Abstract
Membrane proteins exit the endoplasmic reticulum (ER) in COPII-transport vesicles. ER export is a selective process in which transport signals present in the cytoplasmic tail (CT) of cargo membrane proteins must be recognized by coatomer proteins for incorporation in COPII vesicles. Two classes of ER export signals have been described for type I membrane proteins, the diacidic and the dihydrophobic motifs. Both motifs participate in the Sar1-dependent binding of Sec23p–Sec24p complex to the CTs during early steps of cargo selection. However, information concerning the amino acids in the CTs that interact with Sar1 is lacking. Herein, we describe a third class of ER export motif, [RK](X)[RK], at the CT of Golgi resident glycosyltransferases that is required for these type II membrane proteins to exit the ER. The dibasic motif is located proximal to the transmembrane border, and experiments of cross-linking in microsomal membranes and of binding to immobilized peptides showed that it directly interacts with the COPII component Sar1. Sar1GTP-bound to immobilized peptides binds Sec23p. Collectively, the present data suggest that interaction of the dibasic motif with Sar1 participates in early steps of selection of Golgi resident glycosyltransferases for transport in COPII vesicles.
INTRODUCTION
Newly synthesized proteins destined to enter the secretory pathway in higher eukaryotes are cotranslationally translocated across endoplasmic reticulum (ER) membranes. Those completely translocated are eventually secreted, whereas those in which translocation is stopped by a membrane anchor sequence travel as integral membrane components. From the ER, proteins are transported to either the Golgi complex, the lysosomal compartment, or the cell surface by vesicles that bud from each compartment coated with specific cytoplasmic protein complexes (Barlowe et al., 1994; Schekman and Orci, 1996; Rothman and Wieland, 1996; Mellman and Warren, 2000).
The mechanism of protein export from the ER is believed to be a selective process involving the COPII complex (Bannykh et al., 1998; Antonny and Schekman, 2001). Budding of COPII vesicles from ER membranes is initiated by recruiting the small GTPase Sar1 (Barlowe et al., 1993). Sar1 cycles between a GTP-bound form stimulated by the guanine nucleotide exchange factor Sec12p (Barlowe and Schekman, 1993), and a GDP-bound form stimulated by the GTPase-activating activity of Sec23p (Yoshihisa et al., 1993; Antonny et al., 2001). Binding of Sar1-GTP to ER membranes, probably through a hydrophobic motif exposed at its N terminus (Huang et al., 2001; Bi et al., 2002), induces the formation of ER exit sites (Aridor et al., 2001). The interaction between the CTs of integral membrane cargo proteins and Sar1 allows the further interaction and recruitment of the heterodimeric complex Sec23p-Sec24p. These interactions mediate the specific selection and concentration of cargo proteins in COPII transport vesicles, and in turn exclude ER resident proteins (Balch et al., 1994; Aridor et al., 1998; Kuehn et al., 1998; Springer and Schekman, 1998). Other cytosolic proteins, the tetramer Sec13p-Sec31p, participate in vesicle completion (Barlowe et al., 1994; Salama et al., 1997; Lederkremer et al., 2001).
Different amino acid motifs have been proposed as ER export signals (Aridor et al., 2001), which can be classified in two classes: diacidic and dihydrophobic. For the G protein of the vesicular stomatitis virus (VSVG protein), a type I membrane protein, it was demonstrated a Sar1-dependent interaction of a diacidic DXE-containing motif at the C termini of the cytoplasmic tail with the heterodimer Sec23p-Sec24p of the COPII coat (Sevier et al., 2000; Aridor et al., 2001). Similar motifs (EXE and EXD) promote both ER exiting of the rectifying potassium channel proteins Kir1.1, Kir2.1, and Kir4.1 (Stockklausner et al., 2001; Ma et al., 2001) and Golgi complex localization to PRA1 (Abdul-Ghani et al., 2001). Interaction between COPII coat components and a diphenylalanine (FF) or dihydrophobic motif at the C termini of ERGIC-53 (Kappeler et al., 1997; Nufer et al., 2002), and of some members of the p24 protein family (Dominguez et al., 1998; Belden and Barlowe, 2001) was also found necessary for ER exiting of these proteins. In addition, formation of hetero-oligomeric complexes of p24 family members in the ER is a prerequisite for export, probably via entry into budding COPII vesicles (Dominguez et al., 1998; Fullekrug et al., 1999).
Glycosyltransferases, which are type II membrane proteins resident of the Golgi complex, bind Sec23p through their cytoplasmic tails, supporting the notion that they leave the ER in COPII transport vesicles (Dominguez et al., 1998). However, most ER export signals described up to date are not present in the CTs of members of the glycosyltransferase family. Herein, we described a novel class of ER export motif, [RK](X)[RK], at the CT of Golgi resident glycosyltransferases that is required for these type II membrane proteins to exit the ER. The dibasic motif is located proximal to the transmembrane border, and it directly interacts with the COPII component Sar1.
MATERIALS AND METHODS
Molecular Constructs
Expression vectors containing cDNA coding for the full-length versions or the N-terminal domains (Ntds) of GalNAcT, GalT2, and SialT2 fused to the NH2 terminus of enhanced cyan fluorescent protein (CFP) or to enhanced yellow fluorescent protein (YFP) were constructed as described previously (Giraudo et al., 2001). ΔGalT2-CFP and ΔGalNAcT-CFP were generated by polymerase chain reaction (PCR) with sense primers that hybridized at the codon of aa 9 or 8, respectively, and introduced a preceding methionine codon. GalT2, GalNAcT, and SialT2 Ntds mutated at the CTs were generated by PCR with the corresponding sense primer containing the desired mutation. PCR products were subcloned in frame into the enhanced CFP and YFP expression vectors (BD Biosciences Clontech, Palo Alto, CA). SialT21–57RXR25→ AXA-YFP was generated using the QuikChange site-directed mutagenesis kit (Stratagene, La Jolla, CA) according to the manufacturer's instructions. All constructs were controlled by DNA sequencing.
The VSVG-YFP chimeric construct containing the thermosensitive G protein of vesicular stomatitis virus (VSV-G3 ts045) with a spacer sequence at the C termini fused to YFP was kindly provided by P. Keller (Max-Planck Institute, Dresden, Germany) (Keller et al., 2001).
Full-length Iip33 was amplified by PCR from the plasmid containing the corresponding cDNA kindly supplied by M. Jackson (RW Johnson Pharmaceutical Research Institute, San Diego, CA) and subcloned in the pEYFP. Substitution of Iip33 CT for the CT of Gal-T2 was carried out by PCR with a sense primer coding for the sequence of the parent or mutated CT of GalT2.
The Escherichia coli expression constructs pET-11d His-Sar1wt, pET-11d His-Sar1T39N, and pET-11d His-Sar1H79G were kindly provided by W. Balch (Scripps Research Institute, La Jolla, CA).
Cell Culture and Transfection
Chinese hamster ovary (CHO)-K1 cells were grown in DMEM supplemented with 10% fetal calf serum, 100 μg/ml penicillin, and 100 μg/ml streptomycin. Transfections were performed using FuGENE 6 transfection reagent (Roche Diagnostics, Indianapolis, IN) according to manufacturer's instructions.
Protease Protection Assays
Cells were scrapped from the plate in 10 mM Tris-HCl, pH 7.2, and 0.25 M sucrose and membranes collected by pelleting at 13,500 × g for 10 min. Membranes (50 μg of protein) were incubated with proteinase K (20 or 500 μg/ml final concentration) in the presence or absence of 1% Triton X-100 for 30 min at 37°C. The reaction was stopped with 100% (wt/vol) trichloroacetic acid (TCA) added to a final concentration of 15% and left for 15 min on ice. The TCA pellet was collected by centrifuging 5 min at 10,000 rpm for SDS-PAGE.
Fluorescence Microscopy
Cells stably expressing constructs of CFP or YFP fused at the C termini of Ntds of Golgi proteins were grown in DMEM on chambered coverglasses (Lab Tek, Naperville, IL). Imaging processing for pseudocolor, and fluorescence quantitation, were carried out using the MetaMorph 3.0 Imaging System (Universal Imaging, West Chester, PA). Final images were compiled with Adobe Photoshop 5.0.
Fluorescence loss in photobleaching (FLIP) was carried out with an LSM-510 confocal microscope (Carl Zeiss, Jena, Germany) equipped with a culture chamber set at 5% CO2 in air and 37°C, by using a 100× Plan Neofluar objective. CFP and YFP were excited at 458 and 514 nm, respectively. An ER area was selected with the region of interest tool of the laser scanning microscope software (Carl Zeiss) in a focus plane. Images were acquired at the imaging laser power of ∼10–25% of the maximum and 80% of attenuation at 458 and 514 nm. Photobleaching was carried out by successive scanning of selected areas at 100% of the laser power during 1 min. A new image was acquired at the imaging laser power and the bleaching step was repeated. This cycle was repeated every 1 min during 30 min. For fluorescence recovery after photobleaching (FRAP), either a Golgi or ER area was selected in a focus plane so that they were clearly visible. The volume of each organelle was determined, defining the upper and lower Z-limits. The Golgi volume usually comprises ∼1–2 μm above and below the focal plane, whereas the ER volume comprises <1 μm. The Golgi or ER selected volumes were irreversibly photobleached by successive scanning through the Z-series at 100% of the laser power of both wavelengths. Images were taken every minute during 90 min of recovery. Thirty minutes before and during the time course of the bleaching experiments, cells were incubated with 100 μg/ml cycloheximide to ensure that no fluorescence was gained due to protein synthesis.
