Abstract
ADP-ribosylation factors (ARFs) are ubiquitous regulators of virtually every step of vesicular membrane traffic. Yeast Arf3p, which is most similar to mammalian ARF6, is not essential for cell viability and not required for endoplasmic reticulum-to-Golgi protein transport. Although mammalian ARF6 has been implicated in the regulation of early endocytic transport, we found that Arf3p was not required for fluid-phase, membrane internalization, or mating-type receptor-mediated endocytosis. Arf3p was partially localized to the cell periphery, but was not detected on endocytic structures. The nucleotide-binding, N-terminal region, and N-terminal myristate of Arf3p are important for its proper localization. C-Terminally green fluorescent protein-tagged Arf3, expressed from the endogenous promoter, exhibited a polarized localization to the cell periphery and buds, in a cell cycle-dependent manner. Arf3-GFP achieved its proper localization during polarity growth through an actin-independent pathway. Both haploid and homologous diploid arf3 mutants exhibit a random budding defect, and the overexpression of the GTP-bound form Arf3p(Q71L) or GDP-binding defective Arf3p(T31N) mutant interfered with budding-site selection. We conclude that the GTPase cycle of Arf3p is likely to be important for the function of Arf3p in polarizing growth of the emerging bud and/or an unidentified vesicular trafficking pathway.
INTRODUCTION
ADP-ribosylation factors (ARFs) are critical components of several different vesicular trafficking pathways in all eukaryotic cells (see reviews in Moss and Vaughan, 1995; Moss and Vaughan, 1998). Like all members of the Ras-related small GTPase families, interconversion of ARF1 between its active GTP-bound and inactive GDP-bound states requires catalytic assistance. A guanine nucleotide-exchange factor catalyzes the replacement of GDP by GTP (Chardin et al., 1996; Peyroche et al., 1996; Klarlund et al., 1997; Meacci et al., 1997), and a GTPase-activating protein accelerates the hydrolysis of bound GTP to GDP (Cukierman et al., 1995; Ding et al., 1996; Premont et al., 1998). Binding of GTP activates ARF1 and stabilizes its association with target membranes. As a result, coat proteins recruited by ARF1 are translocated from the cytosol to the membrane (Donaldson et al., 1992; Tsai et al., 1992; Palmer et al., 1993). Subsequently, hydrolysis of its bound GTP deactivates ARF1, followed by release of ARF1 and coat proteins from the membrane to the cytosol (Tanigawa et al., 1993; Teal et al., 1994). Thus, movement of ARF1 between membranes and cytosol is one major aspect of its function.
Mammalian ARFs are divided into three classes based on size, amino acid sequence, gene structure, and phylogenetic analysis; ARF1, ARF2, and ARF3 are in class I, ARF4 and ARF5 are in class II, and ARF6 is in class III (Moss and Vaughan, 1998). A role for ARF1 in endoplasmic reticulum (ER) to Golgi and intra-Golgi transport is well established (Rothman, 1996). ARF6 is the most distant relative of ARF1 and is the first member of the ARF subfamily demonstrated to regulate both vesicular trafficking and cytoskeletal organization (D'Souza-Schorey et al., 1995; Peters et al., 1995; Radhakrishna et al., 1996). Later, ARF6 was shown to mediate endocytosis at the apical surface of polarized Madin-Darby canine kidney epithelial cells (Altschuler et al., 1999). In adipocytes, ARF6 was involved in insulin-regulated secretory pathways (Millar et al., 1999; Yang and Mueckler, 1999). A recent report that endogenous ARF6 concentrates near the cleavage furrow after the onset of anaphase suggested that ARF6 has a previously uncharacterized role during mitotic progression (Schweitzer and D'Souza-Schorey, 2002).
To date, five members of the ARF and ARF-like (ARL) family have been characterized in Saccharomyces cerevisiae: yARF1, yARF2, yARF3, yARL1, and yARL3 (Sewell and Kahn, 1988; Stearns et al., 1990; Lee et al., 1994, 1997; Huang et al., 1999). yARF1 and yARF2 resemble mammalian ARF1 in structure and function. These ARFs have been localized to the Golgi complex and a double deletion mutant is not viable (Stearns et al., 1990). Arl1p and Arl3p were localized to the Golgi complex and the perinuclear region, respectively, and were implicated in unidentified vesicular trafficking (Lee et al., 1997; Huang et al., 1999). Our main focus in this work was to analyze the functional significance of Arf3p in yeast. Because of the high homology between mammalian ARF6 and yeast Arf3p, we examined whether the function and localization of this class of ARF would be conserved. Our findings suggest that the GTPase cycle of Arf3p regulates its distribution between a novel intracellular compartment and the cell periphery and that this regulation is also likely to be important for its function in polarity development and/or a still unidentified vesicular trafficking pathway in yeast.
MATERIALS AND METHODS
Strains, Media, and Microbiological Techniques
Table 1 lists the yeast strains used in this study. Yeast culture media were prepared as described by Sherman et al. (1986). YPD contained 1% Bacto-yeast extract, 2% Bacto-peptone, and 2% glucose. SD contained 0.17% Difco yeast nitrogen base (without amino acids and ammonium sulfate), 0.5% ammonium sulfate, and 2% glucose. Nutrients essential for auxotrophic strains were supplied at specified concentrations (Sherman et al., 1986). Sporulation, growth, and mating were carried out as described previously (Lee et al., 1989). Yeast strains were transformed by the lithium acetate method (Ito et al., 1983). Plasmids were constructed according to standard protocols (Sambrook et al., 1989). Gene disruption was carried out as described previously (Lee et al., 1994).
Table 1.
Yeast strains used in this study
| Strain | Genotypea | Source |
|---|---|---|
| YPH250 | MATa ade2, his3, leu2, lys2, trp1, ura3-52, ARF1, ARF2, ARF3, ARL1, ARL3 | Lee et al., 1997 |
| YPH250dr3 | MATa ade2, his3, leu2, lys2, trp1, ura3-52, ARF1, ARF2, arf3, ARL1, ARL3 | This work |
| YPH250dr3p | MATa ade2, his3, leu2, lys2, trp1, ura3-52, ARF1, ARF2, arf3-1, ARL1, ARL3 | This work |
| YPH252dr3 | MATα ade2, his3, leu2, lys2, trp1, ura3-52, ARF1, ARF2, arf3, ARL1, ARL3 | This work |
| YPH252dr3p | MATα ade2, his3, leu2, lys2, trp1, ura3-52, ARF1, ARF2, arf3-1, ARL1, ARL3 | This work |
| YPH250dl1pdr3 | MATa ade2, his3, leu2, lys2, trp1, ura3-52, ARF1, ARF2, arf3, arl1-1, ARL3 | Lee et al., 1997 |
| YPH250dr3pdl3 | MATa ade2, his3, leu2, lys2, trp1, ura3-52, ARF1, ARF2, arf3-1, ARL1, arl3 | Huang et al., 1999 |
| YPH250dr1 | MATa ade2, his3, leu2, lys2, trp1, ura3-52, arf1-1, ARF2, ARF3, ARL1, ARL3 | Lee et al., 1997 |
| YPH252F3GFP | MATα ade2, his3, leu2, lys2, trp1, ura3-52, ARF1, ARF2, ARF3::GFP-KanMax4, ARL1, ARL3 | This work |
ade, adenine-requiring; his, histidine-requiring; trp, tryptophan-requiring; ura, uracil-requiring; leu, leucine-requiring. arl3 represents arl3::hisG-URA3-hisG; arl3-1 represents arl3:hisG; arf3 represents arf3::hisG-URA3-hisG; arf3-1 represents arf3::hisG; arl1 represents arl1::hisG-URA3-hisG; and arl1-1 represents arl1::hisG; and arf1-1 represents arf1::hisG.
Expression and Purification of Recombinant Proteins and Polyclonal Antibody Production
For the His-tag-Arf3p fusion protein, a DNA fragment containing the yARF3 coding region was generated by amplifying of yeast genomic DNA with sequence-specific primers: 5′-primer yARF3.1 (5′-CATATGGGCAATTCAATTTCGAAG-3′) and 3′-primer yARF3.2 (5′-GTATGCGGATCCAACACCAATAAATGCAAT-3′). The primers incorporated unique NdeI and BamHI sites (underlined) at the initiating methionine and six base pairs 3′ of the stop codon, respectively. The polymerase chain reaction (PCR) product was purified and ligated to the expression vector pET15b (Novagen, Madison, WI), yielding pET15byARF3. The His-tagged fusion protein was produced in BL21 (DE3) cells and isolated on Ni2+-NTA resin (QIAGEN, Chatsworth, CA) as described previously (Huang et al., 1999). Denatured purified recombinant yeast Arf3p from SDS-PAGE gel was used as antigen to raise polyclonal antibodies in rabbits essentially as described previously (Harlow and Lane, 1988). The antigen used for preparing antibodies against Ste3p was a recombinant protein representing the C-terminal 187 amino acids of Ste3p. The cDNA was synthesized using primer pairs 5′-CATATGTACTCTAAATTCCTG-3′ and 5′-GGATCCTTAAGGGCCTGCAGTATTTTCTGA-3′. Recombinant protein production and purification and rabbit immunization were carried out as described for yeast Arf3p.
