Abstract
Small molecules that produce nonfunctional protein–protein complexes are an alternative to competitive inhibitors for the inhibition of protein functions. Here we target the activation of the small GTP-binding protein Arf1, a major regulator of membrane traffic, by the Sec7 catalytic domain of its guanine nucleotide exchange factor ARNO. The crystal structure of the Arf1-GDP/ARNO complex, which initiates the exchange reaction, was used to discover an inhibitor, LM11, using in silico screening of a flexible pocket near the Arf1/ARNO interface. Using fluorescence kinetics and anisotropy, NMR spectroscopy and mutagenesis, we show that LM11 acts following a noncompetitive mechanism in which the inhibitor targets both Arf1-GDP and the Arf1-GDP/ARNO complex and produces a nonfunctional Arf-GDP/ARNO complex whose affinity is similar to that of the native complex. In addition, LM11 recognizes features of both Arf and ARNO near the Arf/Sec7 interface, a characteristic reminiscent of the paradigm interfacial inhibitor Brefeldin A. We then show that LM11 is a cell-active inhibitor that impairs Arf-dependent trafficking structures at the Golgi. Furthermore, LM11 inhibits ARNO-dependent migration of Madin–Darby canine kidney (MDCK) cells, demonstrating that ARNO is a target of LM11 in cells. Remarkably, LM11 inhibits the activation of Arf1 but not Arf6 in vitro, pointing to a possible synergy between Arf1 and Arf6 activation by ARNO in cell migration. Our design method shows that flexible regions in protein–protein complexes provide drugable sites with the potential to develop novel tools for investigating and inhibiting signaling pathways.
Keywords: protein–protein interactions, inhibition, GTPase, guanine nucleotide exchage factor, Arf1 factor
The discovery of small molecules that affect protein interactions is of crucial importance for the development of innovative therapeutics (1–3) and for the investigation of molecular pathways in cells (4, 5). However, targeting protein interactions by competitive inhibitors is difficult, because small molecules must compete with large macromolecular partners (3). We recently proposed a concept to inhibit protein functions, which we refer to as “interfacial inhibition” (6, 7). Interfacial inhibitors bind to protein–protein complexes in or near their interface in the course of structural transitions, thereby converting the complexes into abortive conformations rather than preventing the interaction of their components. Nature provides most examples of interfacial inhibitors, of which some are used in human therapeutics, such as colchicine, vinblastine, or camptothecin (7). The critical issue, however, is how to discover or design inhibitors that promote inactive conformations of protein–protein complexes. In this study, we focused on the activation of a small GTP-binding protein (SMG) by its guanine nucleotide exchange factors (GEFs). This reaction is of particular interest, because GEFs define the spatiotemporal specificity of SMG activation by collecting activation signals and stimulating the intrinsically slow GDP/GTP exchange. GEFs are therefore emerging as potential targets in diseases where SMGs are up-regulated, such as cancer or infections (reviewed in refs. 8 and 9). This recently fostered the exploration of novel strategies for discovering competitive inhibitors acting either on the GEF (10–12) or on the SMG (13, 14). An alternative strategy, independent of the nature of the inhibition mechanism, was also devised by using an exchange assay reconstituted in yeast (15).
Here we seek to inhibit the activation of Arf1, a major regulator of cellular traffic (reviewed in ref. 16), by the Sec7 catalytic domain of its GEF ARNO. Remarkably, the first known inhibitor of a GEF is the paradigm interfacial inhibitor Brefeldin A (BFA), which inhibits the activation of Arf1 by a subset of its GEFs (17). BFA takes advantage of the fact that the GDP/GTP exchange reaction involves successive discrete steps, of which the initiating Arf-GDP/Sec7 complex is hijacked by the drug (6, 18) (Fig. 1A). To inhibit the activation of Arf1 by ARNO, which is BFA-insensitive, we target a pocket located near the Arf1/ARNO interface, using the crystal structure of an Arf1-GDP/ARNO complex that was captured by an E to K mutation in the catalytic site of ARNO (ARNOE156K hereafter; refs. 6 and 19). Combining in silico virtual screening to in vitro and cellular assays, we discover and characterize an inhibitor screened to inhibit the function of a SMG by producing a nonfunctional protein–protein complex.
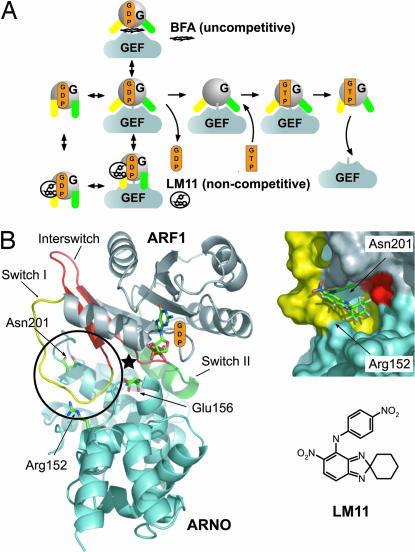
Fig. 1.
The mechanism of Arf activation by its GEFs and its inhibition by LM11. (A) Comparison of the uncompetitive mechanism of BFA and the noncompetitive mechanism of LM11. Representative crystal structures are available for unbound Arf-GDP and Arf-GTP, for the nucleotide-free Arf/Sec7 intermediate, and for the Arf-GDP/Sec7 intermediate, which subdivides in two successive conformations trapped by BFA and by the E to K charge reversal of the catalytic glutamate of the Sec7 domain. (B) The pocket used for the structure-based in silico screen (circle), comprising the switch 1 (yellow) and strand β2 (red) of the interswitch of Arf1 and loop 198–202 (cyan) of ARNO. The approximate location of the binding site for BFA in BFA-sensitive ArfGEFs is shown by an asterisk. A closeup view of the pocket is shown in surface representation with LM11 docked. The chemical structure of LM11 is shown in Inset.
Results
Structure-Based Identification of a Noncompetitive Inhibitor of Arf1 Activation by ARNO.