Chemical Cross-Linking
Cells expressing GalT2 Ntd constructs or the temperature-sensitive mutant VSVG-YFP were incubated at 15°C or 39°C, respectively, to accumulate these constructs in the early secretory compartment (Saraste and Kuismanen, 1984). Urea-stripped microsomes from these cells (Aridor et al., 1998) were incubated with His-tagged recombinant Sar1wt, Sar1T39N, or Sar1H79G (Rowe and Balch, 1995) in the presence or absence of rat liver cytosol. The cross-linking procedure was essentially as described previously (Aridor et al., 2001). Briefly, membranes in 20 mM HEPES, 70 mM KOAc, 1 mM MgOAc, 250 mM sorbitol, and 100 μM of the reversible cross-linker 3,3′-dithiobis[sulfosuccinimidylpropionate] (DTSSP; Pierce Chemical, Rockford, IL) were incubated at 37°C for 15 min, and the reaction was stopped by dilution with Tris-HCl (pH 7.4, 50 mM final concentration). Membranes collected by centrifugation were solubilized with 1% SDS and cleared from insoluble material by 10-min centrifugation at 7000 rpm. The supernatant was supplemented with 50 mM Tris, pH 7.4, 1% Triton X-100, 0.5% deoxycholate, 0.1% SDS, 150 mM NaCl, and protease inhibitors (3 μg/ml leupeptin, 1 mM phenylmethylsulfonyl fluoride, 3 μg/ml aprotinin, 2 μM pestatin). Solubilized membranes were incubated with anti-His monoclonal antibody (Santa Cruz Biotechnology, CA) and protein G-Sepharose beads (Amersham Biosciences, Piscataway, NJ) overnight at 4°C. Immunocomplexes were pelleted by centrifugation at 2500 × g for 10 s, washed five times at 4°C with lysis buffer, and three times with buffer A (150 mM phosphate buffer, pH 7.2, 10 mM EDTA), and denaturated with Laemmli sample buffer. Blots were probed with anti-green fluorescent protein (GFP) (Roche Diagnostics), anti-His, or anti Sec23p (Santa Cruz Biotechnology).
Sar-1 Binding to Synthetic Peptides
Peptides were coupled to Sepharose 6B according to the instructions of Amersham Biosciences. Efficiency of coupling was monitored measuring 2-thiopyridone release at 343 nm. Five nanomoles of Sepharose-bound peptides was used for binding assays. Recombinant His-Sar1H79G or His-Sar1T39N (1 μg of protein) was incubated with the immobilized peptides overnight at 4°C (Dominguez et al., 1998). When indicated, CHO-K1 cytosol was added to the incubate 4 h after Sar1 addition, and the incubation was continued overnight. Beads were collected, washed four times by centrifugation, and suspended in Laemmli sample buffer for Western blot analysis with anti-His and anti-Sec23p antibody.
RESULTS
Glycolipid Glycosyltransferases Lacking Cytoplasmic Tails Fail to Concentrate in the Golgi Complex
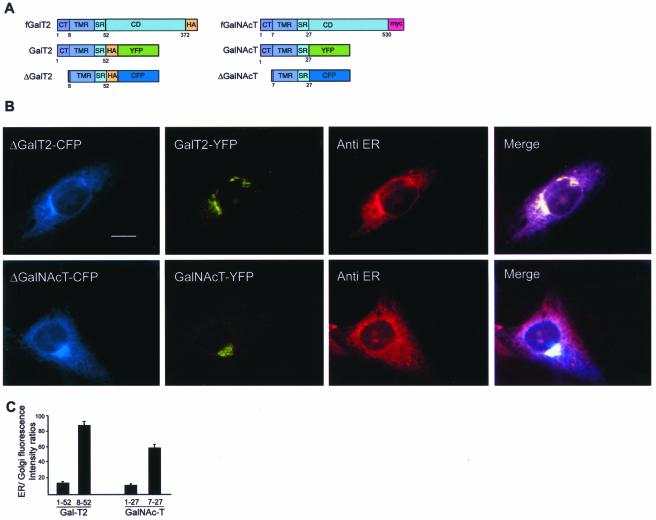
Glycosyltransferases are type II membrane proteins with an Ntd comprising a short CT, a transmembrane region (TMR), and a luminally oriented C-terminal domain bearing the catalytic site (Colley, 1997). The Ntd of glycoprotein (Nilsson et al., 1991) and glycolipid (Giraudo et al., 2001) glycosyltransferases has been found sufficient to move reporter proteins out of the ER and to concentrate them in the Golgi complex (Fullekrug and Nilsson, 1998; Munro, 1998; Giraudo et al., 2001). To characterize molecular determinants that promote exiting of these enzymes from the ER, the Ntds of the glycolipid glycosyltransferases UDP-Gal:GM2 galactosyltransferase, comprising aa 1–52 (GalT2), and of UDP-GalNAc:GM3 N-acetylgalactosaminyltransferase, comprising aa 1–27 (GalNAcT), and of their respective tail-deleted forms, comprising aa 8–52 (ΔGal-T2) or aa 7–27 (ΔGalN-AcT), were fused to spectral variants of the GFP (Figure 1A) and expressed in CHO-K1 cells. The subcellular localization of coexpressed GalT2 and ΔGalT2 (Figure 1B, top row) or GalNAcT and ΔGalNAcT (Figure 1B, lower row) was examined by triple-color imaging. GalT2 and GalNAcT concentrate in the Golgi area (Figure 1B, second panels from left), whereas ΔGalT2 and ΔGalNAcT distributed in a perinuclear, reticular pattern (Figure 1B, first panels from left), colocalizing with the immunostained ER (third panels from left and merging). ER/Golgi fluorescence intensity ratios were quantified for both paired chimaeras, and found to be 5–6 times higher for tail-deleted forms (Figure 1C).
Figure 1.
Cytoplasmic-tail deletions promote ER accumulation of GalNAcT and GalT2. (A) Schematic representation of chimeric constructs used in this study. The CT, TMR, stem region (SR), and lumenal, C-terminal domain (CD) tagged with either HA or c-myc are indicated in the full-length constructs (fGalT2 or fGalNAcT). Deletions of the CTs, and replacement of the CDs by spectral variants of GFP (CFP or YFP) are indicated. An HA epitope was introduced between the GalT2 sequence and the GFP variant. (B) Triple color imaging of single CHO-K1 cells transiently coexpressing ΔGalT2-CFP and GalT2-YFP (top row) or ΔGalNAcT-CFP and GalNAcT-YFP (bottom row), fixed 24 h posttransfection. ER was immunostained with a specific anti-ER antibody and a secondary rhodamine-conjugated antibody. Images were taken with filters for CFP, YFP, or rhodamine (first, second, and third panels from left, respectively). The fourth panel is a merging of these images. Bar, 10 μm. (C) Quantification of images in B. Results are presented as ER/Golgi fluorescence intensity ratio (arbitrary units) of single cells coexpressing the indicated chimaeras. Values are mean ± SD for n = 25 cells.
Tail-deleted Versions Behave as Integral Membrane Proteins with Type II Topology
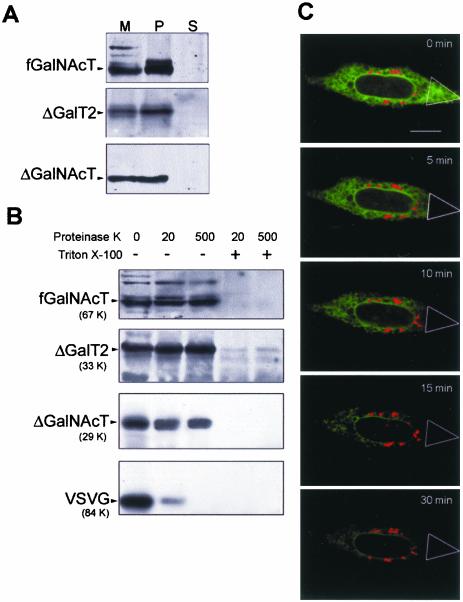
We first examined whether the tail-deleted versions behave as membrane bound polypeptides. Microsomal membrane vesicles were treated with KCl or urea to release membrane-associated proteins, and collected by ultracentrifugation. Similarly to a c-myc tagged, full-length version of GalNAcT (fGalNAcT), Western blots with anti-GFP antibody showed ΔGalT2 and ΔGalNAcT in the pellets, indicating their integral membrane nature. No protein was detected in the supernatants (Figure 2A). We next examined the transmembrane disposition of these integral membrane proteins by determining their accessibility to proteinase K. It was predicted that orientation of the signal-anchor sequence of nascent membrane proteins depends on the difference in the charges of the 15 residues flanking the first signal-anchor, with the more positive portion facing the cytoplasm (Audigier et al., 1987). In the present case, tail deletion and the consequent decrease in positive charges should be taken into account to discard any failure of these polypeptides to acquire the type II membrane topology. Calculation of the positive charge difference [Δ(C-N)] (Hartmann et al., 1989) gave values of –2.5 and –1.5 for, respectively, ΔGalT2 and ΔGalNAcT, which still predicts a type II membrane orientation of these proteins. This prediction was experimentally determined in membranes from cells stably expressing fGalNAcT, which served as an internal standard of type II integral membrane protein, cotransfected with VSVG protein fused to YFP and either ΔGalNAcT or ΔGalT2. Western blot analyses of membranes from fGalNAcT stable transfectant cells coexpressing the VSVG construct with either ΔGalT2 or ΔGalNAcT treated with proteinase K showed that the major fraction of all forms except VSVG was protected from the action of the enzyme (Figure 2B). Results are thus consistent with a lumenal orientation of the C termini of ΔGalNAcT and ΔGalT2 and confirm the prediction that they will insert in the ER membranes with type II topology (Audigier et al., 1987). The type I membrane protein VSVG-YFP, on the other hand, was mostly accessible to the protease as expected from the known cytoplasmic orientation of its C termini (Aridor et al., 2001). Disruption of the permeability barrier of the membrane by 1% Triton X-100 rendered all chimeric proteins accessible to proteinase K.
Figure 2.
