Abstract
Using a cytological assay to monitor the successive chromatin association of replication proteins leading to replication initiation, we have investigated the function of fission yeast Cdc23/Mcm10 in DNA replication. Inactivation of Cdc23 before replication initiation using tight degron mutations has no effect on Mcm2 chromatin association, and thus pre-replicative complex (pre-RC) formation, although Cdc45 chromatin binding is blocked. Inactivating Cdc23 during an S phase block after Cdc45 has bound causes a small reduction in Cdc45 chromatin binding, and replication does not terminate in the absence of Mcm10 function. These observations show that Cdc23/Mcm10 function is conserved between fission yeast and Xenopus, where in vitro analysis has indicated a similar requirement for Cdc45 binding, but apparently not compared with Saccharomyces cerevisiae, where Mcm10 is needed for Mcm2 chromatin binding. However, unlike the situation in Xenopus, where Mcm10 chromatin binding is dependent on Mcm2–7, we show that the fission yeast protein is bound to chromatin throughout the cell cycle in growing cells, and only displaced from chromatin during quiescence. On return to growth, Cdc23 chromatin binding is rapidly reestablished independently from pre-RC formation, suggesting that chromatin association of Cdc23 provides a link between proliferation and competence to execute DNA replication.
INTRODUCTION
Eukaryotes share a common mechanism that coordinates the initiation of DNA replication from a large number of chromosomal origins (reviewed in Bell and Dutta, 2002; Takisawa et al., 2000; Blow and Hodgson, 2002; Diffley and Labib, 2002). Initiation involves the origin recognition complex (ORC), which in yeasts is associated with DNA throughout the cell cycle (Aparicio et al., 1997; Lygerou and Nurse, 1999). Additional proteins associate with ORC during late mitosis/G1, thus establishing competence for initiation during the subsequent S phase. This process, called licensing or pre-replicative complex (pre-RC) formation, involves the initial association of Cdt1 and Cdc6/Cdc18 with ORC and the subsequent assembly of Mcm2–7 proteins onto chromatin (Donovan et al., 1997; Tanaka et al., 1997; Maiorano et al., 2000; Nishitani et al., 2000). S phase initiation itself is triggered by activation of two protein kinases, Cdc7 and cyclin-dependent kinase (CDK). A likely target of Cdc7 is the Mcm2–7 complex (reviewed in Masai and Arai, 2002; Sclafani, 2000), which probably provides helicase activity during replication (Ishimi, 1997), but the targets of CDK remain to be identified.
During replication initiation, further proteins needed for the elongation step of DNA replication, such as Cdc45/Sld3 (Mimura and Takisawa, 1998; Zou and Stillman, 2000; Kamimura et al., 2001; Nakajima and Masukata, 2002) and DNA polymerases α and ε (Mimura and Takisawa, 1998; Mimura et al., 2000), associate with initiation sites and, together with Mcm2–7 proteins, move away from origins (Aparicio et al., 1997; Diffley and Labib, 2002). As S phase progresses, Mcm2–7 proteins dissociate from chromatin and their reassociation with already replicated DNA is blocked, thus restricting replication to a single round per cell cycle. This block to reinitiation is effected by CDK, which blocks the chromatin binding of Mcm2–7 proteins by multiple mechanisms (Nguyen et al., 2001), and by geminin, which inhibits Cdt1 function in Metazoa (McGarry and Kirschner, 1998; Wohlschlegel et al., 2000; Tada et al., 2001).