Affinity Purification of Polyclonal Antibody
Ten micrograms of purified recombinant Arf3p and Ste3p proteins were subjected to SDS-PAGE. After transfer to polyvinylidene difluoride membrane containing Arf3p or Ste3p, the strip was excised and incubated with 5% nonfat dry milk in blocking buffer TBST (25 mM Tris-HCl, pH 7.4, 0.8% NaCl, 0.02% KCl, and 0.1% Tween 20) for 1 h before incubation with anti-Arf3p antiserum (1:10 dilution) or anti-Ste3p antiserum (1:100 dilution) in blocking buffer for 2 h. After washing with TBST three times (5 min each), the strip was incubated with 2.5 ml of elution buffer (0.2 M glycine, 0.25 M NaCl, pH 2.5) for 5 min to elute purified antibodies, which were then neutralized with 2 M Tris to pH 7, and 0.5 ml of 5% BSA was added. Purified antibodies were used at a 1:10 dilution for Western blotting and without dilution for indirect immunofluorescence.
C-Terminal Green Fluorescent Protein (GFP) Tagging of Chromosomal ARF3 Gene
For GFP tagging of Arf3p, we attached heterologous DNA, GFP tag, and selection marker to the Arf3 C termini. By using a PCR-based strategy (Knop et al., 1999), the PCR products created with primers 5′-TACTTGTTAGCCACAATGCAGATTCAACACCAATAAATGCAATGTATCGATGAATTCGAGCTCG-3′ and 5′-GAAGGATTATCCTGGATTTCGAACAACACAAACGTTCCAAAGAAACGTACGCTGCAGGTCGAC-3′ (underlined sequences for homologous recombination) and plasmid pYM12 as template were transformed into YPH252 and selected on a G418 (200-mg/l) plate.
Construction of yARF3 Mutants
Different yARF3 mutants were constructed by amplification of the cloned yARF3 fragment with specific primers. The (T31N) mutation was introduced using a two-step recombinant PCR technique. In the first PCR, overlapping 5′- and 3′-DNA fragments were generated. Primer yARF3.1 and the 5′-T31N primer (AATATTGTGTTCTTACCAGCCTTATCCAG; mutated nucleotides are underlined) were used for amplification of the 5′ fragment (including the first 110 nucleotides of yARF3 open reading frame). The 3′ fragment that contains 477 base pairs was generated using 3′-T31N primer (CTGGTAAGAACACAATATTGTACAAACT) in combination with the yARF3.2 primer. In the second fusogenic PCR, the appropriate pairs of overlapping fragments were combined with the 5′ and 3′-end primers yARF3.1 and yARF3.2 to generate the full-length yARF3(T31N) mutant sequence. The (Q71L) mutant of yARF3 was generated in the same way except using the 5′-Q71L primer 5′-CTCAATCTTTGTAGTCCTCCTACATCCCA-3′ and the 3′-Q71L primer 5′-ATGTAGGAGGACTACAAAGATTGAGAC-3′. The (G2A) mutant was constructed by amplification of the yARF3 fragment with primer pairs 5′-CATATGGCGAATTCATTTCGAAGGTTCTG-3′ and yARF3-HA primer (5′-AGCGTAGTCTGGGACGTCGTATGGGTATTTCTTTGGAACGTTTGTGTTG-3′, hemagglutinin [HA] tags are underlined) to generate an HA epitope-tagged yARF3(G2A) mutant gene. The PCR fragments were then ligated into the pT7Blue plasmid (Novagen), propagated, sequenced, and subcloned into the pVT-101U expression plasmids, a high-copy 2-μ––based expression plasmid containing the ADH1 promoter (Vernet et al., 1987), or the pYEUra3 expression plasmid, a low-copy CEN based expression plasmid containing the GAL1 promoter (BD Biosciences Clontech, Palo Alto, CA).
Yeast Cell Extracts and Immunoblotting
Whole cell extracts were prepared by harvesting of three optical density units (A600) of cells. The yeast cell concentration was assessed by absorbance at 600 nm. Cells were suspended in radioimmune precipitation buffer (50 mM Tris-HCl, pH 8.0, 0.1% SDS, 0.5% deoxycholic acid, 150 mM NaCl, and 1% Nonidet P-40) to a final A600 of 30. Whole cell extracts were then prepared by vortexing with glass beads for 2 min at 4°C and clarified by brief centrifugation. The protein concentration was quantified with a BCA™ Protein Assay Kit (Pierce, Rockford, IL). Proteins separated by SDS-PAGE, were transferred electrophoretically to Immobilon-P membranes (Millipore Corp., Bedford, MA), which were then incubated with antibodies in Tris-buffered saline (pH 7.4) containing 0.1% Tween 20 and 5% nonfat dry milk at room temperature for 60 min. The anti-HA mAb (HA.11, Berkeley Antibody Co., Richmond, CA) and horseradish peroxidase-conjugated goat antimouse IgG + IgM (H + L) were each diluted 1:5000. Bound antibodies were detected with the ECL system (Amersham Pharmacia Biotech) according to the manufacturer's instructions. Primary and secondary antibodies and luminol substrate were removed from the blot using the blot-stripping protocol (Amersham Pharmacia Biotech).
Protein Labeling and Immunoprecipitation
For expression of myristoylated protein, 3H-labeled myristoylated Arf3p was synthesized in yeast and prepared as described by Simon and Aderem (1992). Immunoprecipitation, electrophoresis, and autoradiography were performed essentially as described previously (Lee et al., 1997) by using 10 μl of anti-Arf3p or anti-Arf1p antibodies prepared in our laboratory.
For immunoprecipitation of alkaline phosphatase (ALP), yeast preparation, 35S-protein labeling, immunoprecipitation, electrophoresis, and autoradiography were performed essentially as described previously (Huang et al., 1999), by using antivacuolar alkaline phosphatase (anti-ALP) antiserum prepared in our laboratory.
Endocytosis of Lucifer Yellow and N-(3-Triethylamminiumpropyl)-4-(p-diethylaminophenylhexatrienyl) Pyridinium Dibromide (FM 4-64)
Endocytosis of lucifer yellow (Sigma-Aldrich, St. Louis, MO) was performed as described previously (Huang et al., 1999). Endocytosis of FM 4-64; Molecular Probes, Eugene, OR) was performed as described previously (Vida and Emr, 1995). Fluorescence microscopy was performed with a Nikon Microphot SA microscope. Cells were viewed at a magnification of 1000× and exposed for immunofluorescence photographs for appropriate times.
Analysis of Ste2p and Ste3p Receptor Turnover
Early logarithmic phase (0.5–1.0 × 107 cells/ml) MATa yeast cells carrying plasmid MD53 were grown at 30°C in minimal medium (without Ura) with selection for the Ste2-HA plasmid (kindly provided by Dr. James Konopka, State University of New York, Stony Brook, NY). Cycloheximide, to inhibit translation, was added to a final concentration of 100 μg/ml, and growth continued for 10 min. After removal of 5 ml (t = 0), the cultures were divided into halves. To one half, α-factor (Sigma-Aldrich) was added to a final concentration of 1 × 10–6 M. Five A600 units were removed from the samples at the indicated times thereafter, and cells were collected by centrifugation and dispersed in 100 μl of lysis buffer (8 M urea, 100 mM Tris, and 2% SDS). Glass beads were added and extracts prepared using a vortex mixer for 5 min. The protein concentration was determined with a BCA protein assay kit (Pierce Chemical, Rockford, IL). All samples were diluted to a final protein concentration of 1 μg/μl in SDS-PAGE sample buffer and incubated at 37°C for 10 min before transfer to gels. Ste3p turnover was assayed essentially like that of Ste2p, except that MATα yeast cells were used and endogenous Ste3p was detected using purified anti-Ste3p antibody raised in our laboratory. Twelve micrograms of each sample was subjected to SDS-PAGE, followed by Western blot analysis with monoclonal anti-HA antibody (HA.11; Babco, Richmond, CA).
Subcellular Fractionation by Velocity
Cells were harvested by centrifugation from cultures (50 ml) grown in YPD medium to midexponential phase (A600 = 1). About 50 A600 units of cells were washed by repeated suspension in ice-cold 10 mM NaN3 in double-distilled H2O and centrifugation, incubated with lyticase to form spheroplasts, suspended in 0.2 ml of ice-cold lysis buffer (20 mM Tris-HCl, pH 7.2, 1 mM EDTA, 0.8 M sorbitol) containing the protease inhibitors aprotinin, leupeptin, and pepstatin (each 1 mg/ml), 1 mM benzamidine, and 1 mM phenylmethylsulfonyl fluoride, and disrupted on ice by passing through a 26-gauge needle 20 times and centrifugation with 700 × g for 7 min to remove unbroken cells and debris. For velocity centrifugation, the cleared lysate was subjected to centrifugation (13,000 × g) for 10 min at 4°C to generate the pellet fraction (P13) and the supernatant (S13). The S13 fraction was separated further into membrane (P100) and cytosol (S100) fractions by centrifugation (100,000 × g) for 1 h. Equal proportions of each fraction were subjected to SDS-PAGE and analyzed by Western blot analysis. Different organelle marker proteins were detected with anti-Pma1p (1:5000), anti-Emp47p (1:5000), anti-Arf3p (affinity-purified), anti-Kar2p (1:1000), and anti-Arf1p (1:5000) prepared in our laboratory, and antimitochondrial porin (1:500) and anti-Pgk1p (1:1500), purchased from Molecular Probes.
Extraction of Arf3 from P13 Heavy Membranes and Sucrose Density Gradient
For extraction of Arf3p from heavy membranes, P13 was suspended in buffer containing 0.25 M sucrose, 50 mM HEPES, pH 7.4, 25 mM KCl, 5 mM MgCl2 with protease inhibitors and divided into three parts as previous described (Huang et al., 2002). One part was added to 1.2 ml of the same buffer as mock control, one part into 1.2 ml 1 M NaCl with 50 mM HEPES, pH7.4, and one part to 1.2 ml 0.1M Na2CO3, pH 11, with 50 mM HEPES, pH 7.4. After 30 min of incubation on ice, samples were centrifuged at 150,000 g for 1 h at 4°C. Samples were then analyzed by Western blotting. For the P13 sucrose gradient, P13 was suspended in 800 μl buffer and layered on top of the sucrose gradient consisting of 40%, 45%, 50%, 55%, and 60% sucrose, which was then subjected to centrifugation (150,000 g for 3 h at 4°C in a Beckman SW55 rotor). Twelve fractions were collected manually from the top. Samples were precipitated with 10% trichloroacetic acid, resuspended in SDS sample buffer, separated by SDS-PAGE, and analyzed by Western blotting.
