Abstract
AtPep1, a 23-aa peptide encoded by Arabidopsis PROPEP1, a member of a small, six-member gene family, activates expression of the defense gene PDF1.2 (encoding defensin) and its own precursor gene, PROPEP1, through the jasmonate/ethylene signaling pathway, mediated by a cell-surface receptor, PEPR1. Overexpression of two family members, PROPEP1 and PROPEP2, enhances resistance of Arabidopsis plants against the pathogen Pythium irregulare, and PROPEP2 and PROPEP3 are expressed at highly elevated levels in Arabidopsis in response to pathogen infections and to several pathogen-associated molecules (general elicitors). Here, we report that PDF1.2, PR-1 (pathogenesis protein), and PROPEP genes were differentially expressed in the leaves of intact plants sprayed with methyl jasmonate and methyl salicylate and in excised leaves supplied through cut petioles with peptides derived from the C terminus of each of the encoded proteins. The expression of PDF1.2 and PR-1 elicited by the peptides was blocked in mutant plants deficient in the jasmonate/ethylene and salicylate pathways, and in wild-type plants by treatment with diphenylene iodonium chloride, an inhibitor of hydrogen peroxide production. PROPEP1, PROPEP 2, and PROPEP3 genes appear to have roles in a feedback loop that amplifies defense signaling pathways initiated by pathogens.
Keywords: defensin, jasmonate, plant defense, PR-1, salicylic acid
Innate immunity in plants, as in animals, is defined as the receptor-mediated surveillance system that detects the presence of pathogen-associated molecular patterns (PAMPs) and activates defense genes to provide the first line of host defense against infection (1, 2). Only recently have studies of plant innate immunity, especially in Arabidopsis (3), provided molecular details that have helped define innate immunity in plants. These and others studies have revealed that although the overall strategies of animals and plants for monitoring the presence of pathogens are strikingly similar, the intracellular signaling pathways are quite different (4–6). In plants, the well known general elicitors that activate plant defense genes are now referred to as PAMPs (2, 6), and they are recognized by leucine-rich repeat (LRR) receptor kinases to initiate intracellular signaling activating the expression of defense genes. We recently reported the isolation of a 23-aa peptide from Arabidopsis leaves that is an endogenous elicitor of defensin, which is encoded by PDF1.2, a gene associated with innate immunity in plants (7). The peptide is processed from a precursor protein, PROPEP1 (7), whose gene is a member of a small gene family composed of six annotated genes and one unannotated gene. The receptor for Arabidopsis thaliana (At)Pep1, PEPR1, has been isolated and identified as an LRR receptor kinase (8). We report herein that the six annotated genes are differentially regulated by methyl jasmonate (MeJA) and methyl salicylate (MeSA) and by peptides derived from the six precursors encoded by the genes. The six peptides also differentially regulate the expression of PDF1.2, PR-1 (which encodes pathogenesis protein 1). The data presented here support our previously proposed role for PROPEP genes (7) as endogenous amplifiers of innate immune responses that are initiated by PAMPs.
Results and Discussion
PROPEP Gene Family Members in Arabidopsis Are Differentially Expressed in Leaves in Response to MeJA and MeSA.
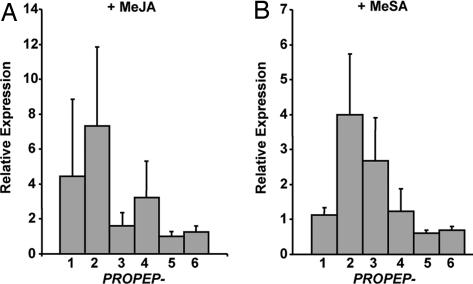
Arabidopsis PROPEP1 (At5g64900) is expressed in the plant's leaves in response to MeJA and ethylene, but expression of other paralogs of the six-member gene family has not been investigated. In Fig. 1A, the expressions of all six paralogs are shown in response to spraying plants with 625 μM MeJA or 2 mM MeSA, and assayed 2 h later. The 2-h time point was chosen from time course analyses in response to both treatments. In response to MeJA, PROPEP1 and PROPEP2 (At5g64890) were highly expressed, with PROPEP4 (At5g09980) being moderately expressed (Fig. 1A). PROPEP3 (At5g64905), PROPEP5 (At5g09990), and PROPEP6 (At2g22000) appear to be unaffected by MeJA.
