Abstract
Background
Bordetella bronchiseptica is a bacterial respiratory pathogen that infects a broad range of mammals, causing chronic and often subclinical infections. Gene expression in Bordetella is regulated by a two-component sensory transduction system, BvgAS, which controls the expression of a spectrum of phenotypic phases transitioning between a virulent (Bvg+) phase and a non-virulent (Bvg-) phase.
Results
Based on the genomic sequence and using the freely available software ArrayOligoSelector, a long oligonucleotide B. bronchiseptica microarray was designed and assembled. This long-oligonucleotide microarray was subsequently tested and validated by comparing changes in the global expression profiles between B. bronchiseptica RB50 and its Bvg- phase-locked derivative, RB54. Data from this microarray analysis revealed 1,668 Bvg-regulated genes, which greatly expands the BvgAS regulon defined in previous reports. For previously reported Bvg-regulated transcripts, the gene expression data presented here is congruent with prior findings. Additionally, quantitative real-time PCR data provided an independent verification of the microarray expression values.
Conclusion
The results presented here provide a comprehensive, genome-wide portrait of transcripts encompassing the BvgAS regulon, while also providing data validating the long-oligonucleotide microarray described here for studying gene expression in Bordetella bronchiseptica.
Background
Bordetellae are Gram negative bacterial respiratory pathogens. Bordetella pertussis and Bordetella parapertussishu, the causative agents of whooping cough, are human-adapted variants of Bordetella bronchiseptica, which naturally infects a broad range of mammals causing chronic and often asymptomatic infections [1]. The majority of gene expression in Bordetella is regulated by a two-component sensory transduction system encoded by the bvg locus. The bvg locus comprises a histidine kinase sensor protein, BvgS, and a DNA-binding response-regulator protein, BvgA. In response to environmental cues, BvgAS controls expression of a spectrum of phenotypic phases transitioning between a virulent (Bvg+) phase and a non-virulent (Bvg-) phase. During the virulent Bvg+ phase, the BvgAS system is fully active and many of the known virulence factors are expressed, such as filamentous hemagglutinin (FHA), pertactin, fimbriae, adenylate cyclase-hemolysin toxin, and dermonecrotic toxin (DNT), as well as a type III secretion system (TTSS) in B. bronchiseptica [2]. Conversely, BvgAS is inactive during the Bvg- phase, resulting in the maximal expression of motility loci, virulence-repressed genes (vrg genes), genes required for the production of urease, and in B. bronchiseptica RB50, a siderophore, alcaligin [3-5]. Previous studies involving phase-locked and ectopic expression mutants demonstrated that the Bvg+ phase promotes respiratory tract colonization by B. pertussis and B. bronchiseptica [6-9], while the Bvg- phase of B. bronchiseptica promotes survival under conditions of nutrient deprivation [6,10].
The signals that activate BvgAS in nature are unknown. However, in the laboratory, BvgAS is active when the bacteria are grown at 37°C and inactive when grown at temperatures below ~26°C or in medium containing MgSO4 or nicotinic acid at concentrations in the millimolar range. Although originally identified as a positive regulator of virulence gene transcription [11], it is now known that BvgAS controls expression of over a hundred different genes whose products are either proven or predicted to participate in a wide variety of cellular activities including many basic physiological functions [12-14]. Additionally, it is now understood that rather than functioning like an ON/OFF switch, BvgAS functions more like a "rheostat" capable of controlling gene expression of many phenotypic phases in response to subtle differences in environmental conditions [10].
The advent of microarray technology has enabled scientists to investigate biological questions, such as those pertaining to bacterial pathogenesis and host-pathogen interactions, in a global fashion. The cDNA microarray represents a popular array type in which double-stranded PCR products are spotted onto glass slides. However, construction of such microarrays presents a number of challenges, largely related to costs associated with amplicon validation, tracking and maintenance. For example, the laborious and problematic tracking of PCR amplicons leads to an estimated 10–30% misidentification [15]. Another limitation of cDNA microarrays is their inability, due to cross-hybridization, to reliably discriminate expression patterns of homologous genes [16]. With oligonucleotide arrays, problems related to clone tracking, homologous gene discrimination, and failed PCR amplicons are avoided, thus making long-oligonucleotide microarrays a more cost- and management- efficient alternative to cDNA microarrays. Here we present the design and assembly of a long-oligonucleotide B. bronchiseptica gene-specific microarray using the currently available genomic sequence generated by the Sanger Institute [17] and the software package ArrayOligoSelector [18]. This long-oligonucleotide microarray was then tested and validated by evaluating changes in the global expression profiles between B. bronchiseptica strain RB50 and its Bvg- phase-locked derivative, RB54.
Results and discussion
To construct a B. bronchiseptica-specific whole genome microarray, the freely available software program, ArrayOligoSelector (AOS) [18], was used to generate 70-mer oligonucleotide probes for every ORF in the Bordetella bronchiseptica RB50 genome [17]. The rationale behind designing and utilizing oligonucleotide probes versus PCR amplicons as probes, and subsequently the 70-mer length of the oligonucleotide probes, was chosen for several reasons. Long oligonucleotides are a highly sensitive alternative to PCR products and provide a means to readily distinguish between genes with high degrees of sequence similarity, which is an issue for the B. bronchiseptica genome [17]. For example, except for the extreme 5' and 3' termini, the fhaB and fhaS genes are nearly identical [19]. Additionally, previous results involving an anchored set of oligonucleotides revealed a strong relationship between the oligonucleotide length and hybridization performance [18].
For each B. bronchiseptica ORF, the AOS program optimizes the oligonucleotide selection on the basis of uniqueness in the genome, sequence complexity, lack of self-binding and GC content. Candidate oligos closest to the 3' end of the gene are then chosen [18]. There are a number of missing array elements due to gene duplications, prophage duplications, and ORF assignments missing from the completed genome annotation [17]. These missing array elements and, in the case of gene duplications, their corresponding represented array elements are listed in Additional File 1- see Supplementary Table S1. A list of the final 4975 oligonucleotide array elements or probes representing the entire B. bronchiseptica genome is given in Additional File 1- see Supplementary Table S2.
To test the usefulness of the newly constructed long-oligonucleotide Bordetella bronchiseptica microarray, a direct comparison between the transcriptional profile of B. bronchiseptica RB50 and RB54, a B. bronchiseptica Bvg- phase locked derivative of RB50, was performed. The rationale behind performing this comparison to validate and test the B. bronchiseptica long-oligonucleotide microarray is that this comparison will globally identify B. bronchiseptica genes regulated by BvgAS. Previous studies, including cDNA microarray studies, have identified 538 genes controlled by BvgAS [2,13,14], thus providing a substantial reportable reference set to validate gene expression data generated from the newly constructed B. bronchiseptica long-oligonucleotide microarray.