Tail-deleted chimaeras are type II membrane proteins and diffuse normally along ER membranes. (A) Membranes from CHO-K1 cells stably expressing fGalNAcT were obtained 24 h after transfection with ΔGalT2-CFP or ΔGalNAcT-CFP. Membranes (M) were homogenized with 1 M KCl (or 2.5 M urea; our unpublished data), separated into pellet (P) and supernatant (S) by centrifugation at 100,000 ×g for 1 h, and Western blotted with anti-GFP antibody. (B) Membranes (50 μg of protein) from cells stably expressing fGalNAcT and cotransfected with VSVG-YFP and either GalT2-CFP or ΔGalT2-CFP or GalNAcT-CFP or ΔGalNAcT-CFP were incubated with the indicated concentrations of proteinase K in the presence or absence of 1% Triton X-100, as indicated. They were then precipitated with TCA and Western blotted with anti-GFP except for fGalNAcT for which anti-c-myc antibody was used. (C) Cells coexpressing GalT2-YFP (red) and ΔGalT2-CFP (green) were supplemented with 100 μg/ml cycloheximide and 2 h later photobleached in the marked area for 1 min at both 458 and 514 nm, and photographed. This procedure was repeated every 1 min as described under MATERIALS AND METHODS to record FLIP. Dual color images were taken at the indicated times. Bar, 10 μm.
Tail-deleted Versions Diffuse Normally along ER Membranes
The possibility that tail-deleted forms may have diffusional constraints in the ER was examined by FLIP and dual-color imaging. Consecutive rounds of scanning of a selected ER area (triangle) of a cell coexpressing GalT2 and ΔGalT2, at both 458 and 514 nm, progressively photobleached the fluorescence of ΔGalT2 in the whole ER (Figure 2C, green). Photobleaching was completed at ∼30 min, at which time the only remaining fluorescence was that of GalT2 located in the Golgi complex region (Figure 2C, red). The diffusion constant of ΔGalT2 calculated from a FRAP experiment (our unpublished data) was ∼0.45 μm2 s–1, which is in the order of those calculated for other ER membrane proteins (Nehls et al., 2000). The mobile fraction calculated was ∼95%, indicating that most of the pool of the construct was free to diffuse. Photobleaching experiments thus excluded the possibility that failure of ΔGalT2 to concentrate in the Golgi complex was due to a severe reduction of its diffusion rate along ER membranes, which could impair its concentration at ER export sites.
A Dibasic Motif in the CT, Proximal to the TMR, Promotes ER Exiting
A Prosite search for type II membrane proteins, in particular glycosyltransferases, revealed a conserved basic amino acid cluster in their CTs, proximal to the border of the TMR (Table 1). Because this cluster is also present in the CTs of GalT2 and GalNAcT its relevance to ER exiting was examined by replacement for alanine residues. Both mutated constructs fused to YFP and the nonmutated counterparts fused to CFP were cotransfected to CHO-K1 cells. A typical experiment carried out with GalT2 shows most GalT2RR7→AA fluorescence (green) displaying an ER characteristic pattern, which contrasts with the Golgi concentrated fluorescence of GalT2 (red) (Figure 3A, prebleaching). The Golgi containing volume was photobleached at both wavelengths, and the fluorescence recovery (FRAP) was monitored along the time. Although GalT2 replenished the fluorescence in the Golgi (Figure 3A, red), GalT2RR7→AA (Figure 3A, green) only diffused along the ER membranes, reestablishing the characteristic reticular pattern of the ER in the selected volume (best noted in the single channel split image at 120 min of FRAP).
Table 1.
Partial data base compilation (ProSite) of sequences of Golgi glycosyltransferase CTs bearing the dibasic motif [RK](X)[RK]
| Transferase | Cytoplasmic tail sequencea | TMR Sequence | Accession no.b | Organism |
|---|---|---|---|---|
| β1, 3GalT | MPLSLFRR | VLLAVLLLVIIWTLFGP | Q9Z0F0 | Mouse |
| β1, 3GalT5 | MAFPKMR | LMYICLLVLGALCLYFSMYSL | Q9Y2C3 | Human |
| β1, 3GalT7 | MSVGRRR | IKLLLGILMMANVFIYFIMEV | Q9NY97 | Human |
| β1, 3GalT8 | MKYLRHRRPN | ATLILAIGAFTLLLFSLLVSP | Q9Y2A9 | Human |
| β1, 4GalT1 | MRLREPLLSGAAMPGASLQRACR | LLVAVCALHLGVTLVYYLAGR | P15291 | Human |
| β1, 4GalT3 | MLRRLLERPC | TLALLVGSQLAVMMYLSLGGF | O60512 | Human |
| β1, 4GalT4 | MGFNLTFHLSYKFR | LLLLLTLCLTVVGWATSNYFVGAI | O60513 | Human |
| β1, 4GalT5 | MRARRGLLRLPRRS | LLAALFFFSLSSSLLYFVYVA | O43286 | Human |
| β1, 4GalT6 | MSVLRRMMRVSNRS | LLAFIFFFSLSSSCLYFIYVA | Q9UBX8 | Human |
| β1, 4GalT7 | MFPSRRKAAQLPWEDGRSGLLSGGLPRKCS | VFHLFVACLSLGFFSLLWLQL | Q9UBV7 | Human |
| GalT MNN10 | MSSVPYNSQLPISNHLEYDEDEKKSRGSKLGLK YKMIYWRKTLCSSLARWRK | LILLISLALFLFIWISDSTIS | P50108 | Yeast |
| α1, 3GalT | MNVKGR | VVLSMLLVSTVMVVFWEYINS | P50127 | Pig |
| β1, 4GlcNAcT | MYLVVCWGRVTGNMISTRHCFSRCKSRSVR | IKATAMLFVAAMLFLALHM | Q09323 | Snail |
| β1, 6GlcNAcT | MLRKLWRRK | LFSFPTKYYFLFLAFSVVTFTVL | Q92180 | Bovine |
| β1, 6GlcNAcT | MLRTLLRRR | LFSYPTKYYFMVLVLSLITFSVL | Q02742 | Human |
| β1, 2GlcNAcTI | MLKKQS | AGLVLWGAILFVAWNALLILFF | P26572 | Human |
| β1, 2GlcNAcT | MRFRIYKRK | VLILTLVVAACGFVLWSSNG | Q10469 | Human |
| β1, 4GlcNAcTIII | MRRYK | LFLMFCMAGLCLISFL | Q09327 | Human |
| β1, 4GalNAcT | MWLGRRA | LCALVLLLACASLGLLYA | Q00973 | Human |
| GalNAcT1 | MRKFAYCK | VVLATSLIWVLLDMFLLL | Q10472 | Human |
| αGalNAcT | MAEVLRTLAGKPKCHALRPMILFLIMLVLVLF | GYGVLSPRSLMPGSLERGFCMAV | P16442 | Human |
| α2, 6SialT1 | MVHINVLKK | FMCVLVVILIALTVCLW | Q92182 | Chick |
| α2, 6SialT1 | MIHTNLKKK | FSCCVLVFLLFAVICV | P15907 | Human |
| α2, 6SialT | MGFLIRRLPKDSRIFR | WLLILTVFSFIITSFSALFGM | Q92183 | Chick |
| α2, 3SialT | MVTVRKRNVK | VFTFAFVLITVTSFLLNY | Q11200 | Chick |
| α2, 3SialT | MVTLRKRTLKVLT | FLVLFIFLTSFFLNYSHTMVA | Q11201 | Human |
| α2, 3SialT | MRRK | TLKYLTFFLLFIFLTSFVLNY | P54751 | Mouse |
| α2, 6SialT | MACILKRK | ALAVSFIALCILLLAMRLA | Q64686 | Rat |
| α2, 8SialT8 | MSPCGRARRQTSRGAMAVLAWKFPRTRLPM | GASALCVVVLCWLYIFPV | Q92185 | Human |
| α2, 8SialT8 | MSPCGRALHTSRGAMAMLARKFPRTRLP | VGASALCVVVLCWLYIFPV | Q64687 | Mouse |
| α2, 3SialT4 | MVSKSRWK | LLAMLALVLVVMVWYSIS | Q11206 | Human |
| α2, 8polysialT1 | MRSIRKR | WTICTISLLLIFY | Q64690 | Mouse |
| α2, 8polysialT | MRSIRKR | WTICTISLLLIFY | Q92187 | Human |
| α1, 2FucT1 | MWTPSRR | QLCLTFLLVCVLSAGSFFF | O09160 | Mouse |
| α1, 2FucT2 | MPSDSCLLSLTVLQRLRAIC | PPLSTFYLFFVIFVVSTIFHC | P97353 | Human |
| α1, 4FucT3 | MYPPGCAKVKCSWHH | CLPGLLLQLLLALCFFSYLRM | Q11126 | Bovine |
| α1, 4FucT3 | MDPLGAAKPQWPWRR | CLAALLFQLLVAVCFFSYL | P21217 | Human |
| α1, 3FucT4 | MGAPWGSPTAAAGGRRGWRRGR | GLPWTVCVLAAAGLTCTALITYA | P22083 | Human |
| α1, 3FucT5 | MDPLGPAKPQWLWRR | CLAGLLFQLLVAVCFFSYL | Q11128 | Human |
| α1, 3FucT7 | MNNAGHGPTRRLRG | LGVLAGVALLAALWLLWLL | Q11130 | Human |
| α1, 3FucTA | MRRPKISLKK | YFYLTLICALLLIFGFSL | Q9VUL9 | Fly |
| Hyaluronan synthase 1 | MRQQDAPKPTPAARR | CSGLARRVLTIAFALLILGLM | Q92839 | Human |
| α1, 2ManT | MALFLSKRLLR | FTVIAGAVIVLLLTLNSNS | P27809 | Yeast |
| α1, 2ManT KTR1 | MAKIMIPASKQPVYKK | LGLLLVAVFTVYVFFHGAQYA | P27810 | Yeast |
| Probable ManT KTR2 | MQICKVFLTQVKK | LLFVSLLFCLIAQTCWLALVPYQ | P33550 | Yeast |
| Probable ManT KTR3 | MSVHHKKKLMPKSALLIRKYQKGIRSS | FIGLIIVLSFLFFMSGS | P38130 | Yeast |
| Probable ManT KTR4 | MRFLSKRILKP | VLSVIILISIAVTVVLYFLTAN | P38131 | Yeast |
| Probable ManT KTR5 | MLLIRRTINAFLGCIH | CNLTATCILIAFVITMYVVLV | P53966 | Yeast |
| ManT KTR6 | MHVLLSKK | IARFLLISFVFVLALMVTIN | P54070 | Yeast |
| Probable ManT KTR7 | MAIRLNPKVRRFLLDKCRQKRYG | FLFLGCIFAILYCMGTWPFFA | P40504 | Yeast |
| α1, 3ManT | MLALRRFILNQRSLRS | CTIPILVGALIIILVLF | P39106 | Yeast |
| α1, 6Man-T | MSRKLSHLIATRKSK | TIVVTVLLIYSLLTFHLSNKRLLSQ | P31755 | Yeast |
The source and accession number is given for each transferase.