Fig. 1.
Inducibility of PROPEP family genes by MeJA and MeSA. (A) Relative fold change in expression of PROPEP family genes in Arabidopsis plants sprayed with 625 μM MeJA compared with untreated control plants as determined by semiquantitative RT-PCR analysis using the β-tubulin gene as a control. (B) Relative expression of PROPEP genes in plants sprayed with 2 mM MeSA versus control plants. Error bars indicate standard deviation from the mean of three experiments.
In plants sprayed with 2 mM MeSA, only PROPEP2 and PROPEP3 were expressed over basal levels (Fig. 1B). These two genes have been shown to be highly expressed in response to fungal, bacterial, and oomycete pathogens, and to elicitors (PAMPs) derived therefrom (7, 9, 10). The high expression of the two genes in response to pathogens and PAMPs and their expression in response to MeJA and MeSA were indicators of their potential importance in the innate immune response. Because it is not known whether any of the genes are tissue-specific or cell-specific, little can be deduced about the causes of the different levels of expression.
AtPep Peptides Differentially Regulate Expression of PROPEP Gene Family Members.
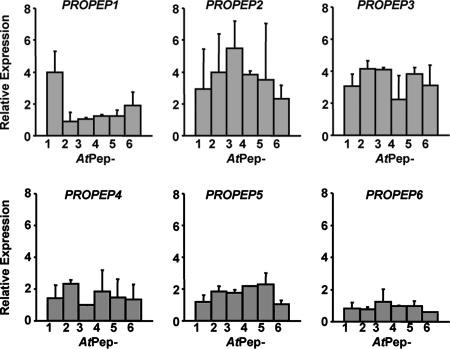
PROPEP1 expression in excised Arabidopsis leaves is induced by supplying low nanomolar concentrations of AtPep1 through cut petioles (7). However, expression of the other five PROPEP gene family members by peptides derived from the C terminus of each gene has not been assayed. The peptide from each gene had been synthesized based on their homology to native AtPep1 (7) and shown to be active at nanomolar concentrations in the alkalinization assay with Arabidopsis suspension-cultured cells. All except AtPep4 competed strongly with radiolabeled AtPep1 for binding to the receptor PEPR1 (8).
In Fig. 2, the expressions of PROPEP1 and the other precursor genes, PROPEP2 (At5g64890), PROPEP3 (At5g64905), PROPEP4 (At5g09980), PROPEP5 (At5g09990), and PROPEP6 (At2g22000), were assayed by supplying each AtPep peptide at 20 nM concentration to excised Arabidopsis leaves for 2 h before assaying mRNA levels by RT-PCR. The 2-h time points were selected because excision (wounding) caused increased background expression levels and complicated interpretations of the data at later time points. Only AtPep1 strongly induced the expression of PROPEP1 (Fig. 2A). However, PROPEP2 and PROPEP3 were strongly induced by all of the peptides (Fig. 2 B and C), and PROPEP4 and PROPEP5 were only weakly expressed (Fig. 2 D and E). PROPEP6 was not expressed in response to any peptide (Fig. 2F). Why the different peptides have differential activities in inducing the expression of the six PROPEP paralogs in planta is not understood, but it may be influenced by the composition of receptor complexes that determine intracellular signaling in leaves.
Fig. 2.
Expression of PROPEP precursor genes in response to AtPep peptides. Relative fold change in expression of PROPEP family genes in leaves supplied with 20 nM AtPep peptides relative to control leaves supplied with water, determined by semiquantitative RT-PCR analysis.
AtPep Peptides Regulate Expression of Pathogen Defense Genes Associated with both Salicylate (SA) and Jasmonate/Ethylene (JA/Et) Signaling Pathways.