Utilizing the B. bronchiseptica long-oligonucleotide microarray, ratio data collected from microarray analysis involving the direct comparison between B. bronchiseptica RB50 and RB54 revealed 1,668 Bvg-regulated genes (identified using SAM with a false discovery rate of <0.1%). This is a substantial increase in the number of Bvg-regulated transcripts compared to the 538 Bvg-regulated genes recently reported by others using cDNA microarray analysis [14]. A complete list of the fold-change expression values from this comparison, along with dye swap experimental data, is provided in Additional File 1- Supplementary Table S3. One possible effect of using the B. bronchiseptica RB50 strain grown at 37°C to represent the Bvg+ phase, as opposed to using a Bvg+ phase-locked strain, is that some Bvg-activated genes could be missed in this analysis. Possible explanations for the large increase in Bvg-regulated transcripts detected in this study, compared to previous reports, may include (i) differences between strains used, for example wild-type versus phase-locked derivatives, (ii) differences between gene expression threshold cut-off values used in analysis, and (iii) for microarray studies, differences between array platforms, such as cDNA microarrays versus oligonucleotide microarrays. Gene expression profiles reported here of demonstrated Bvg-regulated transcripts are consistent with previous results (Figure 1).
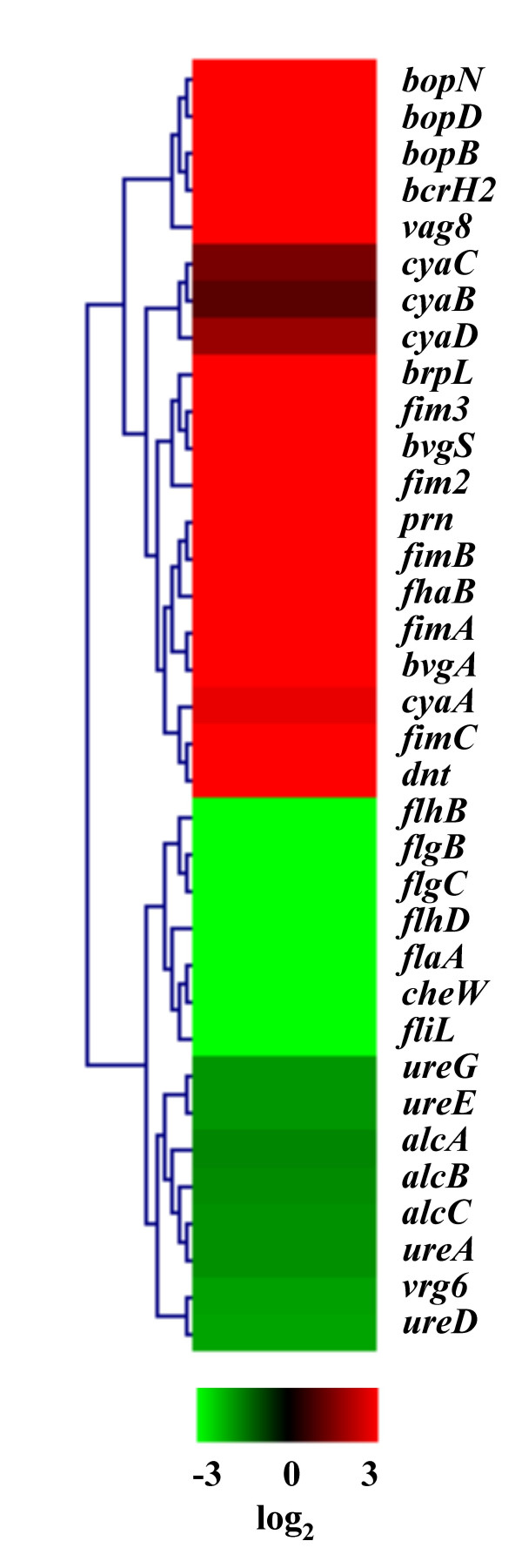
Figure 1.
Expression of demonstrated Bvg-regulated genes. Hierarchical clustering of known B. bronchiseptica Bvg-regulated genes performed using MeV [32]. Expression profiles of genes are in rows. Data are mean centered for each array element and averaged from biological replicates. Red, indicates increased expression in RB50; green, decreased gene expression in RB50 (and increased expression in RB54); black, no significant change in gene expression.
Historically, numerous Bvg-activated genes have been described, such as TTSS genes, fimbrial genes, filamentous hemagglutinin, pertactin, adenylate cyclase-hemolysin toxin, and dermonecrotic toxin [2]. Using the newly constructed oligonucleotide B. bronchiseptica microarray, all of these transcripts were found to be positively regulated by BvgAS (Figure 1). Along with other virulence-related transcripts classically characterized as Bvg-activated, three putative adhesion genes fhaS (+10.37-fold), fhaL (+8.04-fold), and BB0110 (+8.58-fold) were found to be highly Bvg-activated, as well as a putative novel toxin, BB3242 (+50.72-fold), containing an aerolysin and pertussis toxin domain (Table 1 and see Additional File 1- Supplementary Table S3). Seven ATP-binding cassette (ABC) transporters were found to be Bvg-activated, including BB1593 (+83-fold) (Table 1 and see Additional File 1- Supplementary Table S3). In other bacteria, these transporters serve as important virulence factors based upon their role in nutrient uptake and in secretion of toxins and antimicrobial agents [20]. Five autotransporters were highly Bvg-activated, including two virulence-associated transcripts vag8 and sphB1 (Table 1 and see Additional File 1- Supplementary Table S3). The autotransporter sphB1 has been implicated in the maturation process of FHA [21]. The Bvg-activated expression profile of these transporters implicates their importance in pathogenesis. Five genes involved in protein folding and ushering were highly Bvg-activated. These include dsbG (+35.29-fold) and mucD (+2.75-fold), a DegP family serine protease (Table 1 and see Additional File 1- Supplementary Table 3). Homologues of both dsbG and mucD are known to serve roles in virulence in other organisms [22,23]. Twenty-seven transcripts identified as serving roles in global regulatory functions were found to be Bvg-activated, including the conventionally characterized bvgAS and the TTSS regulator brpL (Table 1 and see Additional File 1- Supplementary Table S3). Other Bvg-activated transcripts in this group include the other TTSS regulators (BB1645, BB1646, and BB1642), three proposed two-component response regulatory proteins, and numerous proposed transcriptional regulators. Lastly, BB4228 (+15.70-fold), recently designated BopC and characterized as a novel type-III secretion effector [24], was found to be highly Bvg-activated.
Table 1.