Fission yeast Cdc23 (homologous to Saccharomyces cerevisiae Mcm10/Dna43) is another essential replication protein (Nasmyth and Nurse, 1981; Aves et al., 1998). This factor was identified using screens for mutants affected in DNA replication (Dumas et al., 1982; Solomon et al., 1992) and minichromosome maintenance (Maine et al., 1984). Mcm10 mutants show a reduced efficiency of origin use and replication elongation across origins is impeded (Merchant et al., 1997; Homesley et al., 2000). Mcm10/cdc23 alleles show genetic interactions with a wide range of other replication mutations, suggesting that this factor is involved in both the initiation and elongation steps of DNA replication. These include mutations affecting ORC, Mcm2–7, SpCdc24, ScCdc45, ScDpb11, and subunits of DNA polymerases δ and ε (Tanaka et al., 1999; Homesley et al., 2000; Kawasaki et al., 2000; Liang and Forsburg, 2001; Hart et al., 2002). Physical interactions between Mcm10/Cdc23 and ORC or Mcm2–7 proteins have also been detected (Merchant et al., 1997; Homesley et al., 2000; Izumi et al., 2000; Kawasaki et al., 2000; Hart et al., 2002) and in vivo cross-linking has shown Mcm10 to be associated with DNA at an S. cerevisiae replication origin (Homesley et al., 2000).
Mcm10 shows functional conservation as the S. cerevisiae MCM10 gene can complement a fission yeast cdc23 mutant (Aves et al., 1998), but S. cerevisiae and vertebrate Mcm10 proteins have different properties. Most notably, budding yeast Mcm10 is needed for Mcm2–7 chromatin association (Homesley et al., 2000), whereas in Xenopus, Mcm10 depletion affects a later step in replication initiation, blocking the chromatin association of Cdc45 (Wohlschlegel et al., 2002). To clarify these differences, we have characterized in greater detail the fission yeast Cdc23 homologue. We show that, as in Xenopus, Cdc23 is required for chromatin association of Cdc45 and does not affect earlier stages of replication initiation. Cdc23 is distinct from vertebrate Mcm10, however, in terms of its cell cycle pattern of chromatin association, reflecting differences in how this protein interacts with replication complexes.
MATERIALS AND METHODS
Fission Yeast Strains and Methods
Strains used are shown in Table 1. Media and growth conditions and standard genetic methods were as described by Moreno et al. (1991). Thiamine at 5 μg/ml was used to repress the nmt1 promoter and hydroxyurea (HU) was used at 12 mM. Nitrogen starvation was carried out using EMM medium lacking NH4Cl.
Table 1.
Yeast strains used
| P1046 | mcm2+-YFP::ura4+ade6-M210 leu1-32 ura4-D18 h- | |
| P1051 | mcm2+-CFP::ura4+ade6-M210 leu1-32 ura4-D18 h- | |
| P1054 | mcm7+-CFP::ura4+ade6-M210 leu1-32 ura4-D18 h- | |
| P1082 | cdc23+-CFP::ura4+ade6-M210 leu1-32 ura4-D18 h- | |
| P1083 | sna41+(cdc45)-YFP::ura4+ade6-M210 leu1-32 ura4-D18 h- | |
| P1098 | mcm2+-CFP::ura4+cdc23-IE2-td::kanr (cdc23tstd) | |
| P1100 | sna41+(cdc45)-YFP::ura4 cdc23-IE2-td::kanr (cdc23tstd) h- | |