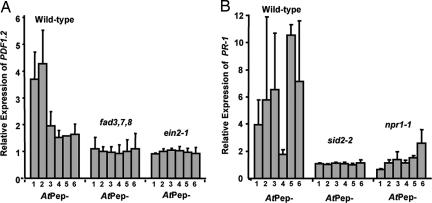
AtPep1 induces the expression of PDF1.2 in excised wild-type Arabidopsis leaves but not in ein2-1 (ethylene-insensitive) (11) or fad3,7,8 (jasmonate-deficient) (12) mutants (7). The other five peptides were initially assayed for their ability to induce the expression of PDF1.2 in wild-type plants, compared with AtPep1. AtPep2 induced PDF1.2 expression more strongly than AtPep1 (Fig. 3A), but the other peptides were only weakly active (Fig. 3A). Induction of PDF1.2 in leaves of ein2–1 and fad3,7,8 mutants by each of the peptides is blocked (Fig. 3A). These experiments indicate that the expression of PDF1.2 in leaves by AtPep peptides is most strongly induced by AtPep1 and AtPep2, and the induction by each requires a functional JA/Et pathway. The other peptides weakly induce PDF1.2 expression, and this induction also appears to require the JA/Et pathway.
Fig. 3.
AtPep-induced PDF1.2 and PR-1 expression is blocked in mutant plants. (A) Relative expression of PDF1.2 in leaves of wild-type Arabidopsis, fad triple-mutant, and ein2-1 plants leaves supplied with 20 nM solution of AtPep peptides through their cut petioles, compared with control leaves supplied with water. (B) Expression of PR-1 in leaves from wild-type Arabidopsis, sid2-2, and npr1-1 mutant plants supplied with 20 nM AtPep peptides through their cut petioles relative to leaves supplied with water. Expression of PDF1.2 and PR-1 was determined by semiquantitative RT-PCR.
The possible effect of AtPep peptides on PR-1 expression had not been assayed previously in Arabidopsis plants. The expression of the gene encoding PR-1 has been a model for studies of the SA defense signaling pathway and has been shown to be blocked in mutants including npr1-1, a SA signaling pathway mutant (13), and sid2-2, a SA biosynthetic mutant (14). In Fig. 3B is shown that with the exception of AtPep4, the expression of PR-1 is strongly induced by supplying excised leaves with 20 nM of each of the AtPep peptides. AtPep4 had previously been shown to be the weakest competitor of the six peptides for binding to the receptor PEPR1 (8). The six peptides were supplied to Arabidopsis leaves from npr1-1 and sid2-2 mutants to determine whether PR-1 expression depended on the SA signaling pathway. PR-1 was poorly expressed in both mutants in response to all of the peptides, indicating that PR-1 induction by each peptide depended on a functional SA pathway.
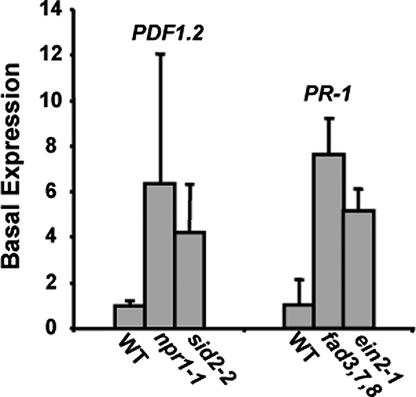
Anomalies were noted in the basal levels of PDF1.2 and PR-1 expression compared with wild-type plants. The sid2-2 and npr1-1 mutant plants had a strikingly increased basal expression of PDF1.2, and the fad3,7,8 and ein2-1 mutant plants highly expressed PR-1 (Fig. 4). These data indicated that there was some type of cross-talk occurring between the two pathways. It appears that when one pathway is blocked, the other pathway may sense a change in the intracellular environment and is activated. Many studies have revealed cross-talk between the JA and SA pathways, and frequently disruptions of one pathway have been reported to lead to up-regulation of the other (15). However, the relationship between the JA and SA pathways is more complicated than simple antagonism, and when applied at lower concentrations, JA and SA have been reported to work synergistically, inducing expression of both PDF1.2 and PR-1 (16).