Representative Bvg+ phase specific genes identified by microarray analysis.
| Function | Gene | Product | Fold-Change ± SEM |
| Cell Adhesin | BB0110 | putative adhesion | 8.58 ± 3.21 |
| BB1658 fim3 | serotype 3 fimbrial subunit precursor | 32.47 ± 11.10 | |
| BB1936 fhaL | adhesion | 8.04 ± 1.63 | |
| BB2312 fhaS | adhesion | 10.37 ± 2.76 | |
| BB2989 fimD | fimbrial adhesion | 11.58 ± 3.96 | |
| BB2992 fimA | fimbrial protein | 33.75 ± 13.88 | |
| BB2993 fhaB | filamentous hemagglutin | 11.37 ± 4.55 | |
| BB3424 | fimbrial protein | 8.92 ± 2.64 | |
| BB3425 fimN | fimbrial subunit protein | 11.95 ± 2.06 | |
| BB3426 fimX | fimbrial protein | 8.92 ± 2.64 | |
| BB3674 fim2 | serotype 2 fimbrial subunit precursor | 21.89 ± 11.68 | |
| Cell Envelope | BB1289 | putative integral membrane protein | 26.29 ± 8.25 |
| BB1368 | putative membrane protein | 3.94 ± 0.56 | |
| BB3119 | putative membrane protein | 19.27 ± 2.17 | |
| BB3157 | putative membrane protein | 16.41 ± 5.54 | |
| BB4029 | putative glycosyl transferase | 4.96 ± 0.32 | |
| BB4266 | putative membrane protein | 6.84 ± 0.82 | |
| BB4269 | putative glycosyl transferase | 2.23 ± 0.42 | |
| BB4284 | putative membrane protein | 2.96 ± 0.14 | |
| Protein Folding and Ushering | BB2320 dsbG | thiol:disulfide interchange protein precursor | 35.29 ± 9.68 |
| BB2991 fimB | chaperone protein | 11.20 ± 7.71 | |
| BB3749 mucD | serine protease; trypsin activity | 2.75 ± 0.41 | |
| BB3803 | putative peptidyl-prolyl cis-trans isomerase | 4.65 ± 1.26 | |
| Global Regulatory Functions | BB0308 | putative transcriptional regulator | 5.67 ± 0.98 |
| BB1607 | putative LysR-family transcriptional regulator | 3.37 ± 0.55 | |
| BB1638 brpL | putative RNA polymerase sigma factor | 50.66 ± 15.58 | |
| BB1642 | putative regulator; catalytic activity | 112.16 ± 97.97 | |
| BB1645 | putative anti-sigma factor; ATP binding | 9.46 ± 3.00 | |
| BB1646 | putative antisigma factor antagonist | 18.19 ± 6.08 | |
| BB2051 | putative regulatory protein | 2.24 ± 0.59 | |
| BB4374 | putative AsnC-family transcriptional regulator | 5.78 ± 7.25 | |
| Metabolism | BB0457 | probable enoyl-CoA hydrates; isomerase activity | 5.72 ± 1.34 |
| BB0458 | probable carboxymuconolactone decarboxylase | 3.42 ± 0.79 | |
| BB1476 | putative gluconate dehydrogenase | 2.88 ± 0.51 | |
| BB1478 | putative carbonic anhydrase precursor | 4.05 ± 0.77 | |
| BB1640 | hypothetical protein; glutamate-cysteine ligase activity | 63.92 ± 18.15 | |
| BB1644 alr | alanine racemase, catabolic | 253.96 ± 97.47 | |
| BB2658 | glutamate decarboxylase | 2.78 ± 0.93 | |
| BB3783 | intracellular PHB depolymerase | 3.23 ± 0.78 | |
| BB4286 nadC | putative nicotinate-nucleotidepyrophosphorylase | 32.86 ± 6.48 | |
| Periplasmic, Exported, Lipoprotein | BB0324 cyaA | bifunctional hemolysin-adenylatecyclase precursor | 6.73 ± 2.52 |
| BB1280 | putative exported protein | 73.44 ± 37.80 | |
| BB1292 | putative exported protein | 272.88 ± 93.88 | |
| BB1641 | putative exported protein | 149.05 ± 97.51 | |
| BB2398 | putative exported protein | 6.77 ± 1.38 | |
| BB2873 | putative membrane protein | 26.85 ± 13.90 | |
| BB3068 | putative exported protein | 10.90 ± 3.11 | |
| BB3242 | putative exported protein | 48.75 ± 9.65 | |
| BB3302 | lipoprotein | 5.09 ± 0.63 | |
| BB3978 dnt | dermonecrotic toxin | 8.63 ± 3.61 | |
| BB4181 | putative lipoprotein | 3.40 ± 0.25 | |
| BB4285 | putative exported protein | 49.73 ± 14.04 | |
| Transport | BB0323 cyaC | cyclolysin-activating lysine-acyltransferase | 2.70 ± 0.27 |
| BB0325 cyaB | cyclolysin secretion ATP-binding protein; ABC transporter | 2.10 ± 0.31 | |
| BB0326 cyaD | cyclolysin secretion protein; transporter activity | 3.48 ± 0.51 | |
| BB0327 cyaE | cyclolysin secretion protein; subtilase activity | 3.66 ± 1.57 | |
| BB0419 sphB1 | autotransporter subtilisn-like protease; substilase activity | 59.34 ± 43.26 | |
| BB1593 | putative ABC transport system, membrane protein | 83.60 ± 28.88 | |
| BB1643 | putative inner membrane transport protein | 39.61 ± 16.68 | |
| BB1864 vag8 | autotransporter | 122.00 ± 79.14 | |
| BB2270 | autotransporter | 4.71 ± 2.07 | |
| BB3826 bfrD | probable TonB-dependent receptor for iron transport | 2.68 ± 0.58 | |
| BB3993 ompQ | outer membrane porin protein OmpQ | 24.34 ± 8.07 | |
| TTSS | BB1616 bopN | putative outer protein N | 504.05 ± 189.58 |
| BB1620 bopD | putative outer protein D | 457.16 ± 172.40 | |
| BB1624 bscI | putative type III secretion protein | 226.35 ± 80.58 | |
| BB1628 bscN | putative ATP synthase in type III secretion | 115.85 ± 37.00 | |
| BB4228 bopCa | hypothetical protein; characterized type III effector | 15.70 ± 3.85 |
DNA microarray analysis was used to measure mRNA levels present in B. bronchiseptica RB50 compared to mRNA levels present in B. bronchiseptica RB54. Differences in mRNA levels are listed as mean fold-changes ± standard error. Fold-changes were calculated by averaging the data from three biological sample sets. aRecently characterized [24].
The overall global transcriptional program observed during the Bvg- phase supports previous data implicating a crucial role for the Bvg-phase in survival during environmental stress conditions [2,25]. Genes annotated to serve global regulatory functions were one of the functional categories with the highest number of Bvg-repressed transcripts. Transcripts in this category include forty-nine transcription factors and/or DNA-binding proteins, and ten two-component systems (Table 2 and see Additional File 1- Supplementary Table 3). Numerous genes involved in transport were identified as Bvg-repressed, including twenty-seven ABC transporters and TonB-dependent receptors, such as bfrG BB1294 (-2.86-fold) and bfeA BB1942 (-2.80-fold) (Table 2 and see Additional File 1- Supplementary Table S3). As mentioned earlier, these transporters serve important biological roles in other organisms, including participation in host-pathogen interactions as known virulence factors [20]. The Bvg-repressed expression profile of these genes highlights nutrient scavenging as a critical ability during Bvg- growth and supports prior data demonstrating that the Bvg- phase is optimized for growth under nutrient limiting conditions [2,25].
Table 2.