| 3830 | leu1-32 ura4-D18 ade6-210 ars1(Mlul)::pRep3X-GFP-atb2::LEU2 h+ | Ding and Smith (1998), Pidoux et al. (2000) |
| P1122 | cdc23+-CFP::ura4+ | |
| P1128 | mcm2+-YFP::ura4+cdc23+-CFP::ura4+ | |
| P1134 | orc6+-CFP::ura4+leu1-32 ade6-M210 ura4-D18 | |
| P1156 | ars1(Mlul)::pRep3X-GFP-atb2::LEU2 sna41+(cdc45)-YFP::ura4+mcm7+-CFP::ura4+ura4-D18 ade6 h- | Derived from 3830 |
| P1162 | mcm4ts(cdc21-M68)-td::ura4+mcm2+-CFP::ura4+sna41+(cdc45)-YFP::ura4+ade6 leu1-32 | Derived from P1023 (Lindner et al., 2002) |
| P1166 | cdc10-129 cdc45+-yfp::ura4+ | |
| P1184 | hsk1-89::ura4+leu1-32 ura4-D18 mcm7+-cfp::ura4+cdc45+-yfp::ura4+ | Derived from N1359 (Takeda et al., 2001) |
| P1214 | cdt1+-CFP::ura4+ura4-D18 h+ | |
| P1220 | mcm2+-CFP::ura4+nmt41X-cdc23-IE2-td::kanr (nmt-cdc23tstd) | |
| P1221 | sna41+(cdc45)-YFP::ura4+nmt41X-cdc23-IE2-td::kanr (nmt-cdc23tstd) | |
| P1226 | nmt41X-cdc23-IE2-td::kanr (nmt-cdc23tstd) cdt1+-CFP::ura4+ | |
| P1266 | cdc23+-CFP::ura4+mcm4(cdc21)tstd::ura4+ | |
| P1276 | sna41+(cdc45)-YFP::kanr nmt1(41X)-cdc13::LEU2 cig1Δ::ura4+cig2Δ::ura4+cdc13Δ::ura4+ura4-D18 leu1-32 ade6-M210 h- | Derived from Fisher and Nurse (1995) |
| P1357 | cdc25-22 cdc23+-CFP::ura4+ | |
| P1358 | cdc22-M45 cdc23+-CFP::ura4+ | |
| P1360 | nda3-311 cdc23+-CFP::ura4+ura4-D18 | |
| P1362 | cdc10-V50 cdc23+-CFP::ura4+ade6 ura4-D18 |
Construction of CFPand YFP-tagged Strains
Oligonucleotide primers used in plasmid construction are shown in Table 2. Cyan fluorescent protein (CFP) or yellow fluorescent protein (YFP)-encoding DNA fragments were amplified from either pECFP-C1 or pEYFP-C1 (Clontech, Palo Alto, CA) using oligos 5′HindIIIATG-CYFP and 3′SacSTOPBamSal-CYFP and inserted into HindIIIand Bam HI-cleaved pSMUG2+ (Lindner et al., 2002) to give either pSMUC2+ (for CFP tagging) or pSMUY2+ (for YFP tagging). Sequences of these plasmids are available at users.ox.ac.uk/~kearsey/plasmids.
Table 2.
Oligos used
| 5′HindIIIATG-CYFP | tttaagctttaatggtgagcaagggcgaggagc |
| 3′SacSTOPBamSal-CYFP | tttgagctctttaggatccgtcgaccttgtacagctcgtccatgccg |
| 5′ApaI-mcm10C | tttgggcccgaaccaagaaaggaagaggagcgat |
| 3′SmaI-mcm10C | tttcccgggagggaactatttctaagtcatcctca |
| 5′XhoI-sna41 | agagctcgaggagaagtttgaaaatgctca |
| 3′SmaI-sna41 | cctccccgggctaatagtgttttgaaggacag |
| 5′XhoI-mcm10ATG | tttctcgagtatgcatgatcccttcattgcagaag |
| 3′SmaI-mcm10N | tttcccgggaggcccttttcctaatgaaacatct |
| 5′EcoRI-nmt1 | tttgaattcggtcgatcgactctagaggatcaga |
| 3′ApaI-nmt1 | tttgggccccatatgatttaacaaagcgactataag |
| 5′ApaI-orc6 | tttgggccctggaaaccattacttatctttgtacc |
| 3′XhoI-orc6 | tttctcgagtgaagcagtaccatctttttcaagctg |
| 5′ApaI-mcm7 | caagtgggcccgccgctgcgaaccccttata |
| 3′SmaI-mcm7 | cttaccccgggcattctccatatgtaaatccg |
| 5′XhoI-mcm2 | acgactcgagacactacaattccttttaatc |
| 3′SmaI-mcm2 | ccaccccgggcaataagatatttagcaaatgttc |
| 5′ApaI-cdt1 | tttgggcccgctattaccccatgttttactattc |
| 3′SmaI-cdt1 | tttcccgggtagaatttgaaagaatagtgatgcta |