Bold and underlined letters correspond to the [RK](X)[RK] motif.
Accession no., Swiss Prot data bank accession number.
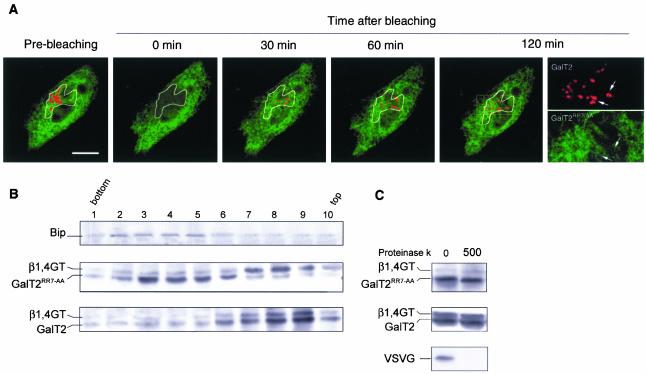
Figure 3.
RR residues in the cytoplasmic tail of GalT2 are critical for its ER exiting. (A) CHO-K1 cells coexpressing GalT2-CFP (red) and Gal-T2RR7→AA-YFP (green) were supplemented with 100 μg/ml cycloheximide 2 h before photobleaching. The Golgi volume was irreversibly photobleached at both 458 and 514 nm. The recovery of fluorescence (FRAP) at the Golgi region was analyzed by dual-color imaging every 5 min for 120 min. Note that GalT2, but not GalT2RR7→AA, replenished the photobleached Golgi. Arrows in the 120-min split image of the squared area indicate regions lacking colocalization of the constructs. Bar, 10 μm. (B) Membranes of a postnuclear supernatant fraction of CHO-K1 cells coexpressing β1,4GT and either GalT2RR7→AA or GalT2 were separated by centrifugation at 200,000 × g for 3 h in a 5–40% OptiPrep gradient. Ten fractions were collected and were Western blotted with anti-BiP (first row) or anti-GFP (second and third rows) antibody. (C) A pool of fractions 3–5 of the gradient of cells expressing Gal-T2RR7→AA and of fractions 7–9 from the gradient of cells expressing GalT2 was diluted with 0.25 M sucrose and pelleted by centrifugation at 100,000 × g for 1 h. Membranes were incubated in the absence and in the presence of 500 μg of proteinase K during 30 min at 37°C and Western blotted with anti GFP antibody. As in Figure 2B, total microsomal membranes from CHO-K1 cells expressing the VSVG protein were also treated as control.
Confocalized images of Figure 3A indicate that most of the mutated form remained in the ER; however, we cannot completely discard that an undefined fraction reached the Golgi area. This possibility is worth consideration if mutation caused some randomization on the topological orientation of the construct in the membranes. If this was the case, it is possible that the properly oriented fraction reaches the Golgi, whereas the fraction with inverted topology remains in the ER. Although data of Figure 2B did not reveal a possible mixed orientation in whole membrane preparations from cells expressing tail deleted versions, we checked in ER membranes from cells expressing the RR→ AA construct whether it was with type II membrane topology. For this, membranes from CHO-K1 cells coexpressing the N-terminal domain (aa 1–81) of the Golgi marker β1,4 Galactosyltransferase (β1,4GT) fused to the NH2 terminus of the CFP and either GalT2 or GalT2RR7→AA fused to the NH2 terminus of the YFP were separated by centrifugation in an OptiPrep density gradient. Figure 3B shows the mutated form displaced toward fractions of the gradient closer to those enriched in the endogenous ER marker BiP (fractions 3–5) than to fractions 7–9 enriched in β1,4GT (Figure 3A). GalT2 on the other hand was heavily concentrated in Golgi fractions 7–9.
We next tested by a protease protection assay whether the mutated form retained in the ER was inserted maintaining the type II membrane topology. Membranes collected from pooled fractions 3–5, enriched in the ER retained fraction of GalT2RR7→AA were incubated with or without proteinase K and the products analyzed by Western blot with anti GFP antibody (Figure 3C). Golgi membranes collected from pooled fractions 7–9 enriched in GalT2, and a whole membrane fraction from cells expressing VSVG (Figure 2B) were used as control for the behavior of, respectively, a normally oriented type II membrane protein and a type I membrane protein, to a protease protection assay. The main conclusion of Figure 3C is that the ER retained GalT2RR7→AA was in the same topological orientation as the Golgi located GalT2 and that its failure to exit the ER cannot be ascribed to a detectable inversion of its topological orientation. Together, results of Figure 3 show that the RR to AA mutation in the cytoplasmic tail of GalT2 impairs the Golgi concentration of the construct by impairing its exit from the ER.
The Dibasic Motif Is Relevant for ER Exiting of Other Members of the Glycosyltransferase Family
Experiments similar to those in Figures 1B and 3A were carried out with Ntds of GalNAcT, CMP-NeuAc:GM3 sialyltransferase (SialT21–57), CMP-NeuAc:LacCer sialyltransferase (SialT11–54) and glycoprotein β1,4Galactosyltransferase (β1,4GT1–81), and results are summarized in Table 2. Substitution in GalT2 of RR residues by other positively charged amino acids (KK), or intercalation of one alanine (KAK or RAR), did not affect the Golgi concentration of the constructs. RR5→ AA substitution in GalNAcT affected its ER exiting. Impairment was only partial in this case, due to the presence of an endogenous GalT2 in CHO-K1 cells that complexes with GalNAcT constructs and carries them to the Golgi (Giraudo et al., 2001) (Figure 3B). Substitution of one of the two R by A in GalNAcT gave intermediate results, with part of the protein in the Golgi and part in the ER, whereas substitutions of any other residue by A were irrelevant. Interestingly, mutation of RR8→ AA or deletion of aa 2–16 in SialT2 had no effect on its Golgi location, whereas mutation of RXR25→ AXA, which is closer to the TMR border, or complete deletion of the CT, resulted in ER location. Mutations in the CTs of SialT1 (RR2→ AA) and β1,4GT (RXR2→ AA) also produced failure of these proteins to exit from the ER. Together, these results indicate that the [RK](X)[RK] motif within the CT is necessary for exiting of the analyzed proteins from the ER, and in addition, that it is most effective when located proximal to the TMR.
Table 2.
Summary of steady state distribution of different CT mutated constructs
| Constructa | CT sequenceb | Distributionc |
|---|---|---|
| GalT2 | MPLSLFRR | Golgi |
| ΔGalT2 | M | ER |
| GalT2RR7→AA | MPLSLFAA | ER |
| GalT2RR7→KK | MPLSLFKK | Golgi |
| GalT2FRR6→RAR | MPLSLRAR | Golgi |
| GalT2FRR6→KAK | MPLSLKAK | Golgi |
| GalNAcT | MWLGRRA | Golgi |
| ΔGalNAcT | M | ER/Golgi |
| GalNAcTW2→A | MALGRRA | Golgi |
| GalNAcTR5→A | MWLGARA | Golgi/ER |
| GalNAcTR6→A | MWLGRAA | Golgi/ER |
| GalNAcTRR5→AA | MWLGAAA | ER/Golgi |
| SialT2 | MSPCGRARRQTSRGAMAVLAWKFPRTRLPM | Golgi |
| Δ2-16SialT2 | MVLAWKFPRTRLPM | Golgi/ER |
| Δ2-29SialT2 | M | ER |
| SialT2RR8→AA | MSPCGRAAAQTSRGAMAVLAWKFPRTRLPM | Golgi/ER |
| SialT2RXR25→AXA | MSPCGRARRQTSRGAMAVLAWKFPATALPM | ER/Golgi |
| SialT11-54 | MRRPSLLIKD | Golgi |
| SialT1RR2→AA | MAAPSLLIKD | ER/Golgi |
| β1,4GT | MRLREPLLSGSAAMPGASLQRACR | Golgi |
| β1,4GTRXR2→AXA | MALAEPLLSGSAAMPGASLQRACR | ER/Golgi |
Constructs fused to spectral variants of GFP.
The border CT/TMR was predicted with the TMpred tool (Hofmann and Stoffel, 1993). Basic clusters are shown in italics. Bold and underlined letters correspond to substituted residues.
The steady-state location of constructs was determined by dual color imaging in cells coexpressing the nonmutated and mutated constructs. Constructs with a dual distribution (Golgi and ER) were expressed as follows: ER/Golgi, localized mostly in the ER; Golgi/ER, localized mostly in the Golgi.
