to the case in which the base is aligned in between the electrode, but there is no stabilizing field.
To examine in more detail the effect of the stabilizing field, we have analyzed a case in which the driving field is turned off as an Adenine base was aligned in the direction perpendicular to the stabilizing field. Then, as a bias of 0.1 V is turned on in between the electrodes, the base starts to align with the stabilizing field and is completely aligned after ∼100 ps. We can thus conclude that as long as the strand is pulled through the pore at a pace slower than ∼100 ps per base, the stabilizing electric field induced by this bias will be sufficient for sequencing purposes. In experiments (20,41) the typical translocation speed is much slower than the reorientation time of ∼100 ps, allowing the bases sufficient time to reorient with the transverse electric field.
Influence of the environment
As we discussed above, water and counterions do influence the electronic structure of nucleotides in solution. Without these species, the nucleotides would have an unscreened charge and electronic transport would be quite different.
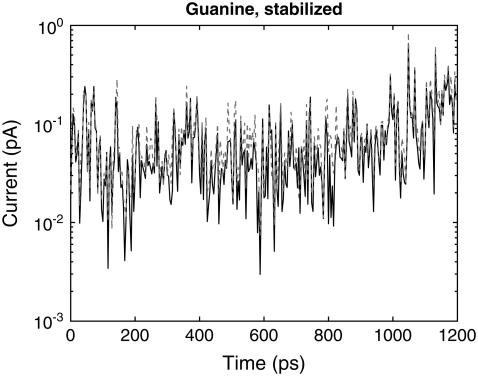
Nevertheless, the fluctuation of water molecules around the nucleotides inside the pore has little direct effect on the electrical current. Fig. 5 shows the current calculated both with and without water, for a Guanine base stabilized in between the electrodes. The presence of water lowers the current on average by 18% but the additional fluctuations in the current due to transport across water molecules are negligible compared to the larger structural fluctuations of the DNA molecule. For the smaller pore geometry used in the previous article (8), an even smaller change of 4% was observed for Adenine as it propagates through the pore unstabilized. This is an expected result, as the smaller pore allows for fewer water molecules to enter, with a corresponding decrease of its contribution to the current. Also, the small effect of water on the current is the result of a slight modification of the nucleotide electronic states. This is opposed to an increase in conductance due to “bridging” effects, which could occur if the pore were larger (42). Considering that structural fluctuations account for orders of magnitude change in conductance, we can conclude that the direct effect of water on conductance can be neglected.
FIGURE 5.
Current, at a bias of 0.1 V, as a function of time for poly(G)15, for a single guanine base stabilized in between the electrodes by the transverse electric field, with (solid black curve) and without (dashed gray curve) water included in the current calculation.
CONCLUSIONS
By combining molecular dynamics simulations with quantum mechanical current calculations, we examined the feasibility of DNA sequencing via transverse electronic transport. Specifically, we have shown that unless the current is sampled with very high frequencies, i.e., >∼1 GHz, the time averaging occurring in the probe apparatus will reduce the fluctuations in the current to such a level that an individual current measurement may be sufficient to sequence the base, assuming that any external sources of noise in the current are small. We also addressed how the transverse field strength, pore diameter, and local environment affect the distinguishability of the bases. Furthermore, we have shown that the electric field induced by the electrodes will effectively stabilize the bases and hence allows for accurate sequencing when 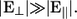 In addition, we have discussed how ions and water effectively screen the charged nucleotides. We have also shown that water in the environment has a negligible direct effect on the electrical current. Our results also emphasize the importance of device calibration.
In addition, we have discussed how ions and water effectively screen the charged nucleotides. We have also shown that water in the environment has a negligible direct effect on the electrical current. Our results also emphasize the importance of device calibration.
It is important to note that there are many issues left to be addressed, like DNA-surface bonding, time for the DNA to find and go through the pore (which will depend on device parameters), and the direct effect of ionic fluctuations on the electronic transport properties of the DNA.
Acknowledgments
We thank D. Branton, M. Ramsey, and P. Wolynes for useful discussions and critical reading of our manuscript.
This research is supported by the National Institutes of Health/National Human Genome Research Institute (J.L. and M.D.) and by the National Science Foundation through its Graduate Fellowship program (M.Z.).
Editor: Dagmar Ringe.