The Arf1-GDP/ARNOE156K complex features a pocket near the Arf1/ARNO interface that comprises residues of Arf1 and ARNO and is remote from the site that binds BFA in BFA-sensitive ArfGEFs (Fig. 1B). As seen by comparing the reaction intermediates trapped by BFA (6), by the E/K mutation (6), and by removal of GDP (20), this pocket persists throughout the exchange reaction but undergoes large conformational changes. We reasoned that small molecules targeting this pocket should stall the exchange reaction and selected 17 candidate inhibitors using in silico structure-based screening of the Chembridge database.
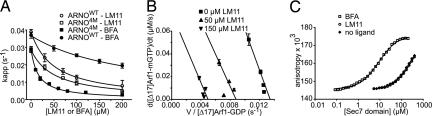
Inhibition activities of the selected compounds (200 μM) were measured by tryptophan or mant-nucleotide fluorescence kinetics using [Δ17]Arf1, a truncated form of Arf whose activation is independent of membranes (19), and the Sec7 domain of human ARNO carrying four mutations that sensitize it to BFA (ARNO4M hereafter) (21, 22). One compound, LM11 (Fig. 1B), reduced the apparent exchange rate constant (kapp) from 0.028 to 0.0078 s−1. We controlled that addition of 0.01% Triton X-100 had no effect (data not shown), indicating that LM11 does not act by aggregation (23). The apparent inhibition constant (kiapp) was 49.7 ± 8.0 μM, a value in the same range as that found for BFA using ARNO4M (15.0 ± 1.1 μM) or BIG1 (22) (Fig. 2A). The Michaelis–Menten formalism has proven a convenient approximation to analyze the mechanism of GEFs and their inhibition (19, 22). Its application to LM11 using Eadie–Hofstee linearization yielded parallel lines (Fig. 2B), which rules out a competitive mechanism, in which LM11 and Arf1 would compete for the same binding site on ARNO. It also excludes a strictly uncompetitive mechanism such as that of BFA in which the compound would bind preferentially to the Arf1/ARNO complex (see Fig. 1A). Thus, LM11 acts by a noncompetitive mechanism.
Fig. 2.
Analysis of the noncompetitive mechanism of LM11. (A) Determination of the apparent inhibition constants (kiapp) of BFA and LM11 measured with [Δ17]Arf1 and either ARNOWT (BFA resistant) or ARNO4M (BFA sensitive) as indicated and increasing concentrations of the drug as in SI Fig. 7C. (B) Eadie–Hoffstee linearization of the Michaelis–Menten saturation curves obtained in the presence of the indicated concentrations of LM11 and by varying [Δ17]Arf1 concentration between 0 and 5 μM. (C) LM11 does not inhibit the formation of the Arf1-GDP/ARNO complex. Anisotropy binding profiles were obtained from the titration of Alexa488-labeled [Δ17]Arf1-GDP with ARNOWT in the absence or in the presence of 100 μM LM11 as indicated. Titration in the presence of 100 μM BFA was performed with the BFA-sensitive ARNO4M mutant. Anisotropy values measured with ARNOWT or ARNO4M in the absence of drug were identical (data not shown).
Defining the Noncompetitive Mechanism of Inhibition of LM11.
Noncompetitive inhibition profiles are complex to characterize, because they cover many possible mechanisms (3). Based on the extensive biochemical knowledge on the spontaneous and Sec7-stimulated exchange reaction (6, 18–22), we set up to identify which steps are affected by the inhibitor.
We first analyzed the effect of LM11 on spontaneous exchange on [Δ17]Arf1. LM11 inhibited mGDP/GTP exchange (kapp = 0.004 ± 0.0002 and 0.0004 ± 0.0004 s−1 without and with 150 μM LM11) but had no effect on methylanthraniloyl GTP (mGTP)/GTP exchange (kapp = 0.0006 ± 0.0001 and 0.0005 ± 0.0002 s−1) [supporting information (SI) Fig. 7A], indicating that it acts before the formation of nucleotide-free Arf. We then investigated whether LM11 would inhibit ARNO-stimulated exchange beyond the formation of the nucleotide-free Arf1/ARNO intermediate. LM11 had no effect on the kinetics of GTP binding to a preformed nucleotide-free Arf1-ARNO complex (kapp = 0.088 ± 0.011 and 0.085 ± 0.009 s−1) (SI Fig. 7B). We conclude from these experiments that LM11 acts upstream of the nucleotide-free intermediate in both the spontaneous and ARNO-stimulated exchange reaction, and that it does not compete with GTP.
To exclude that LM11 acts allosterically by promoting a conformation of either Arf1-GDP or ARNO that cannot form the Arf1-GDP/ARNO complex, we then measured the dissociation constants of this complex by fluorescence anisotropy (Fig. 2C). Using the BFA-sensitive ARNO4M, the apparent Kd was in the 150–200 μM range in the absence of inhibitor and decreased to 8.6 ± 0.7 μM in the presence of BFA (100 μM), in agreement with its uncompetitive mechanism. In contrast, addition of 100 μM LM11 did not modify the apparent Kd, which would have been the case if the formation of the complex was impaired upon binding of the drug. This experiment further rules out unspecific inhibitor aggregation (23). Exclusion of an allosteric effect allows us to conclude that LM11 binds to Arf1-GDP/ARNO and induces a nonproductive complex whose stability is similar to that of the noninhibited complex.
LM11 Targets Arf1-GDP and ARNO in Regions Close to the Arf/ARNO Interface.