Indirect Immunofluorescence
Cells were grown in 25 ml of minimal selective medium with 2% glucose or 0.1% glucose and 2% galactose to a density of 1–2 × 107 cells/ml and prepared for indirect immunofluorescence essentially as described previously (Lee et al., 1997). Affinity-purified Arf3p antibody was used for detection of Arf3p. Anti-vacuolar ATPase 69-kDa subunit (Molecular Probes) was diluted to 1:1000. Anti-actin antibody (ICN Biomedicals, Costa Mesa, CA) was diluted to 1:400. Anti-GFP antibody (Molecular Probes) was diluted to 1:500. Alexa 488 goat anti-rabbit IgG and Alexa 594 goat anti-mouse IgG (Molecular Probes) were used as secondary antibodies at 1:1000 and 1:2000 dilutions, respectively. Nuclei were visualized by staining with H33258 (2 μg/ml), which was included in the mounting solution.
Construction of yARF1yARF3 and yARF3yARF1 Chimeras
The yARF1yARF3 chimeras were prepared using a two-step recombinant PCR technique. In the first PCR, yARF1(1-219) and yARF3(214-552) DNA fragments were generated. yARF1 5′ primer (5′-CATATGGGTTTGTTTGCCTCTAAGTT-3′), and yARF1N′ primer (5′-TTGTCCTCCTACATCCCAGACAGT-3′) were used for amplification of yARF1(1-219). yARF3(214-552) was generated using 3′-yARF3C′ primer (5′-TGGGATGTAGGAGGACAA-3′) in combination with yARF3.2. In the fusogenic PCR, the appropriate pairs of overlapping fragments were combined with the 5′-yARF1 and 3′-yARF3.2 primers to generate the full-length yARF1yARF3 chimeric sequence, which was then purified and subcloned as a HindIII-BamHI fragment into HindIII-BamHI sites of the pVT102U plasmid to yield pVT102UyF1yF3. The yARF3yARF1 chimera encoding the N-terminal 71 amino acids of Arf3p and the C termini of Arf1p was constructed by the same procedure. yARF3(1-213) was amplified with primer pairs yARF3.1 and ARF3N′ (5′-TGTCTTGTCCTCCTACATCCCA-3′), and Arf1p(196–546) was amplified with 3′-yARF1C′ primer (5′-TGGGATGTAGGAGGACAAGACAGA-3′) and antisense primer for yARF1 (5′-ATTCTAGAATTTTTAAGTTGAGTT-3′). The underlined nucleotides of yARF1N′ primer and 3′-yARF1C′ primer were modified from the original yARF1 sequence to match the yARF3 sequence.
Calcofluor White Staining of Cells
For Calcofluor White staining, cells were fixed with 3.7% formaldehyde and stained with 0.1–1 mg/ml Calcofluor White as described by Pringle et al. (1989). Cells were examined and photographed on a Nikon Microphot SA microscope at a magnification of 1000×.
Calcofluor White, used in a Calcofluor White hypersensitivity assay, is a negatively charged, fluorescent dye that can bind to nascent chains of chitin and thus prevents cocrystallization, and the formation of microfibrils, and thereby interferes with cell wall assembly. By this mechanism, Calcofluor White amplifies the effects of cell wall mutations. Yeast was replica-plated to YPD plates containing Calcofluor White (0, 0.5, 1, 1.5 mg/ml) according to Ram et al. (1994).
Latrunculin-A Treatment
For growth of cells in the presence of latrunculin-A (LAT-A), a 500-μl logarithmic culture of YPH252F3GFP was treated with either LAT-A (Calbiochem, San Diego, CA) at a concentration of 100 μMin dimethyl sulfoxide (DMSO) for 15 min at 30°C or DMSO alone (control). The cells were visualized immediately. At the same time point, an aliquot from each culture was fixed and processed for immunostaining with actin and GFP antibodies.
For cells released from G0, a culture enriched in unbudded cells was incubated with LAT-A at 100 μM as described for 4 h (Ayscough et al., 1997) and then visualized under the same conditions as for Arf3-GFP localization studies. Actin cytoskeleton disassembly was verified by immunostaining with actin antibody. The halo assay for assessing LAT-A sensitivity was performed as described previously (Ayscough et al., 1997).
RESULTS
Detection of Endogenous Arf3p in Membrane Fractions
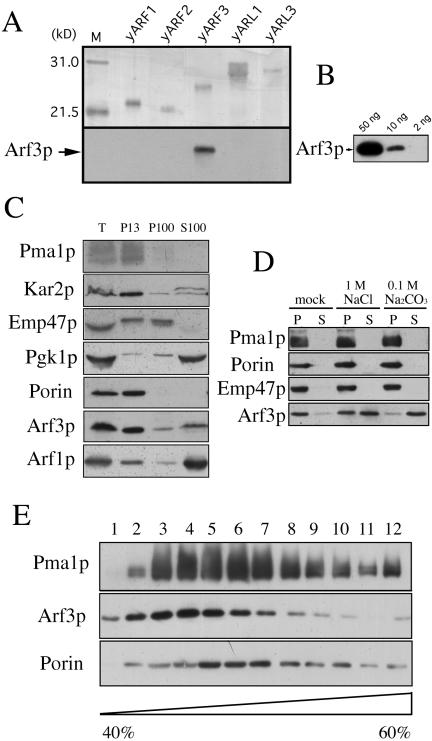
To characterize the yARF3 gene product, we prepared rabbit antiserum against an Escherichia coli-synthesized recombinant, full-length, His-tagged Arf3p fusion protein. Purified recombinant His-tagged yARFs and yARLs (∼30–40 ng) were subjected to Western blot analysis. Immunoblotting with the Arf3p antiserum allowed detection of <10 ng of the protein, whereas no signal was detected with recombinant Arf1p, Arf2p, Arl1p, and Arl3p (Figure 1, A and B). Thus, Arf3p was immunologically distinguishable from Arf1p, Arf2p, Arl1p, and Arl3p. The endogenous Arf3p was detected using affinity-purified antibody against the whole-cell lysate. In wild-type lysate, a protein of ∼20 kDa was recognized, which was not detected in the yarf3 mutant (our unpublished data). The wild-type yeast cell lysate was subjected to differential centrifugation studies. Arf3p, Arf1p, Kar2p (ER marker), Pma1p (plasma membrane marker), Emp47 (Golgi marker), porin (mitochondrial marker), and PGK (cytoplasmic marker) in each fraction were identified by Western blot analysis (Figure 1C). Although some of the endogenous Arf3p was, like Arf1p, in the cytoplasmic fraction (S100), >60% of the Arf3p was found in the heavy membrane fraction (P13), indicating that the membrane association of Arf3p is different from that of Arf1p.
Figure 1.
Detection of Arf3p in membrane fractions. (A) Specific immunoreactivity of anti-Arf3p antibodies. Samples (∼30–40 ng) of purified recombinant his-tagged ARFs and ARLs were subjected to SDS-PAGE on a 15% gel. Top, silver-stained gel; bottom, proteins transferred to polyvinylidene difluoride membranes and immunoblotted with anti-Arf3p antibody, followed by detection using the enhanced chemiluminescence system. (B) Sensitivity of anti-Arf3p antibodies. Fifty, 10, and 2 ng of purified His-tagged recombinant Arf3p protein were subjected to Western blot analysis. (C) Subcellular localization of endogenous Arf3p. Yeast YPH 252 cells were grown in YPD medium. Lysates of spheroplasts from cells were subjected to velocity sedimentation and were separated into P15, P100, and S100 fractions as described in MATERIALS AND METHODS. Proteins in samples of fractions were precipitated, separated by SDS-PAGE, and analyzed by immunoblotting. Arf3p, Arf1p, Emp47p (Golgi marker), porin (mitochondrial marker), Kar2p (an ER marker), and Pma1p (plasma membrane marker) were identified with specific antibodies and detected using the enhanced chemiluminescence system with exposure to Hyper-film MP. Gradient fractions are numbered from the top. (D) Arf3p is weakly associated with the membrane fraction. The heavy membrane-enriched fraction (P13) from wild-type yeast was treated with 1M NaCl, 0.1M Na2CO3, or buffer (mock) on ice for 30 min, followed by centrifugation at 150,000 × g for 1 h. The resulting supernatant and pellet fractions were analyzed by Western blotting by using anti-Arf3p, anti-Pma1, anti-Emp47p, or anti-porin antibodies. (E) P13 was suspended and layered on top of the sucrose gradient (40–60%), which was then subjected to centrifugation. Twelve fractions were collected from the top and samples were precipitated, resuspended in SDS sample buffer, separated by SDS-PAGE, and analyzed by Western blot analysis.
To evaluate stability of the association of Arf3p with membranes, the heavy membrane-enriched fraction (P13) from velocity sedimentation was extracted with 1 M NaCl or with 0.1 M Na2CO3, which solubilizes peripheral, but not integral, membrane proteins (Fujiki et al., 1982). Arf3p, like a peripheral membrane protein, was solubilized by 1 M NaCl or Na2CO3 (pH 11), whereas porin (a mitochondrial outer membrane integral protein), Emp47p (a Golgi membrane integral protein), or Pma1p (a plasma membrane integral protein), remained membrane-associated (Figure 1D).