Cdc23 was C-terminally tagged with CFP by amplifying a cdc23+ fragment with oligos 5′ApaI-mcm10C and 3′SmaI-mcm10C and inserting the fragment into ApaIand SmaI-cut pSMUC2+ to give pSMUC2+cdc23. This plasmid was cut with SpeI to tag the endogenous cdc23+ gene with CFP. Cdc45 was tagged with YFP by amplifying a fragment using oligos 5′XhoI-sna41 and 3′SmaI-sna41, and the resulting fragment was inserted into XhoIand SmaI-cut pSMUY2+ to give pSMUY2+cdc45. pSMUY2+cdc45 was cut with HpaI to direct integration into the cdc45+ (sna41+) locus. Orc6 was tagged with CFP by amplifying a fragment with oligos 5′ApaI-orc6 and 3′XhoI-orc6 and this PCR product was inserted into ApaIand XhoI-cut pSMUC2+ to give pSMUC2+orc6. This was cleaved with SpeI to direct integration into the orc6+ locus. Mcm2 was tagged by using the oligos 5′XhoI-mcm2 and 3′SmaI-mcm2, and the resulting PCR fragment was cloned into XhoIand SmaI-cleaved pSMUC2+ or pSMUY2+. The resulting plasmids were cut with BglII to direct integration into the mcm2+ locus. Mcm7 was tagged with CFP by amplifying the 3′ end of the gene with the oligos 5′ApaI-mcm7 and 3′SmaI-mcm7, and this fragment was inserted in to ApaIand SmaI-cut pSMUC2+ to give pSMUC2+mcm7. This plasmid was linearized with MluI to tag the endogenous mcm7+ gene. Cdt1 was tagged by amplifying a cdt1+ fragment with oligos 5′ApaI-cdt1 and 3′SmaI-cdt1, and this was cloned into ApaIand SmaI-cut pSMUC2+. The resulting pSMUC2+cdt1 plasmid was cut with EcoRI to tag the endogenous cdt1+ gene.
For tagging the cdc45+ gene in the background of the cyclin B shut off strain (nmt1(41X)-cdc13, cdc13Δ cig1Δ cig2Δ; Fisher and Nurse, 1995), we replaced the ura4+ marker in pSMUY+cdc45 with an NgoM IV fragment containing the kanMX6 (kanr) marker to give pSMRY+cdc45-YFP. This was linearized with HpaI to direct integration at the cdc45+ locus, thus generating strain P1276. All constructs were verified by sequencing.
Construction of Degron cdc23 Alleles
The cdc23tstd degron allele was constructed by amplifying the N-terminus–encoding part of the cdc23+ gene using the oligos 5′XhoI-mcm10ATG and 3′SmaI-mcm10N, and this was inserted into a plasmid expressing the DHFR degron from the mcm4+/cdc21+ promoter (XhoIand SmaI-cut pSMUG2+ degron [ura4+-containing] or pSMRG2+degron [kanMX6-containing; Lindner et al., 2002]) to generate pSMUG2+degron+cdc23 or pSMRG2+degron+cdc23. BglII linearization was used for integration of these plasmids at the cdc23+ locus.
To derive a plasmid where the mcm4+ promoter was replaced by the nmt1 thiamine-regulatable promoter, the mcm4+ promoter of pSMUG2+degron or pSMRG2+degron plasmids was removed by EcoRI and ApaI digestion and replaced with an attenuated nmt1 promoter, amplified using the oligos 5′EcoRI-nmt1 and 3′ApaI-nmt1, using pREP41X as template (Basi et al., 1993), thus generating pSMUG2+nmt41degron (ura4+-containing) or pSMRG2+nmt41degron (kanMX6-containing) plasmids. The N-terminus of Cdc23 was removed as a XhoI-SmaI fragment from the pSMUG2+degron+Cdc23 plasmid and inserted into the XhoI/SmaI sites of pSMUG2+nmt41+degron or pSMRG2+nmt41+degron plasmids. BglII linearization was used for integration. All constructs were verified by sequencing.