A CT with Dibasic Amino Acid Motif Promotes Exiting of an ER-Resident Protein
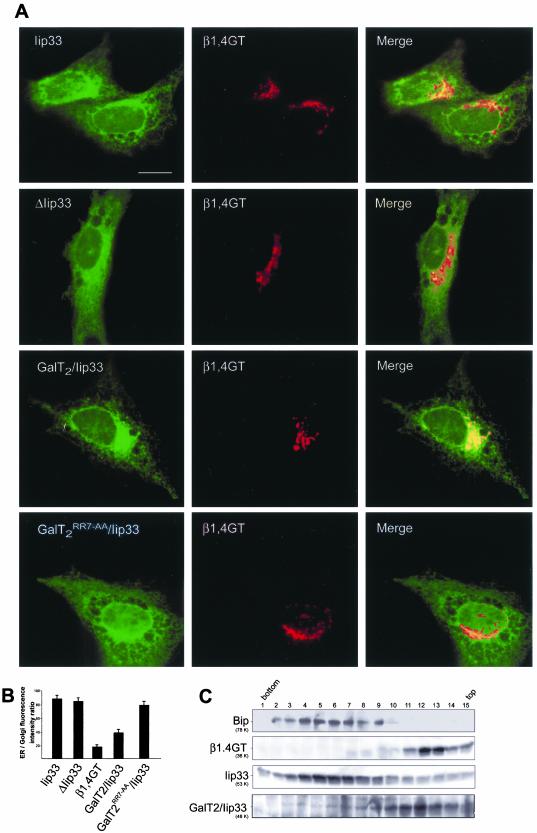
If the [RK](X)[RK] motif acts as an ER exiting signal, it should be able to promote exiting of an ER resident protein. This was confirmed by substitution of the CT of the ER resident, type II membrane protein Iip33 for the CT of GalT2. Iip33, the invariant chain of the major histocompatibility complex class II, exposes a predicted N-terminal domain of 46 aa to the cytoplasm. This domain contains an ER retention signal consisting of three R residues at positions 3–5 (Schutze et al., 1994). The isoform lacking aa 1–15 (Iip31), able to exit the ER, contains another basic cluster (RR) at positions 35–36, 10 aa from the TMR (Strubin et al., 1986). According to our model, complete deletion of the CT would impair exiting of Iip33 from the ER, and its replacement with the GalT2 CT would restore its exiting capability. To test this prediction, Iip33, CT deleted Iip33 (ΔIip33) and tail-substituted/Iip33, (GalT2/Iip33 and GalT2RR7→AA/Iip33) tagged with YFP, were independently cotransfected to CHO-K1 cells with the Golgi marker β1,4GT tagged with CFP. Both, Iip33 and ΔIip33 lacking the ER exiting signal showed ER localization (Figure 4A, first and second rows. respectively). GalT2/Iip33, on the other hand, exit the ER and partially colocalized with β1,4GT in the Golgi (Figure 4A, third row). However, substitution of Iip33 CT with GalT2RR7→AA CT was less effective in promoting its exiting, because most of it distributes with an ER pattern of localization (Figure 4A, fourth row). Quantification of ER/Golgi fluorescence intensity ratios in expressing cells showed this ratio to be ∼5–6 times higher for the ER marker Iip33, and also for ΔIip33 than for the Golgi marker β1,4GT. Confirming the single cell observations, the ratio for GalT2/Iip33 was only ∼2 times higher than β1,4GT, whereas the ratio of GalT2RR7→AA/Iip33 was essentially the same as for Iip33 (Figure 4B).
Figure 4.
The cytoplasmic tail of GalT2 promotes exiting of Iip33 from ER. (A) Dual-color imaging of cells coexpressing β1,4GalT1-81 tagged with CFP at the C termini (β1,4GT, middle panels) and YFP-tagged Iip331-230 (Iip33) or lip3347-230 (ΔIip33) or GalT21-8/Iip3347-230 (GalT2/Iip33) or GalT21-8RR7→ AA-Iip3347-230 (GalT2RR7→AA/Iip33) (left panels in, respectively, top, middle, and bottom row). The typical ER reticular pattern of Iip33 and ΔIip33 contrasts with that of GalT2/Iip33, which partially colocalizes with the Golgi marker β1,4GT. GalT2RR7→AA/Iip33 localization, on the other hand, was essentially the same as that of Iip33. (B) Quantification of images in A. Results are presented as ER/Golgi fluorescence intensity ratio (arbitrary units) of single cells coexpressing the indicated chimaeras. Values are mean ± SD for n = 25 cells. (C) Membranes of a postnuclear supernatant fraction of CHO-K1 cells as in top and middle rows in A were separated by centrifugation at 200,000 ×g for 3 h in a 5–40% OptiPrep gradient. Fifteen fractions were collected and Western blotted with anti-BiP (first row) or anti-GFP (second to fourth rows) antibody. In contrast to Iip33, which cosedimented with the ER marker Bip, Gal-T2/Iip33 was displaced to fractions containing the Golgi membrane marker β1,4GT.
Localization of GalT2/Iip33 was also examined in membrane fractions of cells as in Figure 4A, separated by density gradient centrifugation, and analyzed by Western blotting. ER membranes were distributed along fractions 2–9, as indicated by the presence of the ER marker BiP (Figure 4C, first row), whereas Golgi membranes peaked in fractions 11–15, as indicated by the presence of the marker β1,4GT (Figure 4C, second row). Iip33 cofractionated with membranes of the ER (Figure 4C, third row), but when its CT was replaced by the CT of GalT2 it was displaced to fractions of lesser density showing the highest concentration in fractions 9–13, partially overlapping with both ER- and Golgi-enriched fractions (Figure 4C, fourth row). This result is consistent with those of Figure 4, A and B, and confirms that the peptide with the dibasic motif is able to promote exiting of membrane proteins from the ER.
Sar1 Interacts with the [RK](X)[RK] Motif
The small GTPase Sar1 (Barlowe et al., 1994; Kuge et al., 1994), interacts in the ER with the CTs of cargo proteins, concentrating them at ER export sites from which COPII-transport vesicles originate. Microinjection of the dominant negative (GDP restricted) form of Sar1 (Sar1T39N) inhibits transport resulting in ER accumulation of cargo proteins including those resident of the Golgi (Storrie et al., 1998; Seemann et al., 2000; Ward et al., 2001). We confirmed this for GalT2, GalNAcT and SialT2 Ntds fused to YFP, which accumulated into the ER upon microinjection of recombinant Sar1T39N to the expressing cells (our unpublished results).
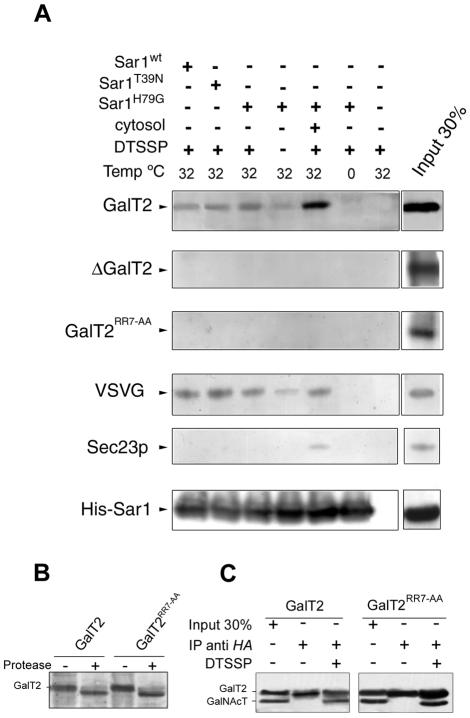
The role of Sar1 in ER→ Golgi transport of the constructs was further examined in urea-stripped microsomes from cells coexpressing either GalT2, or ΔGalT2, or GalT2RR7→AA, and the VSVG protein. Membranes were incubated with recombinant, His-tagged Sar1wt, Sar1T39N (GDP form) or Sar1H79G (GTP form) at 32°C for 15 min. After incubation, the membrane-impermeable and -reversible cross-linker DTSSP was added, and membranes were solubilized with SDS. Sar1 forms were immunoprecipitated with anti-His antibody, and the immunocomplexes were analyzed by Western blot for presence of GalT2 forms. GalT2 (Figure 5A, first row) and VSVG (Figure 5A, fourth row) were cross-linked to all forms of Sar1, indicating binding of Sar1 to these constructs. A small fraction of GalT2 coimmunoprecipitated with Sar1H79G, even in the absence of DTSSP (Figure 5A, first row). No coimmunoprecipitation was observed in the absence of Sar1, or upon incubation on ice. In no case didSar1 coimmunoprecipitate ΔGalT2 (Figure 5A, second row) or GalT2RR7→AA (Figure 5A, third row). The fraction of GalT2 that coimmunoprecipitated with Sar1H79G increased in the presence of rat liver cytosol, suggesting that other cytosolic proteins may enhance this interaction. Sec23p, which participates in the COPII-budding reaction, is a candidate based on its capture from rat liver cytosol as a partner in the Sar1-GalT2 complex (Figure 5A, fifth row).
Figure 5.
Sar1 binds to cytoplasmic tail sequences containing RR residues. (A) Microsomal membranes from cells expressing the constructs indicated at left, fused to YFP, were incubated with the indicated Sar1 forms at 32°C for 15 min in the absence or presence of rat liver cytosol. Microsomes were collected and the reversible cross-linking agent DTSSP was added. Membranes were solubilized with 1% SDS, and recombinant Sar1 was immunoprecipitated with anti-His antibody. The immunocomplexes were run on SDS-PAGE and blotted with anti-GFP and anti-Sec23p antibody. Input 30% stands for 30% of the total protein used for immunoprecipitation. Bottom row shows a representative immunoprecipitate blotted with anti-His antibody. (B) Microsomal membranes from cells expressing GalT2-CFP or GalT2RR7→AA-YFP were treated with 500 μg/ml proteinase K for 30 min, separated on SDS-4–20% gradient PAGE and Western blotted with anti-GFP antibody. (C), Microsomal membranes from cells coexpressing GalT2-CFP or GalT21–52RR7→ AA-YFP and GalNAcT-CFP, cultured at 15°C for 16 h, were treated with DTSSP and solubilized with 1% SDS. GalT2 was immunoprecipitated with anti-HA antibody and immunoprecipitates were Western blotted with anti-GFP antibody.