The N Terminus of Arf3p Is Myristoylated
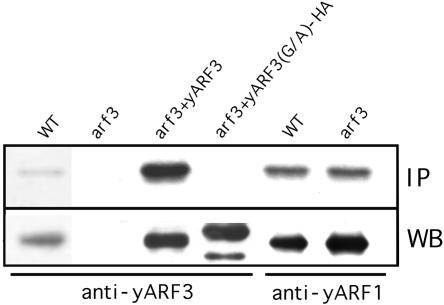
Small GTPases often have lipid modifications that are believed to be important for membrane anchoring. Many members of the ARF family have an amino-terminal myristate, but some do not (Moss and Vaughan, 1995). The amino acid sequence of Arf3p contains a glycine in the second position, and thus Arf3p may also be myristoylated. To determine whether endogenous Arf3p is myristoylated, yeast cells were incubated with cerulenin to inhibit de novo fatty acid biosynthesis (Haun et al., 1993) and were grown in medium containing [3H]myristic acid. All lysates were immunoprecipitated with Arf1p- or Arf3p-specific antibodies. As shown in Figure 2, Arf3p and Arf1p, but not the Arf3p(G2A) mutant, were myristoylated in vivo.
Figure 2.
N-Terminal myristoylation of ARF3. YPH252 yeast wild-type, arf3 mutant, arf3 mutant overexpressing wild-type, or HA-tagged G2A mutant of Arf3p in high-copy (2 μ) pVT101U expression constructs were grown in medium containing [3H]myristic acid. Cell lysates were immunoprecipitated with purified anti-Arf3p or anti-Arf1p antibodies as indicated, and myristate-labeled proteins were detected by fluorography after gel electrophoresis (top). A portion of the same preparations was subjected to Western blot analysis for estimating the quantity of protein precipitated (bottom).
Arf3p Is Not Involved in Known Pathways of Vesicular Trafficking to the Vacuole
To evaluate the possible roles of Arf3p in vesicular trafficking, we examined several distinct exocytotic and endocytotic transport pathways for an Arf3p requirement. We had reported (Lee et al., 1994) that the transport of carboxypeptidase Y (CPY), a soluble vacuolar hydrolase, toward the vacuole was not impaired in an arf3 mutant. Cowles et al. (1997) reported that transport of ALP to the vacuole is different from that of CPY. We therefore investigated whether Arf3p is required for ALP trafficking. Pulse-chase labeling with 35S-labeled cysteine and methionine of wild-type, arf3, and arf1 mutant cells was followed by immunoprecipitation of ALP. Unlike the arf1 mutant, the arf3 mutant had no impairment in processing of ALP (see Supplementary Data), and arf3 cells converted ALP to its mature form with a time course similar to that of wild-type cells. Thus, Arf3p may not be involved in the biosynthetic transport pathways to the vacuole.
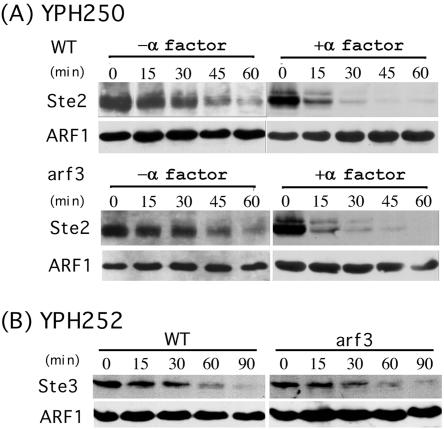
Next, we examined whether Arf3p was involved in the endocytic pathway. To assess fluid-phase endocytosis, the uptake of the dye lucifer yellow was followed. In wild-type cells, this dye can be seen to accumulate in the vacuoles after its internalization. In the arf3 mutant, no apparent effect on lucifer yellow uptake was observed (see Supplementary Data). To determine whether there were defects in membrane internalization trafficking in the endocytosis, we monitored the uptake of a lipophylic dye, FM 4-64 (Vida and Emr, 1995). Wild-type and arf3 cells were incubated on ice for 30 min and then shifted to 30°C to initiate endocytosis of the dye. No significant differences in the uptake or transport of FM 4-64 between wild-type and arf3 mutant cells were observed at 30°C, 15°C, or 37°C (see Supplementary Data). The morphology of the vacuole also seemed to be normal in arf3 mutant cells. To evaluate the constitutive endocytosis and membrane protein turnover, we measured the degradation of α-factor receptor (Ste2p) and a-factor receptor (Ste3p) in the absence of pheromone stimulation. YPH250 (MATa) wild-type and arf3 cells were transformed with plasmid MD53, which contains Ste2-HA. Mid log-phase cultures of cells overexpressing Ste2-HA, YPH252 (MATα) wild-type, or arf3 cells were incubated with cycloheximide for 10 min to block protein synthesis, and cells were collected at different time points. The collected cells were washed with NaN3 to halt membrane trafficking. The degradation of Ste2-HA and Ste3p was evaluated by Western blot analysis. Wild-type and arf3 mutant cells degraded Ste2p and Ste3p at similar rates (Figure 3A, left, and B), indicating that the constitutive membrane protein endocytosis and endosome trafficking toward the vacuole were unaffected in arf3 mutant cells.
Figure 3.
Turnover of Ste2p and Ste3p in arf3 mutant. (A) Cycloheximide (10 μg/ml) was added to mid log cultures of YPH250 wild-type or arf3 mutant cells overexpressing HA-tagged Ste2 and incubated for 10 min at 30°C before addition of α-factor at t = 0. At the indicated times thereafter, samples were removed and cell extracts were subjected to Western blot analysis. Polyclonal anti-HA antibody was used to detect HA-Ste2. (B) Cycloheximide (10 μg/ml) was added to mid log cultures of YPH252 wild-type or arf3 mutant cells. Samples were removed at the indicated times, and cell extracts were subjected to Western blot analysis. Anti-Ste3 antibody was used to detect Ste3p.
Receptor-mediated endocytosis differs from constitutive endocytosis and might use different cellular factors (Davis et al., 1993). To test the possibility that Arf3p is involved in this regulated process, α-factor was added to MATa wild-type or arf3 mutant cells overexpressing HA-epitope-tagged Ste2 after 10 min of incubation with cycloheximide. In the presence of α-factor, the turnover rate of Ste2-HA in both the wild-type and arf3 mutant was increased. The rate of receptor degradation in the wild-type and arf3 mutant remained indistinguishable (Figure 3A, right). Therefore, Arf3p was also not required for receptor-mediated endocytosis. Moreover, mating between a and α type arf3 mutant cells and subsequent sporulation were normal, indicating that ARF3 is not required for the mating and sporulation processes (our unpublished data).
Subcellular Localiztion of GFP-tagged Endogenous Arf3p
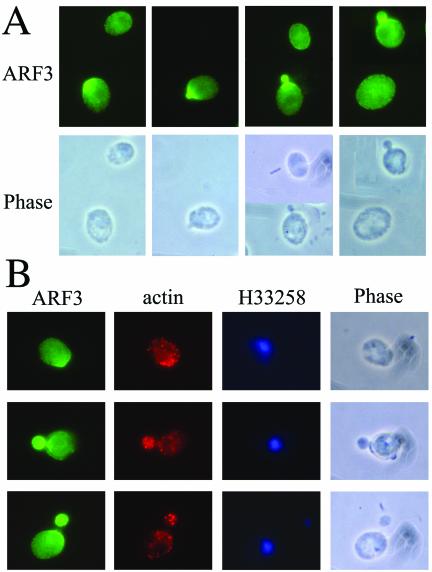
The intracellular localization of Arf3p was examined by immunofluorescence microscopy. Affinity-purified anti-Arf3p antibodies failed to detect Arf3p in wild-type cells, suggesting that endogenous Arf3p was expressed below the limits of detection by this method. We reasoned that GFP tagging of endogenous Arf3p might stabilize Arf3p in cells and enhance its detection. The coding sequence of GFP was precisely inserted in the chromosome immediately upstream of the translational stop codon for ARF3, as described in MATERIALS AND METHODS. Arf3-GFP, expressed from the endogenous ARF3 promoter, is fully functional, because it can support normal bud formation (see below) as well as wild-type Arf3p. Interestingly, Arf3-GFP exhibited a small, fine punctate fluorescence pattern at the cell periphery and also an evenly distributed cytoplasmic staining (Figure 4A). Cells in early phases of the cell cycle displayed concentrated and bright staining of Arf3-GFP at presumptive budding sites and within the tiny buds, suggesting that the localization of Arf3-GFP might be cell cycle dependent. Next, we used indirect immunofluorescence to determine whether Arf3-GFP and actin cortical patches colocalize. Figure 4B shows that Arf3-GFP in fixed cells is located at presumptive budding sites and within the buds, where the actin cortical cytoskeleton is also present. However, apparently there are also actin patches that do not seem to contain Arf3-GFP, and vice versa. We conclude that Arf3-GFP is present during the whole cell cycle and is concentrated toward the incipient bud site during polarity growth. The changes in Arf3-GFP localization occurred at the same points in the cell cycle as did the changes in actin localization, but the two proteins did not seem to colocalize.
Figure 4.
Subcellular localization of Arf3-GFP. (A) Arf3p-GFP concentrated to polarized regions. Arf3 was tagged with a GFP as described in MATERIALS AND METHODS. Cells were fixed and stained with polyclonal anti-GFP antibody. (B) Localization of Arf3-GFP and actin. Cells with Arf3-GFP were fixed and stained with monoclonal anti-actin and polyclonal anti-GFP antibodies. Nucleic acids were stained with H33258.