Representative Bvg- phase specific genes identified by microarray analysis.
| Function | Gene | Product | Fold-Change ± SEM |
| Cell Adhesin | BB1186 | putative hemolysin, homophilic cell adhesion | -2.83 ± 0.18 |
| Cell Envelope | BB0141 wbmD | putative membrane protein | -5.19 ± 0.08 |
| BB2618 flgJ | peptidoglycan hydrolase | -28.12 ± 0.01 | |
| BB2618 lpxB | lipid-A-disaccharide synthase; lipid A biosynthesis | -6.35 ± 0.02 | |
| BB2879 | putative membrane protein | -6.46 ± 0.02 | |
| BB4842 | putative outer membrane protein | -32.02 ± 0.01 | |
| Chemotaxis | BB2543 motA | chemotaxis protein MotA | -26.28 ± 0.02 |
| BB2544 motB | chemotaxis protein MotB | -17.90 ± 0.02 | |
| BB2549 cheR | chemotaxis protein methyltransferase | -10.43 ± 0.03 | |
| BB2552 cheZ | chemotaxis protein CheZ; catalytic activity | -38.44 ± 0.01 | |
| Motility | BB2553 flhB | flagellar biosynthetic protein FlhB | -44.48 ± 0.01 |
| BB2555 flhF | flagellar biosynthesis protein; RNA binding | -32.10 ± 0.00 | |
| BB2559 flgB | flagellar basal-body rod protein FlgB; motor activity | -44.49 ± 0.01 | |
| BB2560 flgC | flagellar basal-body rod protein FlgC; motor activity | -32.10 ± 0.00 | |
| BB2585 fliI | flagellum-specific ATP synthase FliI; ATP binding | -30.75 ± 0.01 | |
| Electron Transport | BB1283 cyoA | ubiquinol oxidase polypeptide II; copper ion binding | -5.11 ± 0.08 |
| BB2800 napA | periplasmic nitrate reductase precursor; iron ion binding | -11.34 ± 0.01 | |
| BB3325 | putative ferredoxin; electron transport activity | -32.43 ± 0.00 | |
| BB3927 | putative cytochrome; electron transport activity | -5.58 ± 0.01 | |
| BB4096 | putative oxidoreductase | -4.90 ± 0.02 | |
| BB4497 cydB | cytochrome D ubiquinol oxidase subunit II | -6.04 ± 0.02 | |
| BB4498 cydA | cytochrome D ubiquinol oxidase subunit I | -5.40 ± 0.07 | |
| BB4945 ivd | isovaleryl-CoA dehydrogenase | -8.90 ± 0.01 | |
| Protein Folding and Ushering | BB0979 | putative universal stress protein | -24.76 ± 0.02 |
| BB2875 | universal stress family protein | -7.45 ± 0.01 | |
| BB3257 fkpB | FkbP-type peptidyl-prolyl cis-trans isomerase | -4.15 ± 0.01 | |
| BB4260 | putative universal stress protein | -9.29 ± 0.01 | |
| Global Regulatory Functions | BB0725 | putative transcriptional regulator | -7.82 ± 0.04 |
| BB1122 | two-component system sensor kinase; signal transducer | -9.65 ± 0.01 | |
| BB1187 | putative LuxR-family transcriptional regulator | -11.60 ± 0.02 | |
| BB2108 | probable two-component response regulator | -18.08 ± 0.01 | |
| BB2323 | putative transcriptional regulator | -11.03 ± 0.03 | |
| BB2540 fliA | RNA polymerase sigma factor; flagellar operon | -16.05 ± 0.01 | |
| BB2542 flhC | flagellar transcriptional activator FlhC; DNA binding | -31.06 ± 0.01 | |
| BB2550 cheB | protein-glutamate methylesterase;two-component regulator | -18.56 ± 0.02 | |
| BB3115 | methyl-accepting chemotaxis signal transducer protein | -10.74 ± 0.02 | |
| BB3866 | probable LysR-family transcriptional regulator | -7.15 ± 0.02 | |
| Metabolism | BB0868 mmsA | methylmalonate-semialdehyde dehydrogenase | -8.72 ± 0.02 |
| BB0978 ggt | gamma-glutamyltranspeptidase precursor | -35.18 ± 0.02 | |
| BB1367 cysG | siroheme synthase, heme biosynthesis | -8.67 ± 0.01 | |
| BB2085 hemC | porphobilinogen deaminase; synthase activity | -16.39 ± 0.01 | |
| BB2147 hemN | coproporphyrinogen III oxidase | -5.84 ± 0.01 | |
| BB4409 hemL | glutamate-1-semialdehyde 2,1-aminomutase | -13.10 ± 0.01 | |
| Transport | BB0976 | lactate permease family protein | -21.29 ± 0.01 |
| BB1174 | putative ABC transport proteins; ATP-binding component | -5.52 ± 0.03 | |
| BB1188 | HlyD-family secretion protein; protein transporter activity | -6.27 ± 0.05 | |
| BB1189 | probable ABC transporter | -7.56 ± 0.06 | |
| BB1191 | putative outer membrane protein; transporter activity | -13.87 ± 0.02 | |
| BB1294 bfrG | putative TonB-dependent receptor | -2.86 ± 0.03 | |
| BB1942 bfeA | ferric enterobactin receptor | -2.80 ± 0.02 | |
| BB2402 | putative sulfate transporter | -6.92 ± 0.02 | |
| BB2433 | multidrug resistance protein | -4.14 ± 0.03 | |
| BB2803 ccmA | putative heme export protein; heme transporter activity | -12.62 ± 0.01 | |
| BB2804 ccmB | heme exporter protein B | -9.57 ± 0.01 | |
| BB2805 ccmC | putative heme export protein | -13.01 ± 0.01 | |
| BB4273 atoE | putative short-chain fatty acids transporter | -7.49 ± 0.01 | |
| BB4495 | probable ATP-binding component; ABC transporter | -33.13 ± 0.01 |
DNA microarray analysis was used to measure mRNA levels present in B. bronchiseptica RB50 compared to mRNA levels present in B. bronchiseptica RB54. Differences in mRNA levels are listed as mean fold-changes ± standard error. Fold-changes were calculated by averaging the data from three biological sample sets.
Consistent with previous results, genes required for the expression of the siderophore alcaligin [26] and urease [5] were preferentially activated in the Bvg- phase, along with a putative hemolysin BB1186 (-2.83-fold) (Table 2 and see Additional File 1- Supplementary Table S3). Also consistent with previous reports, all of the genes known to be involved in chemotaxis, such as cheD (-10.59-fold) and cheW (-15.21-fold), and motility, such as flgB (-44.49-fold) and flgC (-32.10-fold), were Bvg-repressed [3] (Table 2 and see Additional File 1- Supplementary Table S3). Numerous genes involved in electron transport were Bvg-repressed along with numerous genes involved in protein folding and ushering, including a high number of universal stress proteins, congruent with the Bvg- phase role in stress survival [6,25] (Table 2 and see Additional File S1- Supplementary Table S3).