References
- 1.Lander, E. S., et al. 2001. Initial sequencing and analysis of the human genome. Nature. 409:1304–1351. [DOI] [PubMed] [Google Scholar]
- 2.Venter, J. C., et al. 2001. The sequence of the human genome. Science. 291:860–921. [Google Scholar]
- 3.Collins, F. S., E. D. Green, A. E. Guttmacher, and M. S. Guyer. 2003. A vision for the future of genomics research. Nature. 422:835–847. [DOI] [PubMed] [Google Scholar]
- 4.Kasianowicz, J. J., E. Brandin, D. Branton, and D. W. Deamer. 1996. Characterization of individual polynucleotide molecules using a membrane channel. Proc. Natl. Acad. Sci. USA. 93:13770–13773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Braslavsky, I., B. Hebert, E. Kartalov, and S. R. Quake. 2003. Sequence information can be obtained from single DNA molecules. Proc. Natl. Acad. Sci. USA. 100:3960–3964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zwolak, M., and M. Di Ventra. 2005. Electronic signature of DNA nucleotides via transverse transport. Nano Lett. 5:421–424. [DOI] [PubMed] [Google Scholar]
- 7.Gracheva, M. E., A. Xiong, A. Aksimentiev, K. Schulten, G. Timp, and J.-P. Leburton. 2006. Simulation of the electric response of DNA translocation through a semiconductor nanoporecapacitor. Nanotechnology. 17:622–633. [Google Scholar]
- 8.Lagerqvist, J., M. Zwolak, and M. Di Ventra. 2006. Fast DNA sequencing via transverse electronic transport. Nano Lett. 6:779–782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Akeson, M., D. Branton, J. J. Kasianowicz, E. Brandin, and D. W. Deamer. 1999. Microsecond timescale discrimination among polycytidylic acid, polyadenylic acid, and polyuridylic acid as homopolymers or as segments within single RNA molecules. Biophys. J. 77:3227–3233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Deamer, D. W., and M. Akeson. 2000. Nanopores and nucleic acids: prospects for ultrarapid sequencing. Trends Biotechnol. 18:147–151. [DOI] [PubMed] [Google Scholar]
- 11.Meller, A., L. Nivon, E. Brandin, J. Golovchenko, and D. Branton. 2000. Rapid nanopore discrimination between single polynucleotide molecules. Proc. Natl. Acad. Sci. USA. 97:1079–1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vercoutere, W., S. Winters-Hilt, H. Olsen, D. Deamer, D. Haussler, and M. Akeson. 2001. Rapid discrimination among individual DNA hairpin molecules at single-nucleotide resolution using an ion channel. Nat. Biotechnol. 19:248–252. [DOI] [PubMed] [Google Scholar]
- 13.Meller, A., L. Nivon, and D. Branton. 2001. Voltage-driven DNA translocations through a nanopore. Phys. Rev. Lett. 86:34353438. [DOI] [PubMed] [Google Scholar]
- 14.Deamer, D. W., and D. Branton. 2002. Characterization of nucleic acids by nanopore analysis. Acc. Chem. Res. 35:817–825. [DOI] [PubMed] [Google Scholar]
- 15.Meller, A., and D. Branton. 2002. Single molecule measurements of DNA transport through a nanopore. Electrophoresis. 23:2583–2591. [DOI] [PubMed] [Google Scholar]
- 16.Li, J., M. Gershow, D. Stein, E. Brandin, and J. A. Golovchenko. 2003. DNA molecules and configurations in a solid-state nanopore microscope. Nat. Mater. 2:611–615. [DOI] [PubMed] [Google Scholar]
- 17.Nakane, J. J., M. Akeson, and A. Marziali. 2003. Nanopore sensors for nucleic acid analysis. J. Phys.: Condens. Matter. 15:R1365–R1393 [Google Scholar]
- 18.Aksimentiev, A., J. B. Heng, G. Timp, and K. Schulten. 2004. Microscopic kinetics of DNA translocation through synthetic nanopores. Biophys. J. 87:2086–2097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen, P., J. Gu, E. Brandin, Y.-R. Kim, Q. Wang, and D. Branton. 2004. Probing single DNA molecule transport using fabricated nanopores. Nano Lett. 4:2293–2298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fologea, D., M. Gershow, B. Ledden, D. S. McNabb, J. A. Golovchenko, and J. Li. 2005. Detecting single stranded DNA with a solid state nanopore. Nano Lett. 5:1905–1909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Heng, J. B., A. Aksimentiev, C. Ho, P. Marks, Y. V. Grinkova, S. Sligar, K. Schulten, and G. Timp. 2006. The electromechanics of DNA in a synthetic nanopore. Biophys. J. 90:1098–1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li, J., D. Stein, C. McMullan, D. Branton, M. J. Aziz, and J. A. Golovchenko. 2001. Ion-beam sculpting at nanometre length scales. Nature. 412:166–169. [DOI] [PubMed] [Google Scholar]