Cell Peripheral Localization of Arf3p Is Nucleotide Dependent
To investigate whether localization of Arf3p is nucleotide- or N-terminal myristoylation dependent, we constructed expression of nonmyristoylated Arf3p(G2A), GTP binding-defective Arf3p(T31N), and GTP hydrolysis-deficient Arf3p(Q71L) mutants in high-copy (2 μ) pVT101U or low-copy (CEN) pYEUra3 expression vectors as described in MATERIALS AND METHODS. The Arf3p(G2A) had an HA epitope tag at the carboxy terminus to facilitate detection of the recombinant protein. Wild-type Arf3p, Arf3p(G2A), Arf3p(T31N), and Arf3p(Q71L) were expressed in wild-type and arf3 mutant strains, and their expression was confirmed by Western blotting by using anti-Arf3p antibody. Overexpressed Arf3p was myristoylated in cells similar to the endogenous protein (Figure 2). The G2A mutant was immunoprecipitated by anti-Arf3p antibody, but, as expected, was not myristoylated (Figure 2).
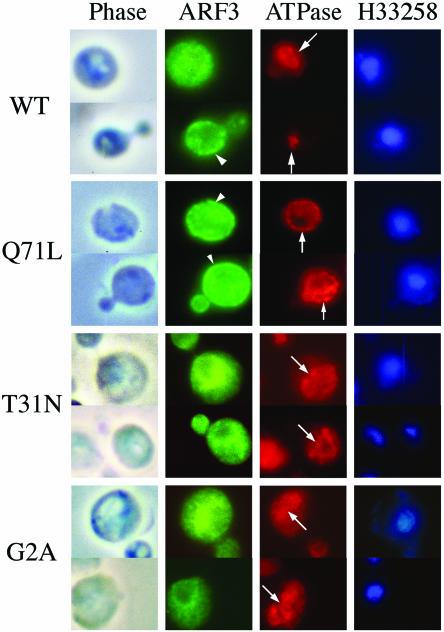
We first examined the intracellular localization of overexpressed Arf3p and its mutants, which were expressed from the high-copy (2 μ) pVT101U expression vector, by immunofluorescence microscopy (Figure 5). Vacuolar ATPase (v-ATPase), a protein associated with the vacuolar membrane, was used as control. Overexpressed Arf3p exhibited a small punctate fluorescence pattern at the cell periphery (Figure 4, arrowhead) or an evenly distributed cytoplasmic staining, although hardly any overexpressed Arf3p was detected in newly forming buds. Arf3p(Q71L) was concentrated in the periphery of the cell, suggestive of plasma membrane localization; however, the GTP-binding-defective Arf3p(T31N) was predominantly localized to an intracellular compartment(s) between the vacuole and perinuclear regions. Results from a sucrose gradient of the P13 heavy membrane-enriched fraction also indicated that the distribution of most of the endogenous Arf3p was similar to that of Pma1p, but not porin (Figure 1E). Furthermore, unmyristoylated Arf3p(G2A), similar to Arf3p(T31N), was also predominantly localized in an unidentified intracellular compartment(s) between the nucleus and the vacuole.
Figure 5.
Subcellular localization of high-expression of Arf3p and its mutants. Indirect immunofluorescence staining of arf3 cells overexpressing wild-type ARF3, ARF3(G2A), ARF3(T31N), or ARF3(Q71L) from high-copy (2 μ) pVTU expression vectors were fixed with formaldehyde, spheroplasted, and treated with affinity-purified anti-Arf3p antibody and monoclonal anti-v-ATPase 69-kDa subunit antibody. Nucleic acids were stained with H33258. Cells were viewed by phase contrast microscopy. Arrows indicate vacuole localization and arrowheads, the plasma membrane.
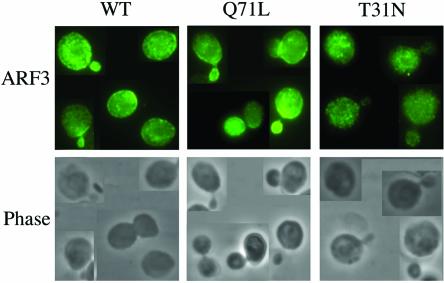
The intracellular localization of Arf3p and its mutants, expressed from the low-copy (CEN) pYEUra3 expression vector, was also examined (Figure 6). Interestingly, similar to Arf3-GFP, the localization of low-level expression of Arf3p wild-type and Arf3p(Q71L) to the cell periphery was cell-cycle dependent, and both were detected in newly forming buds. However, Arf3p(T31N) exhibited predominantly an un-evenly distributed cytoplasmic staining and did not concentrate toward the incipient bud site during polarity growth. The expression of unmyristoylated Arf3(G2A) was too low to be detected under the low-copy (CEN) pYEUra3 expression vector. Our results indicated that the distributions of the GDP-bound and GTP-bound forms of Arf3p were distinct from each other and resembled, in part, those of mammalian ARF6.
Figure 6.
Subcellular localization of low-expression of Arf3p and its mutants. Cells were grown with galactose containing SD medium at 30°C. Indirect immunofluorescence staining of arf3 cells expressing wild-type ARF3, ARF3(T31N), or ARF3(Q71L) from low-copy (CEN) pYEUra3 expression vectors were fixed with formaldehyde, spheroplasted, and treated with affinity-purified anti-Arf3p antibody. Cells were viewed by phase contrast microscopy.
N-Terminal Region of Arf3p Has the Signal for Cell Periphery Localization
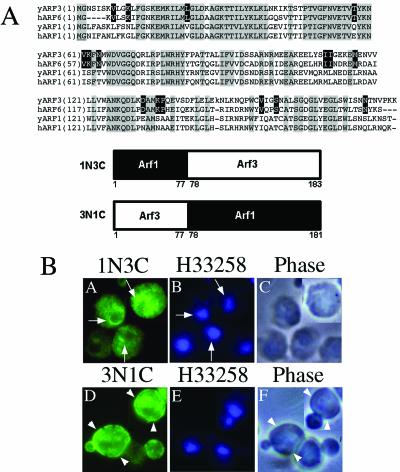
Arf3p shares 54% identity and 75% similarity of amino acid sequences with Arf1p (Figure 7A); however, their cellular localizations are totally different. Yeast Arf1p is detected predominantly in the cytosol, whereas most of the Arf3p is associated with different membrane fractions, depending on the nucleotide bound. To identify the region of the molecule that specifies Arf3p localization, we made chimeras by exchanging the amino- and carboxy-terminal halves of Arf1p and Arf3p (Figure 7A, bottom). The first chimera, ARF1N3C, consisted of residues 1–77 of Arf1p and residues 78–183 of Arf3p. The second, ARF3N1C, comprised residues 1–77 of Arf3p and residues 78–181 of Arf1p. Our anti-ARF3 antibody reacted with the ARF1N3C chimera, but not with the ARF1N3C chimera, and the ARF1N3C chimera was recognized by the anti-ARF1 antibody (our unpublished data). Therefore, the ARF1N3C chimera was overexpressed in arf3 mutant cells and ARF3N1C in the isogenic arf1 mutant cells. Expression of ARF1N3C was detected with the anti-ARF3 antibody and that of ARF3N1C with anti-Arf1p-specific antibody. Immunofluorescence staining revealed that the two chimeras had different localizations. The ARF1N3C chimera was found in a punctuate staining pattern concentrated in the perinuclear regions, whereas the ARF3N1C chimera exhibited a cell periphery localization (Figure 7B). These results indicated that the overall amino acid sequence of Arf3p might prefer a membrane association, and the signal for cell periphery localization should be within the N-terminal 77 amino acids.
Figure 7.
Subcellular localization of Arf1p/Arf3p chimeric proteins. (A) Top, alignment of amino acid sequences of yeast ARF1, yeast ARF3, human ARF1, and human ARF6. The alignment was performed using GeneBee-Net version.2.0. Amino acids identical in all sequences are highlighted with gray blocks, and black background denotes identity in yeast ARF3 and human ARF6. Bottom, construction of the Arf1/Arf3 chimeric proteins. (B) For indirect immunofluorescence staining, samples of the cells were prepared and photographed as described in MATERIALS AND METHODS. Affinity-purified anti-Arf3p antibody (A) or anti-ARF1 antibody (D) were used to detect Arf(3N1C)p or Arf(1N3C)p, respectively. Nucleic acids were stained with H33258, which was included in the mounting solution (B and E). Cells were viewed by phase contrast microscopy (C and F). Arrows indicate perinuclear regions and arrowheads, the plasma membrane.
Arf3-GFP Localization Is Actin Independent
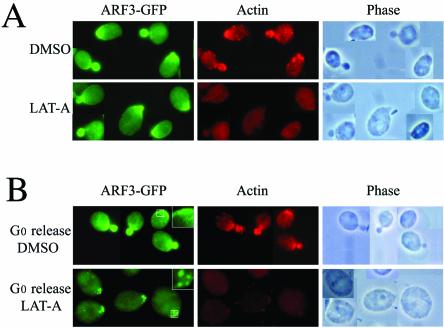
Arf3-GFP exhibits a cell-cycle-dependent pattern of localization, which closely resembles that of actin polymerization. To explore the codependence of the subcellular distribution of Act1p and Arf3p, we used a pharmacological tool, LAT-A, which recently has been shown to allow the temporal dissection of events involved in bud morphogenesis. LAT-A, a macrolide isolated from a marine sponge, is a potent inhibitor of actin cytoskeleton assembly. We examined the consequences of disrupting the actin cytoskeleton with LAT-A on Arf3-GFP localization. Cultures of the yeast strain YPH252F3GFP containing Arf3-GFP under the control of the endogenous ARF3 promoter were grown for 15 min in the presence of LAT-A or the DMSO diluent alone and visualized immediately by microscopy. At the same time point, an aliquot was withdrawn from each culture, fixed, and processed for immunostaining with actin and GFP antibodies (Figure 8A). Whereas the actin cytoskeleton of LAT-A treated cells is virtually absent, the localization of Arf3-GFP is unaffected. Thus, maintenance of the intracellular polarized location of Arf3-GFP is not dependent upon the integrity of the actin cytoskeleton.