Fig. 4.
(Left) Basal expression levels of PDF1.2 in untreated wild-type (WT) Arabidopsis plants compared with the basal levels in untreated npr1-1 and sid2-2 mutant plants. (Right) Basal expression levels of PR-1 in wild-type plants compared with basal levels in fad3,7,8 and ein2-1 mutant plants.
AtPep-Induced PR-1 and PDF1.2 Expression Requires Hydrogen Peroxide Production.
Hydrogen peroxide production is a component of many characterized defense signaling processes (17–20). Supplying diphenylene iodonium chloride, an inhibitor of the NADPH oxidase-generated hydrogen peroxide precursor superoxide (21), to excised leaves, together with AtPep1, totally blocks PDF1.2 expression (7). Supplying diphenylene iodonium chloride to Arabidopsis leaves, along with each of the other five AtPep peptides, blocked expression of PR-1 and PDF1.2 by each peptide, compared with wild-type plants (data not shown). These results indicated that hydrogen peroxide production was essential to AtPep-mediated expression of both the JA/Et and SA defense signaling pathways. Hydrogen peroxide is produced in the apoplast and its role in the innate immune response is not clear (18). The inhibition of hydrogen peroxide production and defense gene activation appears to be related to the early active oxygen burst, and hydrogen peroxide may behave as a “second messenger” in regulating defense-gene transcription in mesophyll cells, as found in tomato leaves (19, 22). The role of hydrogen peroxide in the signaling pathways induced by AtPep peptides should be investigated further.
Overexpression of PROPEP Genes in Transgenic Arabidopsis Plants Produces a Constitutive Expression of both PR-1 and PDF1.2 in Leaves.
Transgenic lines of Arabidopsis expressing 35S::PROPEP1 and 35S::PROPEP2 were shown to have increased resistance toward an oomycete, Pythium irregulare (7). Here, we report that six independent transgenic lines expressing 35S::PROPEP1 or 35S::PROPEP2 express PDF1.2 and PR-1 in leaves at levels higher than those found in wild-type plants (Table 1). These results correlated with the data in Fig. 2 A and B, in which AtPep1 and AtPep2 induce expression of both PDF1.2 and PR-1.
Table 1.
Transgenic plants constitutively expressing PROPEP peptide precursor genes in both PR-1 and PDF1.2
| Transgenic plants | Relative expression levels |
|||
|---|---|---|---|---|
| PROPEP1 | PROPEP2 | PR-1 | PDF1.2 | |
| 35S::PROPEP1 | 17.2 ± 1.7 | 3.2 ± 1.9 | 4.4 ± 0.5 | |
| 35S::PROPEP2 | 76.7 ± 23.8 | 3.4 ± 2.0 | 4.3 ± 1.3 | |
Relative expression of PR-1 and PDF1.2genes in 35S::PROPEP plants as compared with basal levels in untreated wild-type plants is shown. Comparative expression levels were determined by semiquantitative RT-PCR analysis with β-tubulin expression as a control. Results are derived from four different experiments using at least six independent lines per construct.
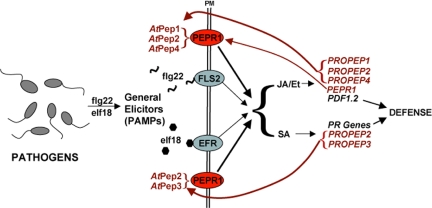
In Fig. 5 is a proposed model that includes the data presented here with recent data for the innate immune responses of Arabidopsis in response to the bacterial PAMPs flg22 and elf18, in which the peptides interact with receptors in Arabidopsis to activate defense genes that regulate both the JA/Et and SA pathways (3, 6, 23, 24), inducing the expression of PDF1.2 and PR protein genes, respectively. How these two pathways are coordinated in response to individual PAMPs has not been explained. However, flg22 and elf18 strongly activate the expression of PROPEP2, PROPEP3, and their receptor, PEPR1 (At1g73080) (3, 24). The expression of these genes would result in the production of AtPep peptides that can amplify signaling for both the JA/Et and SA pathways and expression of defense genes of each pathway. This could explain how PAMPs such as flg22 and elf18 can amplify multiple defense signaling pathways. PROPEP orthologs have been identified in numerous plant species of diverse families, suggesting that a similar amplification mechanism may be found throughout the plant kingdom.