CT Lacking RR Residues Expose Normally to the Cytoplasm
The possibility of a lack of exposure of the CT with RR7→ AA substitution to the cytoplasmic face of the ER, which may impair the interaction with Sar1, was examined in the following two ways. First, membranes from cells expressing GalT2 and GalT2RR7→AA were treated with proteinase K, and generation of the respective products was analyzed in SDS-gradient polyacrylamide gels and Western blot. The formation of a product that migrates slightly faster, resulting from the loss of the short cytoplasmic tail, was similar for both GalT2 and GalT2RR7→AA (Figure 5B), indicating that the accessibility of the CTs of the mutated and nonmutated versions to proteinase K was essentially the same. Second, knowing that GalT2 complexes GalNAcT (Giraudo et al., 2001), the fraction of GalT2 or Gal-T2RR7→AA cross-linked to GalNAcT by DTSSP was determined in membranes from cells cotransfected with GalNAcT and either GalT2 or GalT2RR7→AA. DTSSP cross-links the primary amines if they are close enough. The only primary amines in the CT sequences of GalT2 and GalNAcT are those of the N-terminal methionines. So, the amount of GalNAcT coimmunoprecipitated with GalT2 or Gal-T2RR7→AA by antihemagglutinin (HA) antibody after DTSSP treatment will inform the topological proximity of these N termini. The amount of GalNAcT coimmunoprecipitated by anti-HA antibody was found similar (Figure 5C), indicating that the CT of Gal-T2RR7→AA maintains a topological proximity to the CT of GalNAcT comparable to that of GalT2.
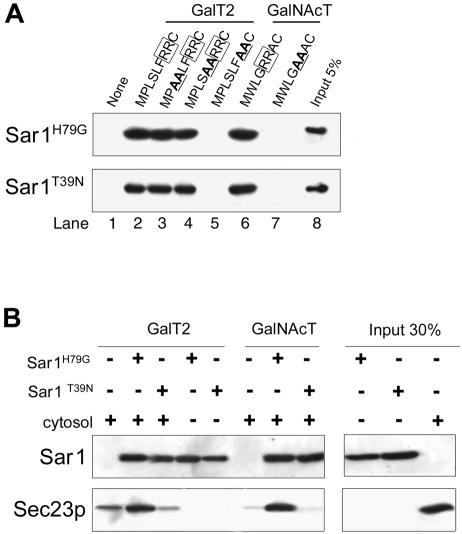
Synthetic Peptides with CT Sequences Bind Sar1
We next examined binding of His-tagged recombinant forms of Sar1 to CT sequences of GalT2 and GalNAcT and their mutated forms, linked to thiopropyl Sepharose 6B through a C-terminal added cysteine. Binding was assessed by determination of bound Sar1 by Western blotting with anti-His antibody. Sar1H79G (Figure 6A, top row), and to a lesser extent Sar1T39N (Figure 6A, bottom row), bound to CT peptides of both GalT2 (lane 2) and GalNAcT (lane 6), indicating that binding was not critically dependent on either the presence of other proteins or the nucleotide state of Sar1. Sar1 binding to RR7→ AA mutant form of GalT2 (lane 5) or RR5→ AA of GalNAcT (lane 7) peptides was below the limit of detection, reinforcing the importance of the dibasic motif for the interaction with Sar1. Other AA substitutions in GalT2 CT sequences (lanes 3 and 4) did not affect binding, and no binding to matrix was observed (lane 1).
Figure 6.
Sar1 binds to synthetic peptides with cytoplasmic tail sequences. (A) Recombinant Sar1H79G (GTP form) and Sar1T39N (GDP form) were incubated with synthetic peptides related to tail sequences of GalT2 (lanes 2–5) and GalNAcT (lanes 6 and 7), coupled to thiopropyl Sepharose 6B beads. After incubation, beads were washed and bound Sar1 analyzed by SDS-PAGE and Western blotting with anti-His antibody. (B) Sepharose bound peptides with the sequence of CT of GalT2 or GalNAcT were incubated with or without the indicated forms of Sar1 as in A and then incubated overnight with CHO-K1 cell cytosol. Beads were collected, washed, separated on SDS-PAGE, and Western blotted with anti-His (top row) or anti Sec23 (bottom row) antibody.
CT Peptides Loaded with Sar1H79G, but Not with Sar1T39N, Bind Sec23p
Immobilized peptides with GalT2 and GalNAcT CT sequences were used to determine whether they were able to recruit Sec23p in vitro, as was evidenced in the experiment in Figure 5A. They were previously loaded with recombinant Sar1H79G or Sar1T39N and then incubated with CHO-K1 cytosol. Recruitment of Sec23p from cytosol was assessed by Western blotting of the Sepharose bound fraction. Peptides not loaded with Sar1 were able to recruit some Sec23p from cytosol, most probably through the binding of endogenous Sar1 (Figure 6B, lanes 1 and 6). Recruitment of Sec23p was clearly increased when GalT2 and GalNAcT CT peptides were overloaded with Sar1H79G (lanes 2 an 7) but not with Sar1T39N (lanes 3 and 8), showing that although both forms of Sar1 bind to CT peptides, only the Sar1(GTP) bound to the CT peptide acquired the conformation able to recruit Sec23p. No Sec23p was detected when cytosol was omitted from the assay (lanes 4 and 5). Together, results of Figures 5 and 6 indicate that CT peptides with the [RK](X)[RK] motif participate in a Sar1 (GTP)-dependent binding to Sec23p.
DISCUSSION
We show herein that deletion of the CT of the Ntds of the glycolipid glycosyltransferases GalT2, GalNAcT, and SialT2 does not affect their ER membrane insertion, acquisition of type II topology or diffusion along ER membranes, but impairs their Golgi concentration. Mutational analyses indicate that the dibasic motif [RK](X)[RK] in CTs, proximal to the TMR, is necessary for these proteins to concentrate in the Golgi complex. This dibasic motif was found in most members of the glycosyltransferase family and conserved among different organisms (Table 1), and was not a common feature of ER resident type II membrane proteins (Teasdale and Jackson, 1996). FRAP experiments showed reduced ER→ Golgi transport rates for tail-deleted or tail-mutated forms of GalT2 or GalNAcT Ntds. The rates of transport calculated for the mutated versions were ∼0.1% min–1, whereas the corresponding values for the nonmutated forms were ∼3.4% min–1, as reported for other glycosyltransferases (Zaal et al., 1999; Dahm et al., 2001). Thus, our results indicate that the CTs, and within them the [RK](X)[RK] motif, are necessary for exiting of these proteins from the ER. Furthermore, complete deletion of the CT of Iip33, which contains a dominant ER retention signal, did not result in ER escape of Iip33. However, the attachment of the CT of GalT2, but not of its RR7→ AA-mutated form, resulted in exiting of Iip33 from the ER, confirming that the dibasic motif [RK](X)[RK] has the capacity to promote ER exiting.
Mutational analyses in the SialT2 Ntd revealed that RR8, located 22 residues away from the TMR, was less effective for promoting ER exiting than RXR25, which is closer to the border of the TMR. Thus, the [RK](X)[RK] motif most effectively promotes ER export when it is proximal to the TMR.
Dibasic sorting signals are also involved in retention/retrieval of ER resident proteins. These are the -K(X)KXX-COOH motif, which we will refer to as -KKXX, and the NH2-M(X)R(X)R-motif, which we will refer to as RR-. The -KKXX motif at the C termini was reported to be sufficient to confer ER localization to many type I membrane proteins (E19, WBP-1, and some members of the p24 protein family) and reporter proteins (Jackson et al., 1990; Jackson et al., 1993; Cosson and Letourneur, 1994). This effect is achieved by interacting with members of the COPI coat (α-, β′-, and ζ-COP) of retrograde transport vesicles (Cosson and Letourneur, 1994; Gaynor et al., 1994; Letourneur et al., 1994; Cosson et al., 1996; Fiedler et al., 1996). The function of -KKXX motif is highly dependent on the identity of the basic amino acids (lysines cannot be replaced by arginines), on the position of the lysine residues (one lysine must be at position –3 and the other at –4 or –5 from the C termini) and on the sequence context in which they are located (Zerangue et al., 2001).
Concerning the RR-signal sequence close to the N terminus, it was reported to be sufficient to confer ER localization to a few type II membrane proteins (Iip33, TRAM, and p63) and reporter proteins (Schutze et al., 1994; Teasdale and Jackson, 1996; Zerangue et al., 1999). It has been proposed that the ER localization is mediated by a direct interaction between the RR-motif and the member of the COPI coat, β-COP (O'Kelly et al., 2002). The position of the RR-residues relative to the N terminus is critical. RR residues in the CT of Iip33 must be at positions 2 and 3, 3 and 4, 4 and 5, 2 and 4, or 3 and 5, ∼40 aa distal to the TMR to act as retention signal; Schutze et al., 1994). Nevertheless, neither the identity of the basic amino acid nor the sequence context seems to be critical for its effect.
So, the location of RR residues along the CTs may define their role as ER retention or exiting motifs. Moreover, for the case of the Simian immunodeficiency virus envelope glycoprotein (a type I membrane protein), KK residues located at position 18 or farther away from the TMR function as ER retrieval signal. However, if these KK residues are located at positions closer to the TMR, the envelope glycoprotein exits the ER and reaches the plasma membrane (Vincent et al., 1998). Further studies will be necessary to know the mechanisms by which similar signals differing in their position along the CTs may compete in defining ER exiting or retention. Masking events by interacting proteins, as reported previously (O'Kelly et al., 2002) may help in defining such a competition.