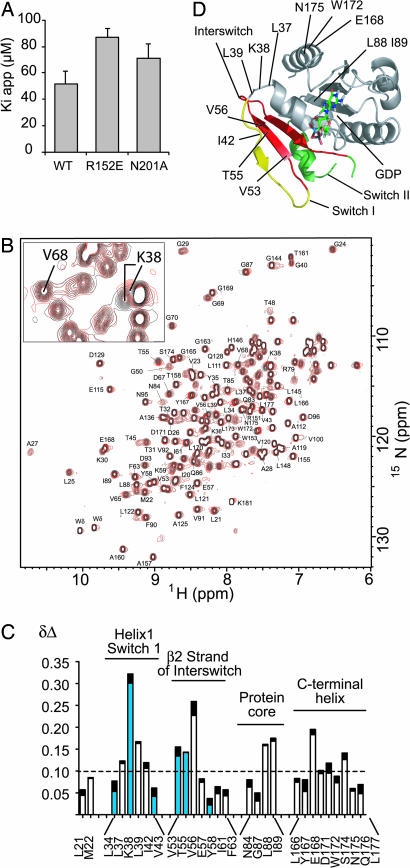
The in silico screen model predicts that LM11 recognizes both ARNO and Arf1-GDP, which is compatible with its inhibitory mechanism. To probe it, we first analyzed the activity of LM11 toward two ARNO constructs carrying mutations in the screened interfacial pocket, R152E or N201A (Fig. 1B), both of which support efficient activation of Arf1 (24). Both mutations resulted in a reduced sensitivity of the exchange reaction to LM11 as seen by the increase of kiapp (Fig. 3A), indicating that LM11 is sensitive to ARNO features in the target pocket.
Fig. 3.
LM11 binds to Arf1-GDP and is sensitive to ARNO mutations. (A) Comparison of kiapp measured for ARNOWT and the R152E and N201A ARNO mutants. (B) Superposition of HSQC spectra of 15N-labeled [Δ17]Arf1-GDP with (red) or without (black) LM11. A zoom is shown in Inset. (C) Chemical-shift variations are indicated as a sum of Δδ1H (black) + Δδ15N (white). Residues that belong to the pocket used for the in silico screen are indicated in blue. (D) Structure of Arf1-GDP, showing residues with HSQC chemical shift variations >0.1 ppm.
We then analyzed the binding of LM11 to 15N-labeled [Δ17]Arf1-GDP using NMR HSQC chemical shift mapping of LM11. Chemical shift variations were localized and of small amplitude, indicating that LM11 binds to Arf1-GDP but does not induce nonspecific denaturation or major conformational changes of the protein. (Fig. 3 B and C; assignments and chemical shifts in SI Tables 1 and 2). No perturbation was observed at either the nucleotide or the BFA binding-sites, confirming that LM11 does not compete with nucleotides and that its binding site is distinct from that of BFA. The largest chemical-shift variations (Δδ1H + Δδ15n > 0.1 ppm) were located in or near the switch 1 region and the interswitch, with a significant overlap with residues comprised in the pocket of the in silico screen (Fig. 3 C and D; SI Table 2). A few additional variations were also detected near the N-terminal helix pocket, which are close enough to the target pocket to respond to the propagation of local reorganization. Altogether, the NMR data indicate that LM11 has a discrete binding site on Arf1-GDP, and that this site is compatible with the screening hypothesis. Because NMR chemical-shift mapping is not sufficient to define the binding site at atomic resolution, details of LM11 interactions with Arf1-GDP and Arf1-GDP/ARNO must now await crystallographic analysis.
Combined, the binding area of LM11 on Arf1-GDP, its sensitivity to ARNO mutations in the target pocket, and its ability to induce a nonproductive Arf1-GDP/ARNO complex qualifies it as an inhibitor targeting an interfacial surface of a protein–protein complex.
Specificity of LM11 in Vitro.
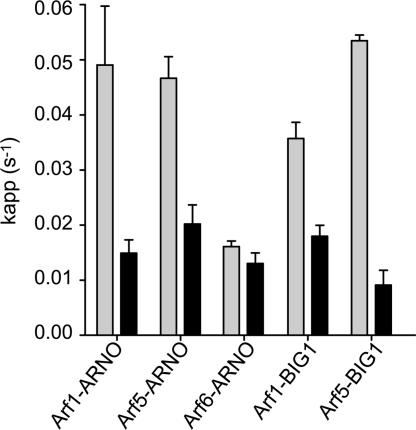
We then analyzed the specificity of LM11 toward different SMG/GEF systems in vitro. We first controlled that LM11 had no effect on the activation of two members of the Rho subfamily by their GEFs, RhoA/Gef337 (0.0037 ± 0.0002 and 0.0028 ± 0.0002 s−1 without and with 150 μM LM11, respectively) and RhoG/TrioD1 (0.0037 ± 0.0002 and 0.003 ± 0.0001 s−1), indicating that it is specific for the Arf/ArfGEF system. We then compared the effect of LM11 on BFA-sensitive and insensitive Sec7 domains (Fig. 4). LM11 inhibited the activation of [Δ17]Arf1 and [Δ14]Arf5 stimulated by the Sec7 domain of BIG1, a BFA-sensitive ArfGEF, which is active on classes I and II Arfs in vitro and functions at the Golgi (25). It also inhibited with similar efficiency ARNOWT, which is naturally insensitive to BFA (26), and its BFA-sensitive version ARNO4M (respectively 49.7 ± 8.0 and 50.1 ± 5.5 μM) (Fig. 2A). Finally, we measured the effect of LM11 on the activation of [Δ17]Arf1, [Δ14]Arf5 and [Δ13]Arf6, as representatives of Arf classes I, II and III, all of which are activated by ARNO in vitro (22) (Fig. 4). Remarkably, LM11 inhibited ARNO-stimulated nucleotide exchange of Arf1 and Arf5, but it had no effect on Arf6, indicating that it distinguishes between the different Arf proteins. Such a specificity to Arf isoforms, previously established for BFA with BFA-sensitive ArfGEFs (22), is consistent with the interfacial mechanism.
Fig. 4.
Specificity of LM11 in vitro. For each complex, 1 μM SMG and 50 nM GEF were incubated in the absence (black) or in the presence of 150 μM of LM11 (gray). The nucleotide exchange reaction was initiated with the addition of 2 μM mGTP and monitored by time-resolved fluorescence.
LM11 Affects Golgi-Derived Trafficking Structures in HeLa Cells.