- 23.Storm, A. J., J. H. Chen, X. S. Ling, H. Zandbergen, and C. Dekker. 2003. Fabrication of solid-state nanopores with single-nanometre precision. Nat. Mater. 2:537–540. [DOI] [PubMed] [Google Scholar]
- 24.Harrell, C. C., S. B. Lee, and C. R. Martin. 2003. Synthetic single-nanopore and nanotube membranes. Anal. Chem. 75:6861–6867. [DOI] [PubMed] [Google Scholar]
- 25.Li, N. C., S. F. Yu, C. C. Harrell, and C. R. Martin. 2004. Conical nanopore membranes. preparation and transport properties. Anal. Chem. 76:2025–2030. [DOI] [PubMed] [Google Scholar]
- 26.Lemay, S. G., D. M. van den Broek, A. J. Storm, D. Krapf, R. M. M. Smeets, H. A. Heering, and C. Dekker. 2005. Lithographically fabricated nanopore-based electrodes for electrochemistry. Anal. Chem. 77:1911–1915. [DOI] [PubMed] [Google Scholar]
- 27.Mannion, J. T., C. H. Reccius, J. D. Cross, and H. G. Craighead. 2006. Conformational analysis of single DNA molecules undergoing entropically induced motion in nanochannels. Biophys. J. 90:4538–4545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Biance, A. L., J. Gierak, E. Bourhis, A. Madouri, X. Lafosse, G. Patriarche, G. Oukhaled, C. Ulysse, J. C. Galas, Y. Chen, and L. Auvray. 2006. Focused ion beam sculpted membranes for nanoscience tooling. Microelectron. Eng. 83:1474–1477. [Google Scholar]
- 29.Ji, Q., Y. Chen, L. L. Ji, X. M. Jiang, and K. N. Leung. 2006. Ion beam imprinting system for nanofabrication. Microelectron. Eng. 83:796–799. [Google Scholar]
- 30.Kale, L., R. Skeel, M. Bhandarkar, R. Brunner, A. Gursoy, N. Krawetz, J. Phillips, A. Shinozaki, K. Varadarajan, and K. Schulten. 1999. NAMD2: greater scalability for parallel molecular dynamics. J. Comput. Phys. 151:283–312. [Google Scholar]
- 31.Foloppe, N., and A. D. MacKerell. 2000. All-atom empirical force field for nucleic acids. I. Parameter optimization based on small molecule and condensed phase macromolecular target data. J. Comput. Chem. 21:86–104. [Google Scholar]
- 32.MacKerell, A. D., and N. K. Banavali. 2000. All-atom empirical force field for nucleic acids. II. Application to molecular dynamics simulations of DNA and RNA in solution. J. Comput. Chem. 21:105–120. [Google Scholar]
- 33.Rappe, A. K., C. J. Casewit, K. S. Colwell, W. A. Goddard, and W. M. Skiff. 1992. UFF, a full periodic-table force-field for molecular mechanics and molecular-dynamics simulations. J. Am. Chem. Soc. 114:10024–10035. [Google Scholar]
- 34.Grun, R. 1979. Crystal-structure of beta-Si3n4—structural and stability considerations between alpha-Si3n4 and beta-Si3n4. Acta Crystallogr. B. 35:800–804. [Google Scholar]
- 35.Jorgensen, W. L. 1981. Pressure-dependence of the structure and properties of liquid normal-butane. J. Am. Chem. Soc. 103:4721–4726. [Google Scholar]
- 36.Di Ventra, M., S. T. Pantelides, and N. D. Lang. 2000. First-principles calculation of transport properties of a molecular device. Phys. Rev. Lett. 84:979. [DOI] [PubMed] [Google Scholar]
- 37.Yang, Z. Q., N. D. Lang, and M. Di Ventra. 2003. Effects of geometry and doping on the operation of molecular transistors. Appl. Phys. Lett. 82:1938. [Google Scholar]
- 38.Rabin, Y., and M. Tanaka. 2005. DNA in nanopores: counterion condensation and coion depletion. Phys. Rev. Lett. 94:148103. [DOI] [PubMed] [Google Scholar]
- 39.Sauer-Budge, A. F., J. A. Nyamwanda, D. K. Lubensky, and D. Branton. 2003. Unzipping kinetics of double-stranded DNA in a nanopore. Phys. Rev. Lett. 90:238101. [DOI] [PubMed] [Google Scholar]
- 40.Mathé, J., H. Visram, V. Viasnoff, Y. Rabin, and A. Meller. 2004. Nanopore unzipping of individual DNA hairpin molecules. Biophys. J. 87:3205–3212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fologea, D., J. Uplinger, B. Thomas, D. S. McNabb, and J. Li. 2005. Slowing DNA translocation in a solid-state nanopore. Nano Lett. 5:1734–1737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lin, J. P., I. A. Balabin, and D. N. Beratan. 2005. The nature of aqueous tunneling pathways between electron-transfer proteins. Science. 310:1311–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]