Fig. 5.
A model for the amplification of signaling pathways for PAMPs and AtPep peptides in Arabidopsis. The PAMPs flg22 and elf18 are perceived by their respective receptors, FLS2 and EFR (25, 26), to initiate signaling through the JA/Et and SA pathways to express the defense protein genes PDF1.2, PR-1, and PROPEP. Peptides derived from the PROPEP genes are transported to the apoplast, where they can interact with the cell-surface receptor PEPR1 to further amplify signaling.
In summary, we present evidence to support the role of peptides derived from PROPEP genes as endogenous elicitors that are generated in response to pathogens and their PAMPs. The peptides have the property of inducing expression of their own genes to initiate a feedback mechanism to amplify the original PAMP signals, thus being PAMP amplifiers. A simplified model is presented in Fig. 5 that incorporates the data reported here and elsewhere (25, 26) into known components of the innate immune response. A similar feedback amplification mechanism is also found in wound signaling in tomato plants, where prosystemin (27) and LepreproHypSys (28) are expressed in response to wounding to generate peptides that amplify the octadecanoid signaling pathway enzymes and therefore JA in vascular bundle cells (29-32). A recent report has suggested that the systemic signal in Arabidopsis that activates the systemic acquired resistance (SAR) pathway may be jasmonic acid or a derivative (33). It is possible that AtPep peptides, as amplifiers of both JA/Et and SA pathways, may play a role in systemic signaling in Arabidopsis and throughout the plant kingdom in species where PROPEP orthologs are found, to both amplify defense signaling by maintaining levels of JA and SA for long-distance signaling.
Materials and Methods
Plant Propagation.
Seeds of the fad3-2 fad7-2 fad8 triple-mutant plant were provided by J. Browse (Washington State University). ein2-1 and npr1-1 mutant seeds were obtained from Arabidopsis Biological Resource Center (Ohio State University, Columbus, OH) and sid2-2 seeds were provided by F. Ausubel (Massachusetts General Hospital, Boston, MA). All mutant seed and wild-type Arabidopsis thaliana (ecotype Columbia) seeds were stored at 4°C until planting.
Wild-type, ein2-1, fad triple-mutant, and npr1-1 plants were planted in twice-autoclaved potting soil with four seeds per 8 cm3 pot. The pots were covered with cheesecloth to retain moisture, and the seeds germinated for 6 days under low light at ≈18°C. The seedlings were moved to growth chambers where they were grown under a 16-h day-length of 250 microeinstein/m2·s of light (1 einstein = 1 mol of photons) at 21°C and watered from the bottom of the pot daily. The sid2-2 seed was planted into twice-autoclaved Metro Mix 360 (Sun Gro Horticulture Distribution, Bellevue, WA) in 4-inch pots. The mix in each pot was covered by a piece of cheesecloth and topped with a thin layer of soil. Four sid2-2 seeds per pot were distributed evenly on the soil and grown under the same conditions as above.
Plant Hormone Treatments.
Plants were sprayed with solutions of either 625 μM MeJA (Bedoukian Research, Danbury, CT) or 2 mM MeSA (Sigma–Aldrich, St. Louis, MO). Both solutions were prepared in 0.1% Triton X-100 (Sigma–Aldrich). The treated plants and control plants were incubated in closed Plexiglas boxes in separate growth chambers.
Excised-Leaf Assays.