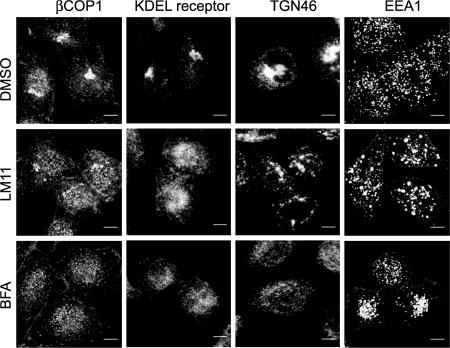
Our preliminary analysis indicates that LM11 inhibits the activation of endogenous Arf proteins in HeLa cells, using a pull-down assay based on the interaction of GTP-loaded Arf proteins with their GGA3 effector (SI Fig. 8). We thus investigated the effect of LM11 on the morphology of trafficking structures at the Golgi, of which Arf1 is a key regulator (25). We first checked that LM11 has no effect on the phalloidin-stained actin cytoskeleton in these cells (data not shown). Endogenous markers whose localization responds to BFA treatment (27) were then visualized by immunofluorescence confocal microscopy in HeLa cells treated with 100 μM LM11 or BFA for 2 h (Fig. 5). As BFA, LM11 caused the dispersion of two cis-Golgi markers, βCOP and the KDEL receptor, from the perinuclear region into small punctate structures throughout the cytoplasm. Arf1 has also been implicated, in a less well established manner, in endosomal trafficking (28, 29). LM11 induced the dispersion of TGN46, which cycles between the trans Golgi network and endosomes, but the resulting punctate structures were significantly larger than those resulting from BFA treatment. A direct effect of LM11 on endosomal structures was then analyzed with EEA1, an early endosome marker, which is present in punctate structures in control cells. Whereas BFA slightly concentrated the punctate structures in the perinuclear region, LM11 accumulated large structures, which resembled those labeled for TGN46. Together, these observations suggest that LM11 affects Arf-dependent trafficking structures at the Golgi as predicted by its impairment of the activation of classes I and II Arfs in vitro. In addition, LM11 has a morphological effect on endosomal structures that differs from that of BFA.
Fig. 5.
Effects of LM11 on Arf1-dependent structures in HeLa cells. Specific markers of membrane traffic, designated on the top, were analyzed by fluorescence confocal microscopy of HeLa cells treated for 2 h with 0.5% of DMSO or 100 μM LM11 or BFA, as indicated on the left. (Scale bar, 8 μm.)
LM11 Inhibits ARNO-Dependent Migration of MDCK Cells.
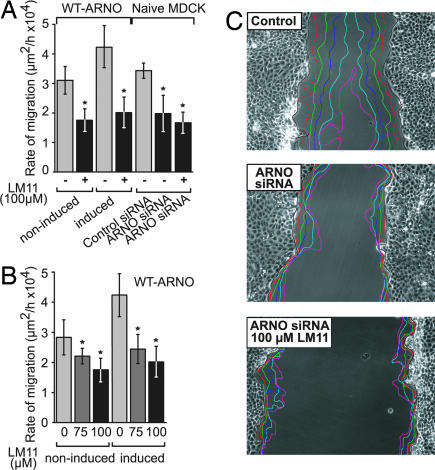
To probe whether LM11 inhibits ARNO in cells, we tested its effect on the migration of MDCK cells, which has been shown to depend on the activation of Arf proteins by ARNO (30, 31). Using a wound closure assay and MDCK cells stably transfected with a doxycycline-repressible wild-type ARNO construct (wt-ARNO), we first showed that LM11 inhibited the rate of migration of noninduced MDCK cells expressing endogenous levels of ARNO (expression repressed with doxycycline, Fig. 6A Left). The effect of LM11 was then measured in cells expressing moderate levels of wt-ARNO (<8-fold over endogenous ARNO, without doxycycline). As previously reported, the rate of migration was increased by 54% by exogenous wt-ARNO expression compared with noninduced wt-ARNO MDCK cells (Fig. 6A; see also SI Movie 1a). LM11 fully inhibited the increase in migration rate resulting from ARNO expression, yielding the same migration rate after LM11 treatment in induced and noninduced wt-ARNO MDCK cells (Fig. 6A; see also SI Movie 1b). This effect was dose-dependent in both cases (Fig. 6B). This suggests that the effect of LM11 in noninduced and wt-ARNO-expressing MDCK cells reflects its inhibition of endogenous and combined exogenous/endogenous ARNO activity, respectively. The activity of LM11 was then analyzed in MDCK cells depleted of ARNO by siRNA (Fig. 6C). Silencing of ARNO inhibited the migration of MDCK cells after wounding to the same extent as LM11 treatment in naive MDCK cells (Fig. 6C). Remarkably, LM11 had no more effect in ARNO-depleted MDCK cells (Fig. 6A). Finally, we compared the effect of LM11 to that of BFA, which disrupts cis- and trans-Golgi structures in MCDK cells >15–20 μM (C. Jackson, personal communication). BFA treatment at this concentration had no effect on the migration rate (data not shown), ruling out an indirect effect of LM11 on migration because of its inhibition of Arf functions at the Golgi. Together, these experiments strongly suggest that the inhibition of MDCK cells migration by LM11 is mediated by its specific inhibition of ARNO.
Fig. 6.
LM11 inhibits ARNO-dependent migration of MDCK cells. (A) Cell sheet migration after scratch wounding analyzed by time-lapse videomicroscopy (SI Movie 1). Data were evaluated by using Student's t test. ∗, P < 0.0001. (B) Dose dependence of LM11 inhibition in noninduced and induced wild-type ARNO-expressing MDCK cells. (C) MDCK cell sheet migration was followed by time-lapse videomicroscopy in the presence of a nonsilencing siRNA (Top), of an ARNO siRNA (Middle), or of 100 μM LM11 and an ARNO siRNA (Bottom). Each colored line represents the hourly progression of the cell monolayer during the first 6 h after wounding.
Discussion
Inhibition of Protein Functions by Targeting a Protein–Protein Complex.