In Situ Chromatin-binding Assay
The chromatin-binding assay was carried out using the procedure in Kearsey et al. (2000) with the modifications in Lindner et al. (2002). For Cdc23-CFP strains, extractions were carried out at 4°C. Images were collected as before (Lindner et al., 2002); at least 100 cells were counted for each data point, and error bars show the range of two experiments. Fluorescence intensities of nuclei were quantitated using a modification of NIH Image macros developed by Dr. Joel Huberman, available at: http://saturn.roswellpark.org/huberman/Quant_Flu_Microscopy/Quant_Flu_Micro.html. Micrococcal nuclease (Sigma, St. Louis, MO) digestion was at a concentration of 2.5 U/ml in an extraction buffer containing 2 mM CaCl2 and 1% Triton X-100. In control reactions, the nuclease was inhibited by adding EGTA to 10 mM. Flow cytometric analysis of samples was carried out as described in Lindner et al. (2002). Filter sets for YFP (41028) and CFP (31044 v2) were from Chroma Inc. (Brattleboro, VT).
Protein Analysis
Protein extracts were made by TCA extraction and analyzed by Western blotting as described previously (Grallert et al., 2000). Cdc23 was detected either using monoclonal 3E1 anti-GFP antibody, or rabbit polyclonal anti-Cdc23. Rabbit polyclonal antibody was raised against N-terminal histidine-tagged full-length Cdc23, produced using expression vector pET-19b in Escherichia coli expression host BL21 (DE3) pLysS, and purified over nickel-charged His-Bind resin (Novagen, Madison, WI). α-Tubulin was detected using Sigma T5168 at a dilution of 1/10,000.
RESULTS
Chromatin Binding of Cdc23 during the Cell Cycle
The chromatin binding of some replication factors such as Mcm2–7 proteins changes during the cell cycle, which reflects important regulatory controls on DNA replication. Mcm10 chromatin association is also regulated in Xenopus, because it binds in a step dependent on pre-RC formation but independent of Cdk2 and Cdc7, and disassociates during S phase together with Mcm2–7 (Wohlschlegel et al., 2002). This behavior appears to be conserved at least in vertebrates, as mammalian Mcm10 associates with nuclear structures in S phase and dissociates in G2 (Izumi et al., 2000, 2001) although in contrast, S. cerevisiae Mcm10 binds to chromatin throughout the cell cycle (Homesley et al., 2000). It is not clear how fission yeast Cdc23 compares with these situations, because a previous report described the protein as being associated with nuclease-resistant nuclear structures (Liang and Forsburg, 2001), and the possibility of changes in nuclear association during the cell cycle has not been examined.
To extend this work we used an “in situ” chromatin-binding assay to analyze Cdc23. This technique, first used with Mcm2–7 proteins (Kearsey et al., 2000; Lindner et al., 2002), uses a detergent wash of permeabilized cells to extract nucleoplasmic, but not chromatin-associated, protein and thus reveals whether a given factor is chromatin associated in single cells, thus avoiding the need for synchronization procedures. We constructed a strain where the sole copy of cdc23+, expressed from its native promoter, is C-terminally tagged with CFP. In growing cells, Cdc23 is nuclear in >95% of binucleate (late M/G1/S phase) as well as uninucleate (G2 phase) cells, even after detergent washing, suggesting that the protein remains chromatin associated during the cell cycle (Figure 1, A and B). The fluorescence intensity is reduced by about half by detergent extraction, suggesting that a fraction of Cdc23 is nucleoplasmic and may be removed by this procedure. Using a doubly tagged strain, cells showing Mcm2-YFP chromatin binding (i.e., in late M, G1, or S phase) also retained Cdc23-CFP, making it unlikely that displacement of this protein is occurring during the short G1 phase (Figure 1A). Furthermore, most cells retain chromatin associated