The effect of CT deletion has been examined in other glycosyltransferases such as α2,6-SialylT (Colley et al., 1992) and β1,4GT (Aoki et al., 1992). The basic amino acid cluster with ER-exiting activity observed in this study was maintained as a short CT remnant in the tail-deleted mutants of those previous studies. This may explain the previous conclusion that the CTs were of minor relevance for concentration of α2,6-SialylT and β1,4-GT in the Golgi. However, given the complexity of the ER-exiting process, the influence of other motifs or combinations of determinants on the ER exiting of different glycosyltransferases is worth considering.
The ER recycling of several Golgi resident proteins has been extensively described and shown to be Sar1 and microtubule dependent (Cole et al., 1998; Storrie et al., 1998; Zaal et al., 1999; Seemann et al., 2000; Ward et al., 2001). In our study, the CT of GalT2 in microsomal membranes of GalT2-expressing cells was cross-linked and coimmunoprecipitated with recombinant forms of Sar1, as it was the VSVG protein known to depend on Sar1 binding for exiting the ER (Aridor et al., 2001). Importantly, GalT2RR7→AA did not cross-link to Sar1, indicating that Sar1 recognizes specifally the motif [RK](X)[RK] of Ntds inserted in ER membranes. Synthetic peptides with CT sequences containing the [RK](X)[RK] motif showed binding activity toward Sar1 in the absence of other proteins. Both GDP and GTP forms of Sar1 bind to the peptides, although their affinities remain to be determined. An examination of the electrostatic potential of the ternary complex Sar1/Sec23p/24p (Bi et al., 2002) with the MEAD program (Bashford and Gerwert, 1992) reveals negatively charged amino acid residues in the membrane facing domain of Sar1 as candidates to interact with the basic cluster in the CTs of the transferases.
Another component of the COPII system, Sec23p, did not bind to the CT sequences, but it did bind the complex of Sar1 bound to the peptides. Noticeably, Sec23p was able to distinguish between GDP and GTP forms of Sar1, with its binding almost exclusive to Sar1GTP-loaded peptides. In summary, from our combined in vivo and in vitro studies, we propose that interaction of Sar1 with the [RK](X)[RK] motif at the CT of members of the glycosyltransferase family, and probably of other type II membrane proteins, is an early event in the selection of these proteins as cargo of COPII-transport vesicles on their way to the Golgi complex.
Acknowledgments
We thank M. Villarreal, and G. Montich for helpful discussions; T. Santa Coloma (Instituto deInvestigaciones Bioquimicas Fundación Campomar, Buenos Aires, Argentina) for help with confocal microscopy; and G. Schachner, S. Deza, and C. Mas for excellent technical assistance. H.J.F.M. is Career Investigator, and C.G.G. fellow, of CONICET. This work was supported in part by grants PMT-PICT 0181 from CONICET, 89/96 from SECYT-UNC (Argentina), 75197-554001 from the International Research Scholar Program of Howard Hughes Medical Institute, and 10087 from the Mizutani Foundation for Glycoscience (Japan).
Article published online ahead of print. Mol. Biol. Cell 10.1091/mbc.E03–02–0101. Article and publication date are available at www.molbiolcell.org/cgi/doi/10.1091/mbc.E03-02-0101.
References
- Abdul-Ghani, M., Gougeon, P.Y., Prosser, D.C., Da-Silva, L.F., and Ngsee, J.K. (2001). PRA isoforms are targeted to distinct membrane compartments. J. Biol. Chem. 276, 6225–6233. [DOI] [PubMed] [Google Scholar]
- Antonny, B., Madden, D., Hamamoto, S., Orci, L., and Schekman, R. (2001). Dynamics of the COPII coat with GTP and stable analogues. Nat. Cell Biol. 3, 531–537. [DOI] [PubMed] [Google Scholar]
- Antonny, B., and Schekman, R. (2001). ER export: public transportation by the COPII coach. Curr. Opin. Cell Biol. 13, 438–443. [DOI] [PubMed] [Google Scholar]
- Aoki, D., Lee, N., Yamaguchi, N., Dubois, C., and Fukuda, M.N. (1992). Golgi retention of a trans-Golgi membrane protein, galactosyltransferase, requires cysteine and histidine residues within the membrane-anchoring domain. Proc. Natl. Acad. Sci. USA 89, 4319–4323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aridor, M., Fish, K.N., Bannykh, S., Weissman, J., Roberts, T.H., Lippincott-Schwartz, J., and Balch, W.E. (2001). The Sar1 GTPase coordinates biosynthetic cargo selection with endoplasmic reticulum export site assembly. J. Cell Biol. 152, 213–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aridor, M., Weissman, J., Bannykh, S., Nuoffer, C., and Balch, W.E. (1998). Cargo selection by the COPII budding machinery during export from the ER. J. Cell Biol. 141, 61–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Audigier, Y., Friedlander, M., and Blobel, G. (1987). Multiple topogenic sequences in bovine opsin. Proc. Natl. Acad. Sci. USA 84, 5783–5787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balch, W.E., McCaffery, J.M., Plutner, H., and Farquhar, M.G. (1994). Vesicular stomatitis virus glycoprotein is sorted and concentrated during export from the endoplasmic reticulum. Cell 76, 841–852. [DOI] [PubMed] [Google Scholar]
- Bannykh, S.I., Nishimura, N., and Balch, W.E. (1998). Getting into the Golgi. Trends Cell Biol. 8, 21–25. [DOI] [PubMed] [Google Scholar]
- Barlowe, C., d'Enfert, C., and Schekman, R. (1993). Purification and characterization of SAR1p, a small GTP-binding protein required for transport vesicle formation from the endoplasmic reticulum. J. Biol. Chem. 268, 873–879. [PubMed] [Google Scholar]
- Barlowe, C., Orci, L., Yeung, T., Hosobuchi, M., Hamamoto, S., Salama, N., Rexach, M.F., Ravazzola, M., Amherdt, M., and Schekman, R. (1994). COPII: a membrane coat formed by Sec proteins that drive vesicle budding from the endoplasmic reticulum. Cell 77, 895–907. [DOI] [PubMed] [Google Scholar]
- Barlowe, C., and Schekman, R. (1993). SEC12 encodes a guaninenucleotide-exchange factor essential for transport vesicle budding from the ER. Nature 365, 347–349. [DOI] [PubMed] [Google Scholar]
- Bashford, D., and Gerwert, K. (1992). Electrostatic calculations of the pKa values of ionizable groups in bacteriorhodopsin. J. Mol. Biol. 224, 473–486. [DOI] [PubMed] [Google Scholar]
- Belden, W.J., and Barlowe, C. (2001). Distinct roles for the cytoplasmic tail sequences of Emp24p and Erv25p in transport between the endoplasmic reticulum and Golgi complex. J. Biol. Chem. 276, 43040–43048. [DOI] [PubMed] [Google Scholar]
- Bi, X., Corpina, R.A., and Goldberg, J. (2002). Structure of the Sec23/24#150;Sar1 pre-budding complex of the COPII vesicle coat. Nature 419, 271–277. [DOI] [PubMed] [Google Scholar]
- Cole, N.B., Ellenberg, J., Song, J., DiEuliis, D., and Lippincott-Schwartz, J. (1998). Retrograde transport of Golgi-localized proteins to the ER. J. Cell Biol. 140, 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colley, K.J. (1997). Golgi localization of glycosyltransferases: more questions than answers. Glycobiology 7, 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colley, K.J., Lee, E.U., and Paulson, J.C. (1992). The signal anchor and stem regions of the β-galactoside 2,6-sialyltransferase may each act to localize the enzyme to the Golgi apparatus. J. Biol. Chem. 267, 7784–7793. [PubMed] [Google Scholar]
- Cosson, P., Demolliere, C., Hennecke, S., Duden, R., and Letourneur, F. (1996). Delta- and zeta-COP, two coatomer subunits homologous to clathrin-associated proteins, are involved in ER retrieval. EMBO J. 15, 1792–1798. [PMC free article] [PubMed] [Google Scholar]
- Cosson, P., and Letourneur, F. (1994). Coatomer interaction with di-lysine endoplasmic reticulum retention motifs. Science 263, 1629–1631. [DOI] [PubMed] [Google Scholar]
- Dahm, T., White, J., Grill, S., Fullekrug, J., and Stelzer, E.H. (2001). Quantitative ER↔Golgi transport kinetics and protein separation upon Golgi exit revealed by vesicular integral membrane protein 36 dynamics in live cells. Mol. Biol. Cell 12, 1481–1498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominguez, M., Dejgaard, K., Fullekrug, J., Dahan, S., Fazel, A., Paccaud, J.P., Thomas, D.Y., Bergeron, J.J., and Nilsson, T. (1998). gp25L/emp24/p24 protein family members of the cis-Golgi network bind both COP I and II coatomer. J. Cell Biol. 140, 751–765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiedler, K., Veit, M., Stamnes, M.A., and Rothman, J.E. (1996). Bimodal interaction of coatomer with the p24 family of putative cargo receptors. Science 273, 1396–1399. [DOI] [PubMed] [Google Scholar]