We recently proposed that signaling protein–protein complexes undergoing conformational motions feature cavities in or near their interfaces that are appropriate for the design or screening of inhibitors (7). In this work, we have taken advantage that the Sec7-stimulated activation of Arf has been described in exceptional detail by crystallographic structures (6) for the structure-based discovery of LM11, a noncompetitive inhibitor that targets the Arf1-GDP/ARNO complex, and we demonstrate that the inhibitor is active in cells toward both Arf1 and ARNO functions. Based on our kinetics, fluorescence anisotropy, mutagenesis, and NMR analysis, we propose that LM11 binds to both Arf-GDP and the Arf-GDP/ARNO complex, yielding a ternary complex of affinity similar to that of the normal reaction intermediate but impaired for the conformational conversion that decreases the affinity for GDP and yields the nucleotide-free complex (Fig. 1A). Our data indicate that LM11 recognizes both components of the complex, suggesting that the expected increase in affinity of the Arf-GDP/ARNO complex because of complex/inhibitor interactions may be compensated for by a less-than-optimal protein/protein interaction between Arf-GDP and ARNO because of the inhibitor. We surmise that LM11 takes advantage of the flexibility of the switch 1 region of Arf1-GDP, which can be observed in unbound Arf1-GDP using NMR dynamics (V. Buosi, C. van Heijenoort, and E. Guittet, personal communication), which results in favorable characteristics for binding a small molecule. In this respect, LM11 probably uses, at the level of a protein–protein complex, adaptive physicochemical characteristics similar to those previously described for unbound proteins within the surface that they use to form protein–protein interactions (32, 33). Remarkably, the effect of LM11 on both trafficking structures and MDCK migration was strong and rapid despite its modest kiapp. This highlights the kinetics component of the inhibitory response, as described for the interfacial inhibitor BFA, in which the inhibitor, by yielding a nonfunctional protein–protein intermediate, stalls the reaction on a sufficiently long timescale for the biological effects to manifest. The discovery of this noncompetitive inhibitor emphasizes that flexible/dynamic regions appearing in protein–protein complexes provide “drugable” sites.
A Tool to Investigate BFA-Insensitive Arf Pathways in Cells.
To date, the substrate specificity of ARNO in vivo has proven difficult to resolve, notably because ARNO and Arf6 are not sensitive to BFA (22, 26). On the one hand, ARNO is active on both Arf1 and Arf6 in vitro with a strong preference for Arf1 (22, 26, 34). On the other hand, ARNO significantly activates Arf6 in cells and is recruited to sites of Arf6 activity at the plasma membrane (30), and it is involved in the migration of MDCK cells (30, 31), a process believed to involve Arf6 preferentially to Arf1. The unique ability of LM11 to selectively inhibit Arf1, but not Arf6, activation by ARNO in vitro, while strongly inhibiting ARNO-dependent migration in vivo, addresses the question of an unexpected involvement of Arf1 activation by ARNO in addition to Arf6 in this process, as suggested in ref. 35. LM11 should now provide a novel tool for future investigations of endogenous BFA-insensitive Arf pathways and of the promiscuity of the ARNO family for Arf1 and Arf6 in cells. In conclusion, our study provides the proof of principle that small molecules that target protein–protein complexes, including low-affinity intermediates, can yield cell-active inhibitors. Such molecules should be instrumental in deciphering cellular pathways and elucidating their specificity, with the potential for development into new therapeutic compounds.
Materials and Methods
Proteins and Reagents.
All Arf and Sec7 constructs and mutants were expressed and purified as described in ref. 22. 15N-labeled [Δ17]Arf1 was expressed as in ref. 36. GST-fused RhoG, RhoA, TRIO (DH1 domain), and GEF337 (DH domain) are a kind gift of A. Debant, A. Blangy, and P. Fort (Centre de Recherche de Biochimie Macromoléculaire, Centre National de la Recherche Scientifique, Montpellier, France). BFA was purchased from Sigma (St. Louis, MO); N-mGTP from Euromedex (Souffelweyersheim, France); alkaline phosphatase from New England Biolabs (Ipswich, MA); molecules selected by virtual screening from Chembridge (San Diego, CA); mouse monoclonal antibodies (Abs) against βCOPI and EEA1 from Sigma and Transduction Laboratories (Lexington, KY); and TRITC-labeled phalloidin and Alexa 488-conjugated secondary Abs from Sigma and Molecular Probes (Leiden, The Netherlands). Mouse Ab against KDEL receptor and sheep Ab against TGN46 were a kind gift from C. Jackson (Laboratoire d'Enzymologie et Biochimie Structurales, Centre National de la Recherche Scientifique).
Virtual Screening.
Structure-based screening for inhibitors was targeted at an interfacial pocket identified with the “binding site” module in Insight II (Accelrys, Cambridge, U.K.). The pocket was first screened for the binding of small fragments with the LUDI module of Insight II. The five fragments with higher scores were used to filter the Chembridge Express-Pick virtual library using the MDL.ISIS/Base software (Elsevier, Amsterdam, The Netherlands), yielding 3,227 commercially available compounds of <500 Da containing at least one fragment. These compounds were then docked into the pocket (defined as all atoms within 10 Å of Asn-201 in ARNO) by using FlexX 1.13.1 (Tripos Associates, Villebon, France). Standard parameters were used for iterative growing and subsequent scoring of FlexX poses as described in ref. 37. Docking positions closer than 5 Å to Asn-201 and with a score smaller than −25 were reranked with X-Score (http://sw16.im.med.umich.edu/software/xtool), which has a more accurate estimation of binding free energies, from which 17 compounds were selected by visual inspection of the top 113 scores.
Kinetics Measurements.