AtPep peptides were assayed for ability to induce defense-related gene expression in excised Arabidopsis leaves as described (7). Briefly, leaves from 3- to 4-week-old plants were excised at the petiole and placed in 800-μl centrifuge tubes containing either 20 nM solutions of each peptide, or for control leaves, distilled water. To block H2O2-dependent gene expression in leaves, 100 μM diphenylene iodonium chloride (Sigma–Aldrich), an inhibitor of H2O2 production (19, 21), was supplied to excised leaves through the cut petiole in small vials with and without AtPep peptides and incubated in Plexiglas boxes in a growth chamber for 2 h under constant light of 200 microeinstein/m2·s at 21°C. To terminate the assays, leaves were frozen in liquid nitrogen.
Semiquantitative RT-PCR Analysis of Relative Gene Expression Levels.
Semiquantitative RT-PCR was performed as described (1). RNA was isolated by using TRIzol reagent according to the manufacturer's instructions (Invitrogen, Carlsbad, CA), and 2 μg of RNA template per reaction was reverse transcribed with the RETROscript kit (Ambion, Austin, TX). For amplification, 2.5 μl of the cDNA generated by the reverse transcription reactions was used as template for 25 μl of PCRs that used Ex Taq Hot Start polymerase and reagents (Fisher Scientific, Pittsburgh, PA).
All primers designed to amplify intron-spanning fragments of the six PROPEP transcripts were used at a concentration of 1 μM. These primers and primers to amplify fragments of the PDF1.2 transcripts (At5g44420), PR-1 transcripts (At2g14610), and β-tubulin transcripts (At5g62690) are as follows: PROPEP1 forward primer, 5′-CTT ATC AGA TCT CAA TGG AGA AAT C-3′, and reverse primer, 5′-CAA TGT AAC TTA AAG TGC CTA ATT ATG-3′; PROPEP2 forward primer, 5′-TCA CCA AAC TAT TGG ATT TCA A-3′, and reverse primer, 5′-GAC TCA ATT TTT GAC TTC TTA ATC C-3′; PROPEP3 forward primer, 5′-CAA CGA TGG AGA ATC TCA GA-3′, and reverse primer, 5′-CTA ATT GTG TTT GCC TCC TTT-3′; PROPEP4 forward primer, 5′-AAC TTA GCT CTC ACG AAG CA-3′, and reverse primer, 5′-AAA AAT AAA GGA CTC GTA GGA GTT-3′; PROPEP5 forward primer, 5′-GAA GAT GCA GCA AGA GAG AG-3′, and reverse primer, 5′-TAG TTA CAT GTC GTA GTC GTT AAC TC-3′; PROPEP6 forward primer, 5′-ATG GAA GTT AAT GGA GAA GAA GA-3′, and reverse primer, 5′-ATT GTT TTG ACC AGG TCG T-3′; PDF1.2 forward primer, 5′-ATG GCT AAG TTT GCT TCC A-3′, and reverse primer, 5′-TTA ACA TGG GAC GTA ACA GAT AC-3′; PR-1 forward primer, 5′-GGA GCT ACG CAG AAC AAC TA-3′, and reverse primer, 5′-AGT ATG GCT TCT CGT TCA CA-3′; and β-tubulin forward primer, 5′-CAA CGC TAC TCT GTC TGT CC-3′, and reverse primer, 5′-TCT GTG AAT TCC ATC TCG TC-3′.
Reactions to analyze PROPEP or PDF1.2 transcript abundance were performed with an initial denaturing/polymerase activating step of 5 min at 94°C followed by 31 repetitions of the following three steps: a 30-s denaturation phase at 94°C, a 30-s annealing period at 55.5°C, and a 1-min elongation step at 72°C. The amplification program was terminated with a 10-min final 72°C elongation phase. PR-1 and β-tubulin reactions were amplified with a shorter program of 29 rather than 31 cycles.