- Fullekrug, J., and Nilsson, T. (1998). Protein sorting in the Golgi complex. Biochim. Biophys. Acta 1404, 77–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fullekrug, J., Suganuma, T., Tang, B.L., Hong, W., Storrie, B., and Nilsson, T. (1999). Localization and recycling of gp27 (hp24gamma3): complex formation with other p24 family members. Mol. Biol. Cell 10, 1939–1955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaynor, E.C., te Heesen, S., Graham, T.R., Aebi, M., and Emr, S.D. (1994). Signal-mediated retrieval of a membrane protein from the Golgi to the ER in yeast. J. Cell Biol. 127, 653–665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giraudo, C.G., Daniotti, J.L., and Maccioni, H.J. (2001). Physical and functional association of glycolipid N-acetyl-galactosaminyl and galactosyl transferases in the Golgi apparatus. Proc. Natl. Acad. Sci. USA 98, 1625–1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartmann, E., Rapoport, T.A., and Lodish, H.F. (1989). Predicting the orientation of eukaryotic membrane-spanning proteins. Proc. Natl. Acad. Sci. USA 86, 5786–5790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmann, K., and Stoffel, W. (1993). TMbase - A database of membrane spanning proteins segments. Biol. Chem. Hoppe-Seyler 374, 166. [Google Scholar]
- Huang, M., Weissman, J.T., Beraud-Dufour, S., Luan, P., Wang, C., Chen, W., Aridor, M., Wilson, I.A., and Balch, W.E. (2001). Crystal structure of Sar1-GDP at 1.7 A resolution and the role of the NH2 terminus in ER export. J. Cell Biol. 155, 937–948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson, M.R., Nilsson, T., and Peterson, P.A. (1990). Identification of a consensus motif for retention of transmembrane proteins in the endoplasmic reticulum. EMBO J. 9, 3153–3162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson, M.R., Nilsson, T., and Peterson, P.A. (1993). Retrieval of transmembrane proteins to the endoplasmic reticulum. J. Cell Biol. 121, 317–333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kappeler, F., Klopfenstein, D.R., Foguet, M., Paccaud, J.P., and Hauri, H.P. (1997). The recycling of ERGIC-53 in the early secretory pathway. ERGIC-53 carries a cytosolic endoplasmic reticulum-exit determinant interacting with COPII. J. Biol. Chem. 272, 31801–31808. [DOI] [PubMed] [Google Scholar]
- Keller, P., Toomre, D., Diaz, E., White, J., and Simons, K. (2001). Multicolour imaging of post-Golgi sorting and trafficking in live cells. Nat. Cell Biol. 3, 140–149. [DOI] [PubMed] [Google Scholar]
- Kuehn, M.J., Herrmann, J.M., and Schekman, R. (1998). COPII-cargo interactions direct protein sorting into ER-derived transport vesicles. Nature 391, 187–190. [DOI] [PubMed] [Google Scholar]
- Kuge, O., Dascher, C., Orci, L., Rowe, T., Amherdt, M., Plutner, H., Ravazzola, M., Tanigawa, G., Rothman, J.E., and Balch, W.E. (1994). Sar1 promotes vesicle budding from the endoplasmic reticulum but not Golgi compartments. J. Cell Biol. 125, 51–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lederkremer, G.Z., Cheng, Y., Petre, B.M., Vogan, E., Springer, S., Schekman, R., Walz, T., and Kirchhausen, T. (2001). Structure of the Sec23p/24p and Sec13p/31p complexes of COPII. Proc. Natl. Acad. Sci. USA 98, 10704–10709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letourneur, F., Gaynor, E.C., Hennecke, S., Demolliere, C., Duden, R., Emr, S.D., Riezman, H., and Cosson, P. (1994). Coatomer is essential for retrieval of dilysine-tagged proteins to the endoplasmic reticulum. Cell 79, 1199–1207. [DOI] [PubMed] [Google Scholar]
- Ma, D., Zerangue, N., Lin, Y.F., Collins, A., Yu, M., Jan, Y.N., and Jan, L.Y. (2001). Role of ER export signals in controlling surface potassium channel numbers. Science 291, 316–319. [DOI] [PubMed] [Google Scholar]
- Mellman, I., and Warren, G. (2000). The road taken: past and future foundations of membrane traffic. Cell 100, 99–112. [DOI] [PubMed] [Google Scholar]
- Munro, S. (1998). Localization of proteins to the Golgi apparatus. Trends Cell Biol. 8, 11–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nehls, S., Snapp, E.L., Cole, N.B., Zaal, K.J., Kenworthy, A.K., Roberts, T.H., Ellenberg, J., Presley, J.F., Siggia, E., and Lippincott-Schwartz, J. (2000). Dynamics and retention of misfolded proteins in native ER membranes. Nat. Cell Biol. 2, 288–295. [DOI] [PubMed] [Google Scholar]
- Nilsson, T., Lucocq, J.M., Mackay, D., and Warren, G. (1991). The membrane spanning domain of beta-1, 4-galactosyltransferase specifies trans Golgi localization. EMBO J. 10, 3567–3575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nufer, O., Guldbrandsen, S., Degen, M., Kappeler, F., Paccaud, J.P., Tani, K., and Hauri, H.P. (2002). Role of cytoplasmic C-terminal amino acids of membrane proteins in ER export. J. Cell Sci. 115, 619–628. [DOI] [PubMed] [Google Scholar]
- O'Kelly, I., Butler, M.H., Zilberberg, N., and Goldstein, S.A. (2002). Forward transport. 14–3-3 binding overcomes retention in endoplasmic reticulum by dibasic signals. Cell 111, 577–588. [DOI] [PubMed] [Google Scholar]
- Rothman, J.E., and Wieland, F.T. (1996). Protein sorting by transport vesicles. Science 272, 227–234. [DOI] [PubMed] [Google Scholar]
- Rowe, T., and Balch, W.E. (1995). Expression and purification of mammalian Sarl. Methods Enzymol. 257, 49–53. [DOI] [PubMed] [Google Scholar]
- Salama, N.R., Chuang, J.S., and Schekman, R.W. (1997). Sec31 encodes an essential component of the COPII coat required for transport vesicle budding from the endoplasmic reticulum. Mol. Biol. Cell 8, 205–217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saraste, J., and Kuismanen, E. (1984). Pre- and post-Golgi vacuoles operate in the transport of Semliki Forest virus membrane glycoproteins to the cell surface. Cell 38, 535–549. [DOI] [PubMed] [Google Scholar]
- Schekman, R., and Orci, L. (1996). Coat proteins and vesicle budding. Science 271, 1526–1533. [DOI] [PubMed] [Google Scholar]
- Schutze, M.P., Peterson, P.A., and Jackson, M.R. (1994). An N-terminal double-arginine motif maintains type II membrane proteins in the endoplasmic reticulum. EMBO J. 13, 1696–1705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seemann, J., Jokitalo, E., Pypaert, M., and Warren, G. (2000). Matrix proteins can generate the higher order architecture of the Golgi apparatus. Nature 407, 1022–1026. [DOI] [PubMed] [Google Scholar]
- Semenza, J.C., Hardwick, K.G., Dean, N., and Pelham, H.R. (1990). ERD2, a yeast gene required for the receptor-mediated retrieval of luminal ER proteins from the secretory pathway. Cell 61, 1349–1357. [DOI] [PubMed] [Google Scholar]
- Sevier, C.S., Weisz, O.A., Davis, M., and Machamer, C.E. (2000). Efficient export of the vesicular stomatitis virus G protein from the endoplasmic reticulum requires a signal in the cytoplasmic tail that includes both tyrosine-based and di-acidic motifs. Mol. Biol. Cell 11, 13–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Springer, S., and Schekman, R. (1998). Nucleation of COPII vesicular coat complex by endoplasmic reticulum to Golgi vesicle SNAREs. Science 281, 698–700. [DOI] [PubMed] [Google Scholar]
- Stockklausner, C., Ludwig, J., Ruppersberg, J.P., and Klocker, N. (2001). A sequence motif responsible for ER export and surface expression of Kir2.0 inward rectifier K(+) channels. FEBS Lett. 493, 129–133. [DOI] [PubMed] [Google Scholar]
- Storrie, B., White, J., Rottger, S., Stelzer, E.H., Suganuma, T., and Nilsson, T. (1998). Recycling of Golgi-resident glycosyltransferases through the ER reveals a novel pathway and provides an explanation for nocodazole-induced Golgi scattering. J. Cell Biol. 143, 1505–1521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strubin, M., Long, E.O., and Mach, B. (1986). Two forms of the Ia antigen-associated invariant chain result from alternative initiations at two in-phase AUGs. Cell 47, 619–625. [DOI] [PubMed] [Google Scholar]
- Teasdale, R.D., and Jackson, M.R. (1996). Signal-mediated sorting of membrane proteins between the endoplasmic reticulum and the Golgi apparatus. Annu. Rev. Cell Dev. Biol. 12, 27–54. [DOI] [PubMed] [Google Scholar]
- Vincent, M.J., Martin, A.S., and Compans, R.W. (1998). Function of the KKXX motif in endoplasmic reticulum retrieval of a transmembrane protein depends on the length and structure of the cytoplasmic domain. J. Biol. Chem. 273, 950–956. [DOI] [PubMed] [Google Scholar]
- Ward, T.H., Polishchuk, R.S., Caplan, S., Hirschberg, K., and Lippincott-Schwartz, J. (2001). Maintenance of Golgi structure and function depends on the integrity of ER export. J. Cell Biol. 155, 557–570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshihisa, T., Barlowe, C., and Schekman, R. (1993). Requirement for a GTPase-activating protein in vesicle budding from the endoplasmic reticulum. Science 259, 1466–1468. [DOI] [PubMed] [Google Scholar]
- Zaal, K.J., et al. (1999). Golgi membranes are absorbed into and reemerge from the ER during mitosis. Cell 99, 589–601. [DOI] [PubMed] [Google Scholar]
- Zerangue, N., Malan, M.J., Fried, S.R., Dazin, P.F., Jan, Y.N., Jan, L.Y., and Schwappach, B. (2001). Analysis of endoplasmic reticulum trafficking signals by combinatorial screening in mammalian cells. Proc. Natl. Acad. Sci. USA 98, 2431–2436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zerangue, N., Schwappach, B., Jan, Y.N., and Jan, L.Y. (1999). A new ER trafficking signal regulates the subunit stoichiometry of plasma membrane K(ATP) channels. Neuron 22, 537–548. [DOI] [PubMed] [Google Scholar]