Figure 8.
Localization of Arf3p-GFP in the presence or absence of LAT-A. Yeast (YPH252F3GFP) with GFP-tagged Arf3p was grown to logarithmic phase, and then divided into two parts: one was treated with DMSO as control and the other was treated with LAT-A dissolved in DMSO for 15 min. (A) Top, DMSO alone did not affect localization of actin patch or ARF3p-GFP. (A) Bottom, LAT-A treatment disassembled the actin cytoskeleton but did not affect Arf3p-GFP polarization. (B) Localization of Arf3-GFP in the presence (bottom) or absence (top) of LAT-A (4 h) in cells exiting G0. Arf3-GFP was still polarized to the presumptive budding site, although F-actin was absent. The areas indicated in the panels were enlarged and are shown in the upper right corners.
To determine whether Arf3-GFP requires the actin cytoskeleton for localization, cultures enriched in unbudded cells (Ayscough et al., 1997) were grown for 4 h in LAT-A and then visualized by microscopy. Under these conditions, the actin cytoskeleton was disassembled; in contrast, the majority of the cells had Arf3-GFP localized at the presumptive budding site (Figure 8B), although Arf3-GFP seemed to be in a larger punctate pattern. This result indicates that Arf3p achieves its location during polarity growth through an actin-independent pathway.
Arf3p Is Not Directly Involved in Actin Cytoskeleton Organization
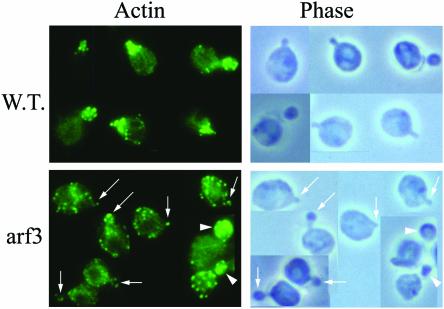
To examine whether Arf3p can affect the actin structure during polarity development, we grew the wild-type and arf3 mutant cells at different temperatures. At 30°C, the wild-type and the arf3 mutant had normal morphology and actin polarization; actin cables were seen, and actin patches were found in the bud (our unpublished data). Interestingly, at a higher temperature (34°C), the arf3 mutant, but not wild-type cells, showed delayed or less actin patch polarization when cells underwent polarizing budding (Figure 9, arrows). The observation that the arf3 elicited actin structural changes only at a higher temperature (34°C) implies that Arf3p does not directly affect the actin structure.
Figure 9.
Arf3 cells are partially defective for early actin patch polarization at 34°C. Wild-type and arf3 cells were grown at 34°C overnight, and then fixed and stained for actin. Top, growth at higher temperature (34°C) did not depolarize actin patch in wild-type yeast; in contrast, with the arf3 mutant, most of the cells showed delayed or incomplete actin patch polarization.
Sensitivity to LAT-A varies among actin cytoskeleton-associated mutants and can be used as an indicator of the effect that the respective proteins have on the stability of the actin cytoskeleton (Ayscough et al., 1997). To test the sensitivity of the arf3 mutant strain to LAT-A, we used a halo assay at different temperatures (30°C, 34°C, and 37°C). The arf3 mutant showed only slightly more sensitivity to LAT-A than did the wild-type stain at higher temperatures (34 and 37°C), indicating that Arf3p may not be directly involved in actin cytoskeleton stability (our unpublished data).
Arf3p Is Involved in Yeast Bud Site Selection
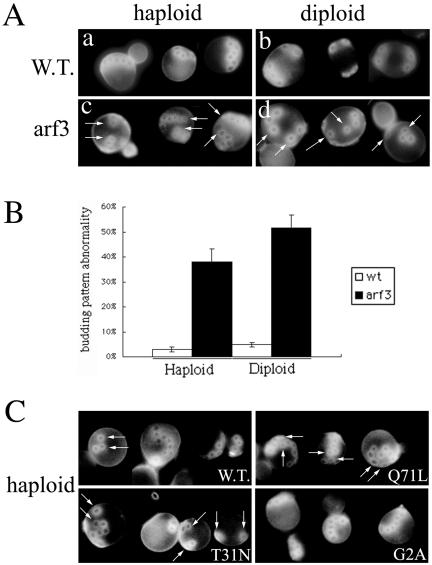
The localization of Arf3p at presumptive budding sites and within the buds suggests that Arf3p may be required for the yeast budding process. The bud site on the mother cell is selected according to haploid- or diploid-specific programs. Haploid cells typically bud in an axial pattern (Figure 10A, a), in which the mother and daughter cells form their new buds directly adjacent to the immediately preceding division site. Diploid cells typically bud in a bipolar pattern (Figure 10A, b), either adjacent to the previous bud site or at the opposite pole of the cell. Calcofluor white staining of chitin in bud scars was used to detect proper yeast budding (Pringle et al., 1989). These patterns can be identified by visualization of the chitinous bud scars on the mother cells. Examining budding patterns in haploid and homologous diploid arf3 mutants, we found that both haploid and diploid arf3 mutants showed abnormal budding patterns (Figure 10A, c and d, arrows). Instead of forming a chain of bud scars, ∼40% of haploid arf3 mutants presented more than three random bud scars, which were not adjacent to their previous bud scars and lost their axial pattern (Figure 10B). In homologous diploid arf3 cells, >50% of diploid arf3 mutants showed more than three randomized bud scars. These findings suggested that Arf3p is involved in proper polarity development.
Figure 10.
Arf3p is involved in bud-site selection. (A) arf3 mutants exhibit abnormal bud pattern. Haploid or diploid wild-type (a and b) and haploid or homologous diploid arf3 mutant (c and d) yeast were fixed and stained with Calcofluor White to visualize bud scars. a and c, haploid cells; b and d, diploid cells. (B) Percentage of abnormal budding pattern in arf3 was also assessed and is shown herein graphically. Haploid or diploid wild-type (white bar) and arf3 (black bars) yeasts were quantified with abnormal budding pattern from three independent experiments. In each experiment, 100 cells with more than three bud scars were counted and analyzed. Error bars represent the range of triplicate determinations. (C) Effects of overexpressing Arf3p and its mutants in budding site selection. Wild-type haploid yeasts overexpressing wild-type ARF3, ARF3(Q71L), ARF3(T31N), or ARF3(G2A) were fixed and stained with Calcofluor White. The abnormal bud sites are indicated by arrows.
We further examined whether overexpression of Arf3p or its mutants could affect bud-site selection. Wild-type yeasts overexpressing Arf3p(Q71L) or Arf3p(T31N) showed a partially abnormal budding pattern (Figure 10C, arrows). In contrast, overexpression of Arf3p or Arf3p(G2A) did not produce an obvious phenotype, although some Arf3p overexpressing cells showed a disconnected chain of bud scars.
To determine whether Arf3p is required for cell wall biosynthesis, we performed Calcofluor White hypersensitivity assays as described in MATERIALS AND METHODS. Both haploid and diploid arf3 mutants did not show more sensitivity to Calcofluor White than did wild-type cells, suggesting that Arf3p may not be involved in cell wall biosynthesis.
DISCUSSION
The mammalian ARF6 GTPase has been implicated to regulate endocytosis, endosome recycling, exocytosis, and cytoskeletal organization. In this study, we showed that although similar in cellular distribution, yeast Arf3p might have different functions from its mammalian homolog ARF6. We showed that Arf3p is not essential for yeast endocytosis and does not play a major role in actin remodeling, but rather that it is important for cell polarity development.
In contrast to the essentially complete cytosolic localization of Arf1p and Arf2p, >50% of Arf3p was recovered in membrane fractions. Overexpressed wild-type human ARF6 was reported to be concentrated at the plasma membrane and to be associated with an endosomal compartment, depending on the nucleotide bound (D'Souza-Schorey et al., 1995; Peters et al., 1995). Our results showed that Arf3p(Q71L), like the activated ARF6(Q67L), was found predominantly in the cell periphery, whereas Arf3p(T31N) occurred in an unidentified intracellular compartment(s) adjacent to the perinuclear region, but not to the vacuole. The localization of Arf3p in internal structures as well as in the cell periphery suggests that it may be cycling between the endocytic and exocytic pathways in a manner similar, but not identical, to mammalian ARF6.
The myristoylation of ARFs was believed to be important for their functions and to be required for membrane anchoring. We showed that Arf3p, like other ARFs, is myristoylated at the glycine residue adjacent to the N-terminal methionine. Arf3p(G2A), a myristoylation-deficient mutant, was, in part, localized to distinct intracellular compartments juxtaposed to the perinuclear regions. Our study of the localization of Arf1p/Arf3p chimeras revealed that the amino acid sequence determining Arf3p association with the cell periphery resided in its N-terminal 77 amino acids. Thus, it occurs that the N-terminal domain, N-terminal myristoylation, and identity of the bound nucleotide were all important for the specific intracellular localization of Arf3p.