Bordetella pertussis only infects humans and, as mentioned above, is responsible for causing an acute upper-respiratory disease known as whooping cough or pertussis. Due to its high degree of similarity to B. pertussis and its broad host range, including animals conveniently used in laboratory studies, B. bronchiseptica is widely used as a model for Bordetella pathogenesis research. Data arising from the recent completion of the comparative sequencing of three different Bordetella strains has revealed a loss or inactivation of a large number of genes in B. pertussis [17]. Thus an intriguing question is how many of the B. bronchiseptica Bvg-regulated genes are intact in B. pertussis. To determine this, the ORF sequence and oligonucleotide probe sequence for every Bvg-regulated gene identified in this study was used to blast the B. pertussis genome sequence. This analysis resulted in identifying 1172 shared genes that are Bvg-regulated in B. bronchiseptica (see Additional File S1- Supplementary Table S4).
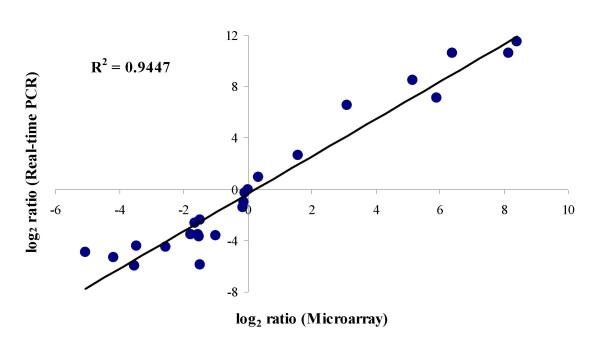
Real-time quantitative PCR was performed to provide an independent assessment of microarray expression measurements for selected genes. Genes were chosen to reflect the full spectrum of fold-changes identified by microarray analysis. Specifically, transcripts identified as having little to no change in gene expression by microarray analysis and selected for qRT-PCR include BB0057 rpoA, BB4989 dnaA, BB1037, and BB3703 eno (Table 3). Bvg-activated genes identified by microarray analysis and selected for qRT-PCR include the TTSS regulatory genes, BB1619 bcrH1 and BB1622 bcrH2 (Table 3). On the polar end of the Bvg spectrum, genes identified as Bvg-repressed and selected for qRT-PCR include BB2522 and BB1315, a putative universal stress protein. Additionally, BB4835 rpoH, sigma-32, identified as Bvg-repressed (-3.09-fold) was also selected for qRT-PCR analysis since it had not been identified as Bvg-regulated by previous cDNA microarray studies [13,14]. Real-time quantitative PCR analysis of this gene set provided data consistent with the quantitative measures by microarray analysis, using the newly constructed B. bronchiseptica oligonucleotide microarray (r2 = 0.94) (Figure 2). Therefore, this real-time quantitative PCR data provides independent verification of the microarray results.
Table 3.
Fold-changes identified by quantitative real-time PCR.
| Gene | Product | qRT-PCR | Microarray |
| BB0057 rpoA | DNA-directed RNA polymerase alpha chain | -2.03 ± 0.01 | -1.09 ± 0.06 |
| BB0110 | putative adhesion | 92.47 ± 0.35 | 8.58 ± 1.21 |
| BB0376 | putative membrane protein | 6.23 ± 0.45 | 3.00 ± 0.72 |
| BB0419 sphB1 | autotransporter subtilisin-like protease | 137.34 ± 6.96 | 59.34 ± 4.26 |
| BB1037 | LysR-family transcriptional regulator | -1.24 ± 0.01 | -1.07 ± 0.15 |
| BB1315 | putative universal stress protein | -64.15 ± 0.35 | -11.43 ± 0.03 |
| BB1593 | putative ABC transport system, membrane protein | 1544.59 ± 8.49 | 83.60 ± 8.88 |
| BB1619 bcrH1 | putative regulatory protein | 2943.53 ± 16.33 | 340.10 ± 7.39 |
| BB1622 bcrH2 | putative regulatory protein | 1532.48 ± 11.25 | 286.50 ± 9.03 |
| BB2108 | probable two-component response regulator | -39.21 ± 1.24 | -18.08 ± 0.01 |
| BB2147 hemN | oxygen-independent coproporphyrinogen III oxidase | -23.10 ± 3.33 | -5.84 ± 0.01 |
| BB2320 dsbG | thiol:disulfide interchange protein precursor | 358.29 ± 5.25 | 35.29 ± 1.68 |
| BB2323 | putative transcriptional regulator | -21.86 ± 3.46 | -11.03 ± 0.01 |
| BB2367 | putative regulatory protein; transcription factor activity | -11.71 ± 2.15 | -2.95 ± 0.11 |
| BB2434 pckG | phosphoenolpyruvate carboxykinase (GTP) | -12.57 ± 1.46 | -2.88 ± 0.04 |
| BB2435 | S-adenosylmethionine-dependent methyltransferase | -5.10 ± 1.24 | -2.76 ± 0.04 |
| BB2522 | conserved hypothetical protein | -58.35 ± 0.35 | -2.82 ± 0.03 |
| BB3447 cysN | sulfate adenylyltransferase subunit 1; (ATP) activity | -12.50 ± 0.15 | -2.00 ± 0.25 |
| BB3703 eno | enolase; phosphopyruvate hydratase activity | -2.68 ± 0.14 | -1.11 ± 0.08 |
| BB4495 | probable ATP-binding component of ABC-transporter | -30.63 ± 0.15 | -33.12 ± 0.01 |
| BB4506 rpoN | probable sigma (54) modulation protein | -11.66 ± 0.56 | -3.38 ± 0.12 |
| BB4835 rpoH | RNA polymerase sigma-32 factor | -6.33 ± 0.22 | -3.09 ± 0.11 |
| BB4989 dnaA | chromosomal replication initiator protein | 1.94 ± 0.01 | 1.27 ± 0.19 |
Quantitative real-time PCR analysis was used to measure mRNA levels present in B. bronchiseptica RB50 compared to mRNA levels present in B. bronchiseptica RB54. Differences in mRNA levels detected by both qRT-PCR and Microarray are listed as mean fold-changes ± standard deviation. qRT-PCR fold-changes were calculated using the relative quantitative (ΔΔCT) method, as detailed in the Methods, from three biological sample sets.
Figure 2.
Comparison of gene expression measurements by microarray hybridization and quantitative real-time PCR. Changes in gene expression were log transformed (in base 2), and the real-time qRT-PCR log2 values (y axis) were plotted against the microarray data log2 values (x axis). The coefficient of determination (R2) is given.