Products of each reaction were separated by electrophoresis and visualized on a Bio Imaging System by using GeneSnap version 6.00.26 software (Syngene, Frederick, MD). Gel images were analyzed with GeneTools analysis software version 3.02.00 (Syngene). The relative intensity of each band was calculated by normalization to a tubulin band to yield a numerical ratio. The semiquantitative PCR method was performed in duplicate for every sample, and the original RNA extraction experiments were performed in triplicate to calculate average ratios.
Construction of Vectors and Plant Transformation.
Cassettes containing PROPEP genes under the control of the cauliflower mosaic virus 35S promoter were generated with the pART-7/pBART binary vector system (34), as described (7), and used to transform wild-type Arabidopsis (ecotype Columbia) plants (35).
Acknowledgments
We thank Julia Gothard and Sue Vogtman for growing our plants. This research was supported by National Science Foundation Grant IBN 0090766 (to C.A.R.); the Charlotte Y. Martin Foundation; and Washington State University College of Agriculture, Human, and Natural Resources Sciences.
Abbreviations
- MeJA
methyl jasmonate
- MeSA
methyl salicylate
- SA
salicylate
- JA/Et
jasmonate/ethylene
- PDF1.2
defensin
- PR
pathogenesis protein
- PAMP
pathogen-associated molecular pattern
- PEPR1
AtPep1 receptor.
Footnotes
The authors declare no conflict of interest.
References
- 1.Janeway CA, Jr, Medzhitov R. Annu Rev Immunol. 2002;20:197–216. doi: 10.1146/annurev.immunol.20.083001.084359. [DOI] [PubMed] [Google Scholar]
- 2.Brunner F, Rosahl S, Lee J, Rudd JJ, Geiler C, Kauppinen S, Rasmussen G, Scheel D, Nürnberger T. EMBO J. 2002;21:6681–6688. doi: 10.1093/emboj/cdf667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zipfel C, Robatzek S, Navarro L, Oakeley EJ, Jones JDG, Felix G, Boller T. Nature. 2004;428:764–767. doi: 10.1038/nature02485. [DOI] [PubMed] [Google Scholar]
- 4.Nürnberger T, Lipka V. Mol Plant Pathol. 2005;6:335–345. doi: 10.1111/j.1364-3703.2005.00279.x. [DOI] [PubMed] [Google Scholar]
- 5.Ausubel FM. Nature Immunol. 2005;6:973–979. doi: 10.1038/ni1253. [DOI] [PubMed] [Google Scholar]
- 6.Zipfel C, Felix G. Curr Opin Plant Biol. 2005;8:353–360. doi: 10.1016/j.pbi.2005.05.004. [DOI] [PubMed] [Google Scholar]
- 7.Huffaker A, Pearce G, Ryan CA. Proc Natl Acad Sci USA. 2006;103:10098–10103. doi: 10.1073/pnas.0603727103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yamaguchi Y, Pearce G, Ryan CA. Proc Natl Acad Sci USA. 2006;103:10104–10109. doi: 10.1073/pnas.0603729103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Toufighi K, Brady SM, Austin R, Ly E, Provart NJ. Plant J. 2005;43:153–163. doi: 10.1111/j.1365-313X.2005.02437.x. [DOI] [PubMed] [Google Scholar]
- 10.Craigon DJ, James N, Okyere J, Higgins J, Jotham J, May S. Nucleic Acids Res. 2004;32:D575–D577. doi: 10.1093/nar/gkh133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Guzman P, Ecker JR. Plant Cell. 1990;2:513–523. doi: 10.1105/tpc.2.6.513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McConn M, Browse J. Plant Cell. 1996;8:403–416. doi: 10.1105/tpc.8.3.403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cao H, Bowling SA, Gordon AS, Dong X. Plant Cell. 1994;6:1583–1592. doi: 10.1105/tpc.6.11.1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wildermuth MC, Dewdney J, Wu G, Ausubel FM. Nature. 2001;414:562–565. doi: 10.1038/35107108. [DOI] [PubMed] [Google Scholar]