We speculate that Arf3p, like ARF6, may have a role(s) in exocytosis or endocytosis. However, study of ALP and CPY trafficking (Lee et al., 1994) clearly indicated that Arf3p is not involved in known pathways of vesicular trafficking from the ER and Golgi complex to the vacuole. In addition, the fluid phase, membrane internalization, and mating-type receptor-mediated endocytosis were not impaired in the arf3 mutant, suggesting that Arf3p is not essential for the known endocytosis pathway. Although ARF6 has been implicated in modulating the cortical actin cytoskeleton, our data suggest that Arf3p does not directly affect the actin structure. First, the arf3 mutant is only partially defective for actin patch polarization at temperatures above 34°C. Second, the arf3 mutant was only slightly more sensitive to LAT-A than the wild-type strain, only at temperatures above 34°C. This result is consistent with a recent report showing that ARF6 is probably not involved in the partial depolymerization of the cortical actin cytoskeleton that enables recruitment of the reserve pool of secretory granules to the plasma membrane (Vitale et al., 2002). Although we favor the notion that Arf3p has no direct effect on actin cytoskeleton stability, because the arf3 mutant only partially affected actin structural changes at higher temperatures, the possibility that Arf3p mediates some other subtle modifications of the actin cytoskeleton cannot be excluded. Whether there is an unknown temperature-sensitive protein that can coordinate with Arf3p in synergistic effects on actin cytoskeleton stability remains to be determined.
Cells in early phases of the cell cycle displayed bright concentration of Arf3-GFP at presumptive budding sites and within the tiny buds, suggesting that the localization of Arf3-GFP might be cell-cycle dependent. Similar to Arf3-GFP, wild-type Arf3p and Arf3p(Q71L) expressed at low level also localized in the cell periphery and were detected in newly forming buds. It is known that Bud1/Rsr1 GTPase, which helps to direct the organization of the actin cytoskeleton to the growth site (Kang et al., 2001), is present over the entire cell surface not just at the incipient bud site (Michelitch and Chant, 1996; Park et al., 1997). In contrast, Bud2p and Bud5p, which are the Bud1p GTPase-activating protein and the GTP/GDP exchange factor, respectively, localize at incipient bud sites (Park et al., 1999; Kang et al., 2001), raising the possibility that local activation of Bud1p regulators is responsible for deposition of the Bud1-associated complexes at that site. Whether Arf3p can interact with its regulators or possibly with lipid at polarized sites and execute its function needs further investigation.
The changes in the cellular distribution of Arf3-GFP during the cell cycle are very similar to the pattern of changes that the actin cortical patches undergo during the cell cycle, but the two proteins did not seem to colocalize. Arf3p localization was unaffected when the inhibitor of actin assembly LAT-A caused disassembly of the actin cytoskeleton, suggesting that Arf3p does not require actin to achieve or maintain its polarized localization. Other proteins involved in cell polarity development, including Myo2p, calmodulin, and Bud6/Aip3p, also achieve polarized localization by an actin-independent pathway (Ayscough et al., 1997). In addition, the proteins Cdc42p and Bem1p establish polarity even though the actin cytoskeleton is totally disassembled (Ayscough et al., 1997). Thus, it seems that yeast cells have evolved an actin-independent pathway for positioning of polarity development proteins.
When cells were released from G0 (unpolarized) and treated with LAT-A, the actin cytoskeleton became disassembled and the majority of the cells had Arf3-GFP at the prebud sites. Interestingly, in yeast treated with LAT-A, Arf3-GFP occurred in larger granules, whereas in those treated with DMSO (control) there was a fine punctate pattern with increasing concentration toward the prebud sites. These data indicate that Arf3p achieves its location during polarity growth through an actin-independent pathway. In addition, the transition of Arf3-GFP from a larger granule to a fine punctate distribution in a gradient toward the polarized sites may depend on the local actin cytoskeleton rearrangement or other unidentified processes that can be affected by LAT-A. Whether Arf3p, like ARF6, can direct secretory granules into a small release-ready granule pool and a larger pool, i.e., regulate the transit of granules to the plasma membrane, needs to be determined.
Polarized localization of ARF3-GFP during the early cell cycle led us to suspect a role for Arf3p in polarity development. We showed that both haploid and homologous diploid arf3 mutants had abnormalities in bud site selection. Wild-type yeast overexpressing Arf3p(Q71L) or Arf3p(T31N) also exhibited a partially abnormal budding pattern. These dominant negative phenotypes might result from interference with the function of Arf3p or from depletion of essential growth factors. Polarized growth involves a hierarchy of events such as selection of the bud-site, polarization of the cytoskeleton to the selected site, and transport of secretory vesicles containing components required for bud growth. Ni and Snyder (2001) reported that in diploid yeasts, many classes of genes, including those involved in actin-cytoskeleton organization, cell polarity, vesicular transport, and cell wall synthesis, participated in bud-site selection. Although the importance of Arf3p in bud site selection was not described in their study, it is possible that these differences may be due to the use of a different yeast strain with different phenotype penetrance with differences in the capacity of other molecules to compensate for the lack of Arf3p function. Whether Arf3p, like Rsr1p/bud1p, can constitute a GTPase signaling module that may help direct bud formation components to the selected cell division site needs to be investigated further (Park et al., 1997, 1999).
An emerging theme in the function of Arf family members is regulation of the membrane lipid composition. However, there are still many gaps in our understanding of the specific role(s) of individual ARF proteins in membrane trafficking. The precise role of Arf3p in membrane trafficking remains to be elucidated. It is possible that Arf3p, like ARF6, serves to regulate the outward flow of membrane, thereby controlling the shape and structural organization of the plasma membrane. Our results provide further evidence of the complexity of the physiological function(s) of Arf3p in polarity development. It is important to define better the mechanisms by which Arf3p exerts its function(s) in helping to define the site of the polarizing growth of the emerging bud and/or an unidentified membrane trafficking pathway.
Supplementary Material
Acknowledgments
We thank Dr. James Konopka for the Ste2-HA plasmid. We thank Drs. Randy Haun, Martha Vaughan, and Joel Moss for critical review of this manuscript. This work was supported by grants from the National Science Council, Republic of China (NSC-90-2320-B-002-003), Program for Promoting Academic Excellence of University EDU-91-FA01-1-4, and the Yung-Shin Biomedical Research Funds (YSP-86-019) to F.-J.S.L.
Article published online ahead of print. Mol. Biol. Cell 10.1091/mbc.E03–01–0013. Article and publication date are available at www.molbiolcell.org/cgi/doi/10.1091/mbc.E03-01-0013.
Abbreviations used: ARF, ADP-ribosylation factor; ER, endoplasmic reticulum; PCR, polymerase chain reaction.
Online version of this article contains supplementary data for some figures. Online version is available at www.molbiolcell.org.
References
- Altschuler, Y., Liu, S., Katz, L., Tang, K., Hardy, S., Brodsky, F., Apodaca, G., and Mostov, K. (1999). ADP-ribosylation factor 6 and endocytosis at the apical surface of Madin-Darby canine kidney cells. J. Cell Biol. 147, 7–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayscough, K.R., Stryker, J., Pokala, N., Sanders, M., Crews, P., and Drubin. D.G. (1997). High rates of actin filament turnover in budding yeast and roles for actin in establishment and maintenance of cell polarity revealed using the actin inhibitor latrunculin-A. J. Cell Biol. 137, 399–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chardin, P., Paris, S., Antonny, B., Robineau, S., Beraud-Dufour, S., Jackson, C.L., and Chabre, M. (1996). A human exchange factor for ARF contains Sec7- and pleckstrin-homology domains. Nature 384, 481–484. [DOI] [PubMed] [Google Scholar]
- Cowles, C.R., Snyder, W.B., Burd, C.G., and Emr, S.D. (1997). Novel Golgi to vacuole delivery pathway in yeast: identification of a sorting determinant and required transport component. EMBO J. 16, 2769–2782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cukierman, E., Huber, I., Rotman, M., and Cassel, D. (1995). The ARF1 GTPase-activating protein: zinc finger motif and Golgi complex localization. Science 270, 1999–2002. [DOI] [PubMed] [Google Scholar]
- D'Souza-Schorey, C., Li, G., Colombo, M.I., and Stahl, P.D. (1995). A regulatory role for ARF6 in receptor-mediated endocytosis. Science 267, 1175–1178. [DOI] [PubMed] [Google Scholar]
- Davis, N.G., Horecka, J.L., and Sprague, G.F. (1993). Cis- and transacting functions required for endocytosis of the yeast pheromone receptors. J. Cell Biol. 122, 53–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding, M., Vitale, N., Tsai, S.C., Adamik, R., Moss, J., and Vaughan, M. (1996). Characterization of a GTPase-activating protein that stimulates GTP hydrolysis by both ADP-ribosylation factor (ARF) and ARF-like proteins. J. Biol. Chem. 271, 24005–24009. [DOI] [PubMed] [Google Scholar]
- Donaldson, J.G., Cassel, D., Kahn, R.A., and Klausner, R.D. (1992). ADP-ribosylation factor, a small GTP-binding protein, is required for binding of the coatomer protein β-COP to Golgi membranes. Proc. Natl. Acad. Sci. USA 89, 6408–6412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujiki, Y., Hubbard, A.L., Fowler, S., and Lazarow, P.B. (1982). Isolation of intracellular membranes by means of sodium carbonate treatment: application to endoplasmic reticulum. J. Cell Biol. 93, 97–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harlow, E., and Lane, D. (1988). Antibodies: A Laboratory Manual, Cold Spring Harbor, NY: Cold Spring Harbor Laboratory.