All kinetics experiments were performed with Arf proteins truncated of their N-terminal helix and loaded with GDP before the experiments. Activation of [Δ17]Arf1 was monitored by either tryptophan fluorescence (emission/excitation wavelengths of 292/340 nm), fluorescence of the N-methylanthraniloyl fluorophore (mGDP or mGTP) (360/440 nm), or FRET (292/440 nm). All measurements were performed at 37°C in 50 mM Tris (pH 8)/50 mM NaCl/2 mM MgCl2/2 mM 2-mercaptoethanol. Inhibitors were incubated for 5 min before initiating the reaction with GTP or mGTP. Fluorescence data were fitted by using the program Origin 6.1 (Microcal, Northampton, MA). Spontaneous mGDP/GTP exchange of [Δ17]Arf1 was measured with [Δ17]Arf1 (1 μM) loaded with mGDP (10 μM) in the presence of EDTA with or without LM11 (150 μM) and followed by the addition of GTP (200 μM). Spontaneous mGTP/GTP exchange was measured as for the mGDP/GTP exchange. The nucleotide-free complex was formed by elimination of GDP by alkaline phosphatase (20 units) and 2 mM EDTA as described in ref. 38; binding of GTP to the complex (0.2 μM) was then initiated with 1 μM mGTP in the presence or absence of 150 μM LM11. All values are means ± SD of at least three independent experiments.
Fluorescence Anisotropy.
[Δ17]Arf1-GDP was labeled with Alexa 488 at pH 8.3 at 4°C for 2 h, conditions that favor unique labeling of the N terminus (labeling ratio 72%). Fluorescence anisotropy profiles were obtained in the serial dilution format on a Beacon 2000 (Panvera, Madison, WI) polarization instrument. Experiments were performed at 4°C by using Alexa 488-labeled [Δ17]Arf1-GDP at a fixed concentration (10 nM) in 50 mM Tris, pH 8/200 mM NaCl/2 mM MgCl2/100 μM GDP/10% glycerol/2 mM 2-mercaptoethanol. Anisotropy values were similar at 4°C and 37°C (data not shown). Data points were taken at equilibrium starting from 120 μl of a solution of ARNO [initial concentration 200 μM (BFA) or 400 μM (LM11, no ligand)] containing 10 nM Alexa 488-labeled [Δ17]Arf1-GDP with or without 100 μM inhibitor. For each subsequent measurement, 30 μl was removed from the initial solution and replaced by 30 μl containing 10 nM Alexa 488-labeled [Δ17]Arf1-GDP with or without 100 μM of inhibitor. Anisotropy data were fit by using the Bioeqs program (39).
NMR Spectroscopy.
2D and 3D NMR spectra (2D-HSQC, 3D-HSQC NOESY, and 3D-HSQC TOCSY) were recorded on a Bruker (Billerica, MA) Avance 500 NMR spectrometer by using a 5-mm TXI z-grad cryo-probe at 300 K. The sample contained 1 mM 15N[Δ17]Arf1 in 50 mM Tris·HCl/150 mM NaCl/50 μM GDP/0.5 mM 2-mercaptoethanol, pH 7.0 with 5% 2H2O for the lock. A mixing time of 100 msec was used for the NOESY and a 20-msec spin-lock was used for the TOCSY. Acquisition size for 3D spectra was 400(1H) × 70(15N) × 1024(1H) with 8 scans. All data were processed with the Bruker UXNMR package and analyzed with CINDY (40). We compared our data to the published chemical-shift assignments for 15N[Δ17]Arf1-GDP (36), of which 9 missing assignments were added, and 21 were corrected (SI Table 1). For chemical-shift mapping of LM11, 2D-HSQC spectra of 15N[Δ17]Arf1-GDP (210 μM in 50 mM Tris·HCl/30 mM NaCl/50 μM GDP/0.5 mM 2-mercaptoethanol/5% 2H2O at pH 8.0) were recorded without or with 400 μM LM11 at 300 K with a Bruker Avance 600 NMR spectrometer equipped with a 5-mm TXI z-gradient cryoprobe. In all experiments, the 1H carrier was centered on the water resonance, and a WATERGATE sequence was incorporated to suppress the solvent resonance. Spectral widths of 13 ppm for 1H and 34 ppm for 15N were used.
Immunofluorescence Microscopy.
HeLa cells were grown and fixed as described in ref. 41. Incubation with the first Ab was performed overnight at 4°C in PBS containing 0.25% BSA, 0.01% Tween 20, and 0.01% saponin. Abs against TGN46 and EEA1 were diluted 1:200, and βCOPI was used at 1:100 and KDEL receptor at 1:500 dilution. Staining with secondary Ab was carried out for 20 min at room temperature by using Alexa 488-conjugated goat anti-mouse (1:800) or donkey anti-sheep (1:500) IgG and TRITC-labeled phalloidin (1:100). Images were collected with a Leica (Rueil-Malmaison, France) TCS SP2 upright laser-scanning confocal microscope with an oil 63× (N.A. 1.32) objective. Different fluorochromes were detected sequentially in frame-interlace mode with the acousto-optical tunable filter system using laser lines 488 nm (Alexa 488) and 514 nm (TRITC). Serial sections were acquired satisfying the Nyquist criteria for sampling and processed using the ImajeJ 1.35 software (http://rsb.info.nih.gov/ij).
Wound-Healing/Cell Sheet Migration Assay.
wt-ARNO fused to a N-terminal c-myc tag was expressed in MDCK cells under control of the tetracycline-repressible transactivator. MDCK cells stably transfected with wt-ARNO vector were plated on plastic dishes coated with collagen I at 3 μg/ml to form monolayers. Confluent monolayers were wounded by scraping with a tip, rinsed with media to remove dislodged cells, and placed back into MEM with 5% FBS with or without 20 ng/ml doxycycline, to repress or allow, respectively, further transgene expression, and with or without LM11. Cell sheet migration into the cleared wound area (≈350-μm width × ≈22-mm length) was recorded by using a Zeiss (Le Peck, France) Axiovert 200M inverted microscope equipped with a thermostated incubation chamber maintained at 37°C under 5% CO2. Digital images were acquired every 5 min for 16 h by using a CoolSnap HQ CCD camera. The wounded area was measured on each time frame, and the increment of the area recovered by cells over time was determined by using Metamorph software. The migration rate (μm2/h) was calculated between two sequential frames separated by a time interval of 1 h during 8 h or until total wound closure. The average migration rates are means ± SD of at least three separate experiments. Similar migration and inhibition rates were obtained with nontransfected MDCK and noninduced wt-ARNO-MDCK cell monolayers (data not shown).