- 15.Kunkel BN, Brooks DM. Curr Opin Plant Biol. 2002;5:325–331. doi: 10.1016/s1369-5266(02)00275-3. [DOI] [PubMed] [Google Scholar]
- 16.Mur LA, Kenton P, Atzorn R, Miersch O, Wasternack C. Plant Physiol. 2006;140:249–262. doi: 10.1104/pp.105.072348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Laloi C, Apel K, Danon A. Curr Opin Plant Biol. 2004;7:323–328. doi: 10.1016/j.pbi.2004.03.005. [DOI] [PubMed] [Google Scholar]
- 18.Torres MA, Jones JD, Dangl JL. Plant Physiol. 2006;141:373–378. doi: 10.1104/pp.106.079467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Orozco-Cárdenas ML, Narváez-Vásquez J, Ryan CA. Plant Cell. 2001;13:179–191. [PMC free article] [PubMed] [Google Scholar]
- 20.Coego A, Ramirez V, Gil MJ, Flors V, Mauch-Mani B, Vera P. Plant Cell. 2005;17:2123–2137. doi: 10.1105/tpc.105.032375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.O'Donnell VB, Tew DG, Jones OTG, England PJ. Biochem J. 1993;290:41–49. doi: 10.1042/bj2900041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Browse J. Vitam Horm (San Francisco) 2005;72:431–456. doi: 10.1016/S0083-6729(05)72012-4. [DOI] [PubMed] [Google Scholar]
- 23.Gómez-Gómez L, Felix G, Boller T. Plant J. 1999;18:277–284. doi: 10.1046/j.1365-313x.1999.00451.x. [DOI] [PubMed] [Google Scholar]
- 24.Zipfel C, Kunze G, Chinchilla D, Caniard A, Jones JDG, Boller T, Felix G. Cell. 2006;125:749–760. doi: 10.1016/j.cell.2006.03.037. [DOI] [PubMed] [Google Scholar]
- 25.Gómez-Gómez L, Boller T. Trends Plant Sci. 2002;7:251–257. doi: 10.1016/s1360-1385(02)02261-6. [DOI] [PubMed] [Google Scholar]
- 26.Kunze G, Zipfel C, Silke R, Niehaus K, Boller T, Felix G. Plant Cell. 2004;16:3496–3507. doi: 10.1105/tpc.104.026765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McGurl B, Pearce G, Orozco-Cárdenas ML, Ryan CA. Science. 1992;255:1570–1573. doi: 10.1126/science.1549783. [DOI] [PubMed] [Google Scholar]
- 28.Pearce G, Ryan CA. J Biol Chem. 2003;278:30044–30050. doi: 10.1074/jbc.M304159200. [DOI] [PubMed] [Google Scholar]
- 29.Narváez-Vásquez J, Ryan CA. Planta. 2004;218:360–369. doi: 10.1007/s00425-003-1115-3. [DOI] [PubMed] [Google Scholar]
- 30.Narváez-Vásquez J, Pearce G, Ryan CA. Proc Natl Acad Sci USA. 2005;102:12974–12977. doi: 10.1073/pnas.0505248102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wasternack C, Stenzel I, Hause B, Hause G, Kutter C, Maucher H, Neumerkel J, Feussner I, Miersch O. J Plant Physiol. 2006;163:297–306. doi: 10.1016/j.jplph.2005.10.014. [DOI] [PubMed] [Google Scholar]
- 32.Schilmiller AL, Howe G. Curr Opin Plant Biol. 2005;8:369–377. doi: 10.1016/j.pbi.2005.05.008. [DOI] [PubMed] [Google Scholar]
- 33.Truman W, Bennett MH, Kubigsteltig I, Turnbull C, Grant M. Proc Natl Acad Sci USA. 2007;104:1075–1080. doi: 10.1073/pnas.0605423104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gleave AP. Plant Mol Biol. 1992;20:1203–1207. doi: 10.1007/BF00028910. [DOI] [PubMed] [Google Scholar]
- 35.Clough SJ, Bent AF. Plant J. 1998;16:735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]