- Haun, R.S., Tsai, S.C., Adamik, R., Moss, J., and Vaughan, M. (1993). Effect of myristoylation on GTP-dependent binding of ADP-ribosylation factor to Golgi. J. Biol. Chem. 268, 7064–7068. [PubMed] [Google Scholar]
- Huang, C.F., Buu, L.M., Yu, W.L., and Lee, F.J. (1999). Characterization of a novel ADP-ribosylation factor-like protein (yARL3) in Saccharomyces cerevisiae. J. Biol. Chem. 274, 3819–3827. [DOI] [PubMed] [Google Scholar]
- Huang, C.F., Chen, C.C., Tung, L., Buu, L.M., and Lee, F.J. (2002). The yeast ADP-ribosylation factor GAP, Gcs1p, is involved in maintenance of mitochondrial morphology. J. Cell Sci. 115, 275–282. [DOI] [PubMed] [Google Scholar]
- Ito, H., Fukuda, Y., Murata, K., and Kimura, A. (1983). Transformation of intact yeast cells treated with alkali cations. J. Bacteriol. 153, 163–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang, P.J., Sanson, A., Lee, B., and Park, H.O. (2001). A GDP/GTP exchange factor involved in linking a spatial landmark to cell polarity. Science 292, 1376–1378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klarlund, J.K., Guilherme, A., Holik, J.J., Virbasius, J.V., Chawla, A., and Czech, M.P. (1997). Signaling by phosphoinositide-3,4,5-trisphosphate through proteins containing pleckstrin and Sec7 homology domains. Science 275, 1927–1930. [DOI] [PubMed] [Google Scholar]
- Knop, M., Siegers, K., Pereira, G., Zachariae, W., Winsor, B., Nasmyth, K., and Schiebel, E. (1999). Yeast functional analysis reports. Epitope tagging of yeast genes using a PCR-based strategy: more tags and improved practical routines. Yeast 15, 963–972. [DOI] [PubMed] [Google Scholar]
- Lee, F.J., Huang, C.F., Yu, W.L., Buu, L.M., Lin, C.Y., Huang, M.C., Moss, J., and Vaughan, M. (1997). Characterization of an ADP-ribosylation factor-like 1 protein in Saccharomyces cerevisiae. J. Biol. Chem. 272, 30998–31005. [DOI] [PubMed] [Google Scholar]
- Lee, F.J., Lin, L.W., and Smith, J.A. (1989). N alpha acetylation is required for normal growth and mating of Saccharomyces cerevisiae. J. Bacteriol. 171, 5795–5802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, F.J., Stevens, L.A., Kao, Y.L., Moss, J., and Vaughan, M. (1994). Characterization of a glucose-repressible ADP-ribosylation factor 3 (ARF3) from Saccharomyces cerevisiae. J. Biol. Chem. 269, 20931–20937. [PubMed] [Google Scholar]
- Meacci, E., Tsai, S.C., Adamik, R., Moss, J., and Vaughan, M. (1997). Cytohesin-1, a cytosolic guanine nucleotide-exchange protein for ADP-ribosylation factor. Proc. Natl. Acad. Sci. USA 94, 1745–1748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michelitch, M., and Chant, J. (1996). A mechanism of Bud1p GTPase action suggested by mutational analysis and immunolocalization. Curr. Biol. 6, 446–454. [DOI] [PubMed] [Google Scholar]
- Millar, C.A., Powell, K.A., Hickson, G.R., Bader, M.F., and Gould, G.W. (1999). Evidence for a role for ADP-ribosylation factor 6 in insulin-stimulated glucose transporter-4 (GLUT4) trafficking in 3T3–L1 adipocytes. J. Biol. Chem. 274, 17619–17625. [DOI] [PubMed] [Google Scholar]
- Moss, J., and Vaughan, M. (1995). Structure and function of ARF proteins: activators of cholera toxin and critical components of intracellular vesicular transport processes. J. Biol. Chem. 270, 12327–12330. [DOI] [PubMed] [Google Scholar]
- Moss, J., and Vaughan, M. (1998). Molecules in the ARF orbit. J. Biol. Chem. 273, 21431–21434. [DOI] [PubMed] [Google Scholar]
- Ni, L., and Snyder, M. (2001). A genomic study of the bipolar bud site selection pattern in Saccharomyces cerevisiae. Mol. Biol. Cell 12, 2147–2170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer, D.J., Helms, J.B., Beckers, C.J., Orci, L., and Rothman, J.E. (1993). Binding of coatomer to Golgi membranes requires ADP-ribosylation factor. J. Biol. Chem. 268, 12083–12089. [PubMed] [Google Scholar]
- Park, H.O., Bi, E., Pringle, J.R., and Herskowitz, I. (1997). Two active states of the Ras-related Bud1/Rsr1 protein bind to different effectors to determine yeast cell polarity. Proc. Natl. Acad. Sci. USA 94, 4463–4468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park, H.O., Sanson, A., and Herskowitz, I. (1999). Localization of Bud2p, a GTPase-activating protein necessary for programming cell polarity in yeast to the presumptive bud site. Genes Dev. 13, 1912–1917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters, P.J., Hsu, V.W., Ooi, C.E., Finazzi, D., Teal, S.B., Oorschot, V., Donaldson, J.G., and Klausner, R.D. (1995). Overexpression of wild-type and mutant ARF1 and ARF6: distinct perturbations of nonoverlapping membrane compartments. J. Cell Biol. 128, 1003–1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peyroche, A., Paris, S., and Jackson, C.L. (1996). Nucleotide exchange on ARF mediated by yeast Gea1 protein. Nature 384, 479–481. [DOI] [PubMed] [Google Scholar]
- Premont, R.T., Claing, A., Vitale, N., Freeman, J.L., Pitcher, J.A., Patton, W.A., Moss, J., Vaughan, M., and Lefkowitz, R.J. (1998). β2-Adrenergic receptor regulation by GIT1, a G protein-coupled receptor kinase-associated ADP ribosylation factor GTPase-activating protein. Proc. Natl. Acad. Sci. USA 95, 14082–14087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pringle, J.R., Preston, R.A., Adams, A.E.M., Stearns, T., Drubin, D.G., Haarer, B.K., and Jones, E.W. (1989). Fluorescence microscopy methods for yeast. Methods Cell Biol. 31, 357–435. [DOI] [PubMed] [Google Scholar]
- Radhakrishna, H., Klausner, R.D., and Donaldson, J.G. (1996). Aluminum fluoride stimulates surface protrusions in cells overexpressing the ARF6 GTPase. J. Cell Biol. 134, 935–947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ram, A.F.J., Wolters, A., Hoopen, R.T., and Klis, F.M. (1994). A new approach for isolating cell wall mutants in Saccharomyces cerevisiae by screening for hypersensitivity to calcodlour white. Yeast 10, 1019–1030. [DOI] [PubMed] [Google Scholar]
- Rothman, J.E. (1996). The protein machinery of vesicle budding and fusion. Protein Sci. 5, 185–194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook, J., Fritsch, E.F., and Maniatis, T. (1989). Molecular Cloning: A Laboratory Manual, Cold Spring Harbor, NY: Cold Spring Harbor Laboratory.
- Schweitzer, J.K., and D'Souza-Schorey, C. (2002). Localization and activation of the ARF6 GTPase during cleavage furrow ingression and cytokinesis. J. Biol. Chem. 277, 27210–27216. [DOI] [PubMed] [Google Scholar]
- Sewell, J.L., and Kahn, R.A. (1988). Sequences of the bovine and yeast ADP-ribosylation factor and comparison to other GTP-binding proteins. Proc. Natl. Acad. Sci. USA 85, 4620–4624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman, F., Fink, G.R., and Hicks, J.B. (1986). Methods in Yeast Genetics, Cold Spring Harbor, NY: Cold Spring Harbor Laboratory.
- Simon, S.M., and Aderem, A. (1992). Myristoylation of proteins in the yeast secretory pathway. J. Biol. Chem. 267, 3922–3931. [PubMed] [Google Scholar]
- Stearns, T., Kahn, R.A., Botstein, D., and Hoyt, M.A. (1990). ADP ribosylation factor is an essential protein in Saccharomyces cerevisiae and is encoded by two genes. Mol. Cell. Biol. 10, 6690–6699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanigawa, G., Orci, L., Amherdt, M., Ravazzola, M., Helms, J.B., and Rothman, J.E. (1993). Hydrolysis of bound GTP by ARF protein triggers uncoating of Golgi-derived COP-coated vesicles. J. Cell Biol. 123, 1365–1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teal, S.B., Hsu, V.W., Peters, P.J., Klausner, R.D., and Donaldson, J.G. (1994). An activating mutation in ARF1 stabilizes coatomer binding to Golgi membranes. J. Biol. Chem. 269, 3135–3138. [PubMed] [Google Scholar]
- Tsai, S.C., Adamik, R., Haun, R.S., Moss, J., and Vaughan, M. (1992). Differential interaction of ADP-ribosylation factors 1, 3, and 5 with rat brain Golgi membranes. Proc. Natl. Acad. Sci. USA 89, 9272–9276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vernet, T., Dignard, D., and Thomas, D.Y. (1987). A family of yeast expression vectors containing the phage f1 intergenic region. Gene 52, 225–233. [DOI] [PubMed] [Google Scholar]
- Vida, T.A., and Emr, S.D. (1995). A new vital stain for visualizing vacuolar membrane dynamics and endocytosis in yeast. J. Cell Biol. 128, 779–792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitale, N., Chasserot-Golaz, S., Bailly, Y., Morinaga, N., Frohman, M.A., and Bader, M.-F. (2002). Calcium-regulated exocytosis of dense-core vesicles requires the activation of ADP-ribosylation factor (ARF)6 by ARF nucleotide binding site opener at the plasma membrane. J. Cell Biol. 159, 79–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, C.Z., and Mueckler, M. (1999). ADP-ribosylation factor 6 (ARF6) defines two insulin-regulated secretory pathways in adipocytes. J. Biol. Chem. 274, 25297–25300. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.