SiRNA oligonucleotides were designed by using the Canis familiaris ARNO sequence and purchased from Eurogentec (Angers, France). A control nonsilencing siRNA duplex was used. MDCK cells were transfected with 150 pmol ARNO or control siRNA oligonucleotides by using Lipofectamine 2000. At 2 days after transfection, MDCK cells monolayers were wounded, and the level of ARNO or cytohesin1 (negative control) was measured at the end of the wound-healing experiment by immunoblot after immunoprecipitation of the cell lysates by using anti-ARNO (A18) and anticytohesin 1 (139) Abs (kind gift from S. Bourgoin, Centre Hospitalier, Université de Québec, Ste-Foy, PQ, Canada) with Trueblot kit (SI Fig. 9). Transfection efficiency was determined by epifluorescence microscopy by using Rhodamine-labeled ARNO siRNA. Cell sheet migration was recorded and analyzed as described above for inducible MDCK cells.
Supplementary Material
Acknowledgments
This work was supported by the Association pour la Recherche Contre le Cancer, the ACI-Biologie cellulaire, Moléculaire et Structurale of the French Ministère de la Recherche (J.C.) and the Agence Nationale de la Recherche (J.C. and A.C.). J.V. was supported by a grant from the Fondation pour la Recherche Médicale. We thank the staff at the Imaging and Cell Biology Facility [Centre National de la Recherche Scientifique (CNRS), Gif-sur-Yvette, France] for assistance with the confocal microscopes; A. Cavé and Y.S. Yang (Centre de Biochimie Structurale, CNRS, Montpellier, France) for help with NMR data acquisition and treatment; B. Olofsson and E. Dransart [Laboratoire d'Enzymologie et Biochimie Structurales (LEBS), CNRS] for help with confocal microscopy; F. Luton [Institut de Pharmcologie Moleculaire et Cellulaire (IPMC), CNRS, Sophia-Antipolis, France] for the gift of ARNO-expressing MDCK cells; Gregoire Malandain (INRIA, Sophia Antipolis, France) for producing the videos; C. van Heijenoort, V. Buosi and E. Guittet (Institut de Chimie des Substances Naturelles, CNRS) and C. Jackson (LEBS, CNRS) for sharing unpublished data; B. Antonny (IPMC, CNRS, Sophia-Antipolis, France), and A. Di Nardo (Ecole Normale Supérieure, CNRS, Paris, France) for critical reading of the manuscript.
Abbreviations
- SMG
small GTP-binding protein
- GEF
guanine nucleotide exchange factor
- BFA
Brefeldin A
- mGTP
methylanthraniloyl GTP
- MDCK
Madin–Darby canine kidney.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0700773104/DC1.
References
- 1.Toogood PL. J Med Chem. 2002;45:1543–1558. doi: 10.1021/jm010468s. [DOI] [PubMed] [Google Scholar]
- 2.Berg T. Angew Chem Int Ed Engl. 2003;42:2462–2481. doi: 10.1002/anie.200200558. [DOI] [PubMed] [Google Scholar]
- 3.Arkin MR, Wells JA. Nat Rev Drug Discov. 2004;3:301–317. doi: 10.1038/nrd1343. [DOI] [PubMed] [Google Scholar]
- 4.McCormick F. Curr Opin Biotechnol. 2000;11:593–597. doi: 10.1016/s0958-1669(00)00149-x. [DOI] [PubMed] [Google Scholar]
- 5.Pelish HE, Peterson JR, Salvarezza SB, Rodriguez-Boulan E, Chen JL, Stamnes M, Macia E, Feng Y, Shair MD, Kirchhausen T. Nat Chem Biol. 2006;2:39–46. doi: 10.1038/nchembio751. [DOI] [PubMed] [Google Scholar]
- 6.Renault L, Guibert B, Cherfils J. Nature. 2003;426:525–530. doi: 10.1038/nature02197. [DOI] [PubMed] [Google Scholar]
- 7.Pommier Y, Cherfils J. Trends Pharmacol Sci. 2005;26:138–145. doi: 10.1016/j.tips.2005.01.008. [DOI] [PubMed] [Google Scholar]
- 8.Rossman KL, Der CJ, Sondek J. Nat Rev Mol Cell Biol. 2005;6:167–180. doi: 10.1038/nrm1587. [DOI] [PubMed] [Google Scholar]
- 9.Zeghouf M, Guibert B, Zeeh JC, Cherfils J. Biochem Soc Trans. 2005;33:1265–1268. doi: 10.1042/BST0331265. [DOI] [PubMed] [Google Scholar]
- 10.Schmidt S, Diriong S, Mery J, Fabbrizio E, Debant A. FEBS Lett. 2002;523:35–42. doi: 10.1016/s0014-5793(02)02928-9. [DOI] [PubMed] [Google Scholar]
- 11.Mayer G, Blind M, Nagel W, Bohm T, Knorr T, Jackson CL, Kolanus W, Famulok M. Proc Natl Acad Sci USA. 2001;98:4961–4965. doi: 10.1073/pnas.091100698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hafner M, Schmitz A, Grune I, Srivatsan SG, Paul B, Kolanus W, Quast T, Kremmer E, Bauer I, Famulok M. Nature. 2006;444:941–944. doi: 10.1038/nature05415. [DOI] [PubMed] [Google Scholar]
- 13.Gao Y, Dickerson JB, Guo F, Zheng J, Zheng Y. Proc Natl Acad Sci USA. 2004;101:7618–7623. doi: 10.1073/pnas.0307512101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Desire L, Bourdin J, Loiseau N, Peillon H, Picard V, De Oliveira C, Bachelot F, Leblond B, Taverne T, Beausoleil E, et al. J Biol Chem. 2005;280:37516–37525. doi: 10.1074/jbc.M507913200. [DOI] [PubMed] [Google Scholar]