Given that BvgAS is the major virulence regulator in Bordetella, the information presented in this report should allow researchers to design future experiments targeting some newly identified Bvg-regulated genes and study their role in the pathogenesis of this organism. A recent study demonstrated that Bordetella bronchiseptica, Bordetella parapertussis and Bordetella pertussis all contain a higher number of transcription factors or regulatory elements than the genome size of each strain would predict [27]. The microarray analysis presented here identifies, for the first time, an additional 138 predicted global regulator elements as being a part of the BvgAS regulon, which could be described as a modulon. Moreover, the data indicates that a high number of B. bronchiseptica regulators are under control, either directly or indirectly, of BvgAS, thus highlighting an organized hierarchical network, with multiple layers of control, governing transcriptional regulation in B. bronchiseptica. Distinct phenotypic changes occurring at each end of the Bvg regulatory continuum have been described since 1960 [28]. Since then, the molecular basis underlying these morphological phases has been directly linked to the coordinate expression of the BvgAS regulon. The results presented here provide a comprehensive, genome-wide portrait of transcripts encompassing the BvgAS regulon, while also providing data validating the usefulness of utilizing the long-oligonucleotide microarray described here for studying gene expression in B. bronchiseptica.
Conclusion
Long-oligonucleotide microarrays represent a highly sensitive, cost and management efficient, alternative to cDNA microarrays. Using the complete Bordetella bronchiseptica genome sequence and ArrayOligoSelector software, a 70-mer oligonucleotide Bordetella bronchiseptica microarray was designed and assembled. This long-oligonucleotide microarray was then tested and validated by comparing changes in the global expression profiles between Bordetella bronchiseptica RB50 and its Bvg- phase-locked derivative, RB54. Data from this microarray analysis revealed 1,668 Bvg-regulated genes, which dramatically increases the number of Bvg-regulated transcripts identified to date. Additionally, gene expression profiles of previously demonstrated Bvg-regulated transcripts are consistent with previous results and quantitative real-time PCR data provided an independent verification of the microarray expression values. The results presented here provide a comprehensive, genome-wide portrait of transcripts encompassing the BvgAS regulon, and provide data validating the use of the long-oligonucleotide microarray described here for studying gene expression in Bordetella bronchiseptica.
Methods
Microarray fabrication
Using ArrayOligoSelector [18], one 70-mer oligonucleotide probe was selected to represent each B. bronchiseptica RB50 ORF [17]. The 70-bp oligonucleotide probes were synthesized (Illumina, San Diego, CA), resuspended in 3×SSC to a final concentration of 60 pmol/μL, and spotted onto poly-L-lysine-coated microscope slides, using a BioRobotics Microgrid II microarrayer (Genomic Solutions, Ann Arbor, MI) as described elsewhere [29]. Each probe was printed three times per slide. All oligonucleotide sequences are listed (see Additional File 1- Supplementary Table S2).
Bordetella Culture and RNA Isolation
B. bronchiseptica strain RB50 was isolated from a naturally infected New Zealand White rabbit [6] and RB54 is a Bvg- phase-locked derivative of RB50 [10]. B. bronchiseptica strains were maintained on Bordet-Gengou (BG) agar (Difco) containing 10% defibrinated sheep blood for determination of colony morphology and hemolytic activity. Three independent biological replicates of B. bronchiseptica RB50 and RB54, picked from well-isolated colonies on BG plates, were initially grown in Stainer-Scholte (SS) broth [30] supplemented with 40 μg/mL streptomycin. To ensure similar inocula and growth phase among all biological replicates, bacteria were then subcultured at a starting optical density at 600 nm (OD600) of 0.02 into a 250-mL flask containing 50 mL SS broth and grown at 37°C with shaking at 275 rpm for 24 hours. At this time the OD600 for the RB50 cultures were approximately 1 and the OD600 for the RB54 cultures were approximately 3.5. Total cellular RNA was extracted using Trizol reagent (Invitrogen, Carlsbad, CA), treated with RNase-free DNase I (Invitrogen, Carlsbad, CA), and purified using RNeasy columns (Qiagen, Valencia, CA) according to manufacturers' recommended protocols.
Preparation of labeled cDNA
A 2-color hybridization format was used for the microarray analysis. For each biological replicate, RNA extracted from B. bronchiseptica RB50 was used to create Cy5-labeled cDNA and RNA extracted from B. bronchiseptica RB54 was used to create Cy3-labeled cDNA. Conversely, dye-swap experiments were performed analogously, in which RNA extracted from B. bronchiseptica RB50 was used to create Cy3- labeled cDNA and B. bronchiseptica RB54 was used to create Cy5- labeled cDNA. Fluorescently-labeled cDNA copies of the total RNA pool were prepared by direct incorporation of fluorescent nucleotide analogs during a first-strand reverse transcription (RT) reaction as follows: 5 μg total RNA and 4.4 μg of random oligonucleotide hexamers were incubated 2 minutes at 98°C, cooled on ice, combined with SuperScript III RTase buffer, 0.5 mM dATP, dGTP, dCTP, 0.2 mM dTTP, 1.5 nmol Cy3- or Cy5-dUTP (Amersham), and 2 μL SuperScript III RTase (in a volume of 26 μL). This mixture was incubated 10 minutes at 25°C and 120 minutes at 50°C. The two differentially labeled reactions to be compared were mixed and buffer exchange, purification, and concentration was accomplished by microcon-10 (Amicon) filtration.
Microarray hybridization and data analysis
Oligonucleotide microarrays were first prehybridized for 1 hour in 5×SSC, 1% BSA and 0.1% SDS at 42°C, followed by washing with H2O and then with isopropanol and dried by centrifugation for 5 minutes at 50×g. Following prehybridization, 45 μL hybridization solution (labeled cDNA, 5 μg tRNA, 2×SSC, 25% formamide, 0.1% SDS) was applied to oligonucleotide microarrays and incubated in a humidified chamber overnight at 50°C. Arrays were subsequently removed from humidified chambers and quickly submerged and washed in 1×SSC and 0.05% SDS for approximately 2 minutes, followed by two additional washes for 2 minutes each in fresh 0.06×SSC. Slides were then dried by centrifugation for 5 minutes at 50×g, scanned using a GenePix 4000B microarray scanner, and analyzed with GenePix Pro software (Axon Instruments, Union City, CA). Spots were assessed visually to identify those of low quality and arrays were normalized so that the median of ratio across each array was equal to 1.0. Automatically and manually flagged spots, spots with the sum of medians (635/532) signal intensity less than or equal to 100, and spots with signal intensity below threshold (sum of median intensities plus one standard deviation above the mean background) were filtered out prior to analysis. Ratio data from the three biological replicates were compiled and normalized based on the total Cy3% intensity and Cy5% intensity to eliminate slide to slide variation. Gene expression data were then normalized to 16S rRNA. The statistical significance of the gene expression changes observed was assessed by using the significant analysis of microarrays (SAM) program [31]. A one-class unpaired SAM analysis using a false discovery rate of 0.063% (<0.1%) was preformed. Hierarchical clustering of microarray data using Euclidean Distance metrics and Average Linkage clustering was performed using MeV software from TIGR [32].
Quantitative real-time PCR
DNase-treated total RNA (1 μg) from each biological replicate was reverse transcribed using 300 ng of random oligonucleotide hexamers and SuperScript III RTase (Invitrogen, Carlsbad, CA) according to manufacturer's protocol. The resulting cDNA was diluted 1:1000 and 1 μL used in quantitative PCR reactions containing 300 nM primers and 2XSYBR Green PCR Master Mix (Applied Biosystems) using an Applied Biosystems 7300 real-time PCR detection system (Applied Biosystems, Foster City, CA). Primers were designed using Primer Express software (Applied Biosystems, Foster City, CA) and are listed in Additional File 1- Supplementary Table 4. To confirm the lack of DNA contamination, reactions without reverse transcriptase were performed. Dissociation curve analysis was performed for verification of product homogeneity. Threshold fluorescence was established within the geometric phase of exponential amplification and the cycle of threshold (Ct) determined for each reaction. The cycle of threshold (Ct) from all three biological replicates for each strain (RB50 and RB54) was compiled and the 16S RNA amplicon was used as an internal control for data normalization. Fold change in transcript level was determined using the relative quantitative method (ΔΔCT) [33].
Microarray accession numbers
Microarrays have been deposited in ArrayExpress under accession number E-MEXP-961.
Authors' contributions
TLN performed B. bronchiseptica culturing, microarray design, microarray construction, microarray analysis, cluster analysis, quantitative real-time PCR analysis, data analysis, and drafted the manuscript. TLN read and approved the final manuscript.
Supplementary Material
Table S1. Missing array elements due to gene duplications, prophage duplications, and ORF assignments missing from the completed genome annotation. In the Gene Duplication category, the ORF number representing the array element for both genes is given. Table S2. Oligonucleotide sequence for each ORF/represented array element. Table S3. Fold-Change expression values from a direct comparison between the transcriptional profile of B. bronchiseptica RB50 and RB54. DNA microarray analysis was used to measure mRNA levels present in B. bronchiseptica RB50 compared to mRNA levels present in B. bronchiseptica RB54. Differences in mRNA levels are listed as mean fold-changes + standard error. Fold-changes were calculated by averaging the data from three biological sample sets. The fluorescent labels were exchanged in dye-swap experiments performed on all three biological replicates. ORF, Name, Product, Function, and General Category headings were parsed from both Sanger annotation files [17] and Cummings et al. [14]. Data presented in the SAM, Score(d), q-value, and localdr(%) columns were assessed by using the significant analysis of microarrays (SAM) program [31]. A one-class unpaired SAM analysis using a false discovery rate of 0.063% (<0.1%) was preformed. Table S4. Quantitative real-time PCR primers.
Acknowledgments
Acknowledgements
The author would like to thank Peggy Cotter for the generous gift of the B. bronchiseptica Bvg- phase-locked derivative strain, RB54, and Brooke Peterson-Burch for his assistance with the installation and initial running of the AOS program. The author would also like to thank Eric Nicholson, Susan Brockmeier and Karen Register for critical review of the manuscript. Mention of trade names or commercial products in this article is solely for the purpose of providing specific information and does not imply recommendation or endorsement by the U.S. Department of Agriculture.
References
- Preston A, Parkhill J, Maskell DJ. The bordetellae: lessons from genomics. Nature reviews. 2004;2:379–390. doi: 10.1038/nrmicro886. [DOI] [PubMed] [Google Scholar]
- Cotter PA, Jones AM. Phosphorelay control of virulence gene expression in Bordetella. Trends in microbiology. 2003;11:367–373. doi: 10.1016/S0966-842X(03)00156-2. [DOI] [PubMed] [Google Scholar]
- Akerley BJ, Monack DM, Falkow S, Miller JF. The bvgAS locus negatively controls motility and synthesis of flagella in Bordetella bronchiseptica. Journal of bacteriology. 1992;174:980–990. doi: 10.1128/jb.174.3.980-990.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akerley BJ, Miller JF. Flagellin gene transcription in Bordetella bronchiseptica is regulated by the BvgAS virulence control system. Journal of bacteriology. 1993;175:3468–3479. doi: 10.1128/jb.175.11.3468-3479.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMillan DJ, Shojaei M, Chhatwal GS, Guzman CA, Walker MJ. Molecular analysis of the bvg-repressed urease of Bordetella bronchiseptica. Microbial pathogenesis. 1996;21:379–394. doi: 10.1006/mpat.1996.0069. [DOI] [PubMed] [Google Scholar]
- Cotter PA, Miller JF. BvgAS-mediated signal transduction: analysis of phase-locked regulatory mutants of Bordetella bronchiseptica in a rabbit model. Infect Immun. 1994;62:3381–3390. doi: 10.1128/iai.62.8.3381-3390.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akerley BJ, Cotter PA, Miller JF. Ectopic expression of the flagellar regulon alters development of the Bordetella-host interaction. Cell. 1995;80:611–620. doi: 10.1016/0092-8674(95)90515-4. [DOI] [PubMed] [Google Scholar]
- Martinez de Tejada G, Cotter PA, Heininger U, Camilli A, Akerley BJ, Mekalanos JJ, Miller JF. Neither the Bvg- phase nor the vrg6 locus of Bordetella pertussis is required for respiratory infection in mice. Infect Immun. 1998;66:2762–2768. doi: 10.1128/iai.66.6.2762-2768.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merkel TJ, Stibitz S, Keith JM, Leef M, Shahin R. Contribution of regulation by the bvg locus to respiratory infection of mice by Bordetella pertussis. Infect Immun. 1998;66:4367–4373. doi: 10.1128/iai.66.9.4367-4373.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cotter PA, Miller JF. A mutation in the Bordetella bronchiseptica bvgS gene results in reduced virulence and increased resistance to starvation, and identifies a new class of Bvg-regulated antigens. Molecular microbiology. 1997;24:671–685. doi: 10.1046/j.1365-2958.1997.3821741.x. [DOI] [PubMed] [Google Scholar]
- Weiss AA, Falkow S. Genetic analysis of phase change in Bordetella pertussis. Infect Immun. 1984;43:263–269. doi: 10.1128/iai.43.1.263-269.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deora R. Differential regulation of the Bordetella bipA gene: distinct roles for different BvgA binding sites. Journal of bacteriology. 2002;184:6942–6951. doi: 10.1128/JB.184.24.6942-6951.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hot D, Antoine R, Renauld-Mongenie G, Caro V, Hennuy B, Levillain E, Huot L, Wittmann G, Poncet D, Jacob-Dubuisson F, Guyard C, Rimlinger F, Aujame L, Godfroid E, Guiso N, Quentin-Millet MJ, Lemoine Y, Locht C. Differential modulation of Bordetella pertussis virulence genes as evidenced by DNA microarray analysis. Mol Genet Genomics. 2003;269:475–486. doi: 10.1007/s00438-003-0851-1. [DOI] [PubMed] [Google Scholar]
- Cummings CA, Bootsma HJ, Relman DA, Miller JF. Species- and strain-specific control of a complex, flexible regulon by Bordetella BvgAS. Journal of bacteriology. 2006;188:1775–1785. doi: 10.1128/JB.188.5.1775-1785.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson A, Mazumder A, Stewart M, Balasubramanian S. Technology for microarray analysis of gene expression. Current opinion in biotechnology. 1998;9:609–614. doi: 10.1016/S0958-1669(98)80138-9. [DOI] [PubMed] [Google Scholar]
- Hughes TR, Mao M, Jones AR, Burchard J, Marton MJ, Shannon KW, Lefkowitz SM, Ziman M, Schelter JM, Meyer MR, Kobayashi S, Davis C, Dai H, He YD, Stephaniants SB, Cavet G, Walker WL, West A, Coffey E, Shoemaker DD, Stoughton R, Blanchard AP, Friend SH, Linsley PS. Expression profiling using microarrays fabricated by an ink-jet oligonucleotide synthesizer. Nature biotechnology. 2001;19:342–347. doi: 10.1038/86730. [DOI] [PubMed] [Google Scholar]
- Parkhill J, Sebaihia M, Preston A, Murphy LD, Thomson N, Harris DE, Holden MT, Churcher CM, Bentley SD, Mungall KL, Cerdeno-Tarraga AM, Temple L, James K, Harris B, Quail MA, Achtman M, Atkin R, Baker S, Basham D, Bason N, Cherevach I, Chillingworth T, Collins M, Cronin A, Davis P, Doggett J, Feltwell T, Goble A, Hamlin N, Hauser H, Holroyd S, Jagels K, Leather S, Moule S, Norberczak H, O'Neil S, Ormond D, Price C, Rabbinowitsch E, Rutter S, Sanders M, Saunders D, Seeger K, Sharp S, Simmonds M, Skelton J, Squares R, Squares S, Stevens K, Unwin L, Whitehead S, Barrell BG, Maskell DJ. Comparative analysis of the genome sequences of Bordetella pertussis, Bordetella parapertussis and Bordetella bronchiseptica. Nature genetics. 2003;35:32–40. doi: 10.1038/ng1227. [DOI] [PubMed] [Google Scholar]
- Bozdech Z, Zhu J, Joachimiak MP, Cohen FE, Pulliam B, DeRisi JL. Expression profiling of the schizont and trophozoite stages of Plasmodium falciparum with a long-oligonucleotide microarray. Genome biology. 2003;4:R9. doi: 10.1186/gb-2003-4-2-r9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Julio SM, Cotter PA. Characterization of the filamentous hemagglutinin-like protein FhaS in Bordetella bronchiseptica. Infection and immunity. 2005;73:4960–4971. doi: 10.1128/IAI.73.8.4960-4971.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson AL, Chen J. ATP-binding cassette transporters in bacteria. Annu Rev Biochem. 2004;73:241–268. doi: 10.1146/annurev.biochem.73.011303.073626. [DOI] [PubMed] [Google Scholar]
- Coutte L, Antoine R, Drobecq H, Locht C, Jacob-Dubuisson F. Subtilisin-like autotransporter serves as maturation protease in a bacterial secretion pathway. The EMBO journal. 2001;20:5040–5048. doi: 10.1093/emboj/20.18.5040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenson TH, Weiss AA. DsbA and DsbC are required for secretion of pertussis toxin by Bordetella pertussis. Infection and immunity. 2002;70:2297–2303. doi: 10.1128/IAI.70.5.2297-2303.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yorgey P, Rahme LG, Tan MW, Ausubel FM. The roles of mucD and alginate in the virulence of Pseudomonas aeruginosa in plants, nematodes and mice. Molecular microbiology. 2001;41:1063–1076. doi: 10.1046/j.1365-2958.2001.02580.x. [DOI] [PubMed] [Google Scholar]
- Kuwae A, Matsuzawa T, Ishikawa N, Abe H, Nonaka T, Fukuda H, Imajoh-Ohmi S, Abe A. BopC is a novel type III effector secreted by Bordetella bronchiseptica and has a critical role in type III-dependent necrotic cell death. The Journal of biological chemistry. 2006;281:6589–6600. doi: 10.1074/jbc.M512711200. [DOI] [PubMed] [Google Scholar]
- Porter JF, Parton R, Wardlaw AC. Growth and survival of Bordetella bronchiseptica in natural waters and in buffered saline without added nutrients. Appl Environ Microbiol. 1991;57:1202–1206. doi: 10.1128/aem.57.4.1202-1206.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giardina PC, Foster LA, Musser JM, Akerley BJ, Miller JF, Dyer DW. bvg Repression of alcaligin synthesis in Bordetella bronchiseptica is associated with phylogenetic lineage. Journal of bacteriology. 1995;177:6058–6063. doi: 10.1128/jb.177.21.6058-6063.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madan Babu M, Teichmann SA, Aravind L. Evolutionary dynamics of prokaryotic transcriptional regulatory networks. Journal of molecular biology. 2006;358:614–633. doi: 10.1016/j.jmb.2006.02.019. [DOI] [PubMed] [Google Scholar]
- Lacey BW. Antigenic modulation of Bordetella pertussis. The Journal of hygiene. 1960;58:57–93. doi: 10.1017/s0022172400038134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisen MB, Brown PO. DNA arrays for analysis of gene expression. Methods Enzymol. 1999;303:179–205. doi: 10.1016/s0076-6879(99)03014-1. [DOI] [PubMed] [Google Scholar]
- Stainer DW, Scholte MJ. A simple chemically defined medium for the production of phase I Bordetella pertussis. Journal of general microbiology. 1970;63:211–220. doi: 10.1099/00221287-63-2-211. [DOI] [PubMed] [Google Scholar]
- Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proceedings of the National Academy of Sciences of the United States of America. 2001;98:5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saeed AI, Sharov V, White J, Li J, Liang W, Bhagabati N, Braisted J, Klapa M, Currier T, Thiagarajan M, Sturn A, Snuffin M, Rezantsev A, Popov D, Ryltsov A, Kostukovich E, Borisovsky I, Liu Z, Vinsavich A, Trush V, Quackenbush J. TM4: a free, open-source system for microarray data management and analysis. BioTechniques. 2003;34:374–378. doi: 10.2144/03342mt01. [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT Method. Methods (San Diego, Calif) 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Missing array elements due to gene duplications, prophage duplications, and ORF assignments missing from the completed genome annotation. In the Gene Duplication category, the ORF number representing the array element for both genes is given. Table S2. Oligonucleotide sequence for each ORF/represented array element. Table S3. Fold-Change expression values from a direct comparison between the transcriptional profile of B. bronchiseptica RB50 and RB54. DNA microarray analysis was used to measure mRNA levels present in B. bronchiseptica RB50 compared to mRNA levels present in B. bronchiseptica RB54. Differences in mRNA levels are listed as mean fold-changes + standard error. Fold-changes were calculated by averaging the data from three biological sample sets. The fluorescent labels were exchanged in dye-swap experiments performed on all three biological replicates. ORF, Name, Product, Function, and General Category headings were parsed from both Sanger annotation files [17] and Cummings et al. [14]. Data presented in the SAM, Score(d), q-value, and localdr(%) columns were assessed by using the significant analysis of microarrays (SAM) program [31]. A one-class unpaired SAM analysis using a false discovery rate of 0.063% (<0.1%) was preformed. Table S4. Quantitative real-time PCR primers.