- 15.Blangy A, Bouquier N, Gauthier-Rouviere C, Schmidt S, Debant A, Leonetti JP, Fort P. Biol Cell. 2006;98:1387–1398. doi: 10.1042/BC20060023. [DOI] [PubMed] [Google Scholar]
- 16.D'Souza-Schorey C, Chavrier P. Nat Rev Mol Cell Biol. 2006;7:347–358. doi: 10.1038/nrm1910. [DOI] [PubMed] [Google Scholar]
- 17.Klausner RD, Donaldson JG, Lippincott-Schwartz J. J Cell Biol. 1992;116:1071–1080. doi: 10.1083/jcb.116.5.1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Peyroche A, Antonny B, Robineau S, Acker J, Cherfils J, Jackson CL. Mol Cell. 1999;3:275–285. doi: 10.1016/s1097-2765(00)80455-4. [DOI] [PubMed] [Google Scholar]
- 19.Beraud-Dufour S, Robineau S, Chardin P, Paris S, Chabre M, Cherfils J, Antonny B. EMBO J. 1998;17:3651–3659. doi: 10.1093/emboj/17.13.3651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Goldberg J. Cell. 1998;95:237–248. doi: 10.1016/s0092-8674(00)81754-7. [DOI] [PubMed] [Google Scholar]
- 21.Robineau S, Chabre M, Antonny B. Proc Natl Acad Sci USA. 2000;97:9913–9918. doi: 10.1073/pnas.170290597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zeeh JC, Zeghouf M, Grauffel C, Guibert B, Martin E, Dejaegere A, Cherfils J. J Biol Chem. 2006;281:11805–11814. doi: 10.1074/jbc.M600149200. [DOI] [PubMed] [Google Scholar]
- 23.McGovern SL, Caselli E, Grigorieff N, Shoichet BK. J Med Chem. 2002;45:1712–1722. doi: 10.1021/jm010533y. [DOI] [PubMed] [Google Scholar]
- 24.Cherfils J, Ménétrey J, Mathieu M, Le Bras G, Robineau S, Béraud-Dufour S, Antonny B, Chardin P. Nature. 1998;392:101–105. doi: 10.1038/32210. [DOI] [PubMed] [Google Scholar]
- 25.Donaldson JG, Honda A, Weigert R. Biochim Biophys Acta. 2005;1744:364–373. doi: 10.1016/j.bbamcr.2005.03.001. [DOI] [PubMed] [Google Scholar]
- 26.Chardin P, Paris S, Antonny B, Robineau S, Beraud-Dufour S, Jackson CL, Chabre M. Nature. 1996;384:481–484. doi: 10.1038/384481a0. [DOI] [PubMed] [Google Scholar]
- 27.Volpicelli-Daley LA, Li Y, Zhang CJ, Kahn RA. Mol Biol Cell. 2005;16:4495–4508. doi: 10.1091/mbc.E04-12-1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bonifacino JS, Jackson CL. Cell. 2003;112:141–142. doi: 10.1016/s0092-8674(03)00038-2. [DOI] [PubMed] [Google Scholar]
- 29.Shen X, Xu KF, Fan Q, Pacheco-Rodriguez G, Moss J, Vaughan M. Proc Natl Acad Sci USA. 2006;103:2635–2640. doi: 10.1073/pnas.0510599103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Santy LC, Casanova JE. J Cell Biol. 2001;154:599–610. doi: 10.1083/jcb.200104019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Santy LC, Ravichandran KS, Casanova JE. Curr Biol. 2005;15:1749–1754. doi: 10.1016/j.cub.2005.08.052. [DOI] [PubMed] [Google Scholar]
- 32.DeLano WL, Ultsch MH, de Vos AM, Wells JA. Science. 2000;287:1279–1283. doi: 10.1126/science.287.5456.1279. [DOI] [PubMed] [Google Scholar]
- 33.Arkin MR, Randal M, DeLano WL, Hyde J, Luong TN, Oslob JD, Raphael DR, Taylor L, Wang J, McDowell RS, Wells JA, Braisted AC. Proc Natl Acad Sci USA. 2003;100:1603–1608. doi: 10.1073/pnas.252756299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Macia E, Chabre M, Franco M. J Biol Chem. 2001;276:24925–24930. doi: 10.1074/jbc.M103284200. [DOI] [PubMed] [Google Scholar]
- 35.Cohen LA, Honda A, Varnai P, Brown FD, Balla T, Donaldson JG. Mol Biol Cell. 2007;18:2244–2253. doi: 10.1091/mbc.E06-11-0998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Seidel RD, 3rd, Amor JC, Kahn RA, Prestegard JH. J Biol Chem. 2004;279:48307–48318. doi: 10.1074/jbc.M402109200. [DOI] [PubMed] [Google Scholar]
- 37.Guichou JF, Viaud J, Mettling C, Subra G, Lin YL, Chavanieu A. J Med Chem. 2006;49:900–910. doi: 10.1021/jm050716a. [DOI] [PubMed] [Google Scholar]
- 38.Beraud-Dufour S, Paris S, Chabre M, Antonny B. J Biol Chem. 1999;274:37629–37636. doi: 10.1074/jbc.274.53.37629. [DOI] [PubMed] [Google Scholar]
- 39.Grillo AO, Brown MP, Royer CA. J Mol Biol. 1999;287:539–554. doi: 10.1006/jmbi.1999.2625. [DOI] [PubMed] [Google Scholar]
- 40.Ponchon L, Dumas C, Fesquet D, Padilla A. J Biomol NMR. 2004;28:299–300. doi: 10.1023/B:JNMR.0000013691.50297.11. [DOI] [PubMed] [Google Scholar]
- 41.Dransart E, Morin A, Cherfils J, Olofsson B. J Biol Chem. 2005;280:4674–4683. doi: 10.1074/jbc.M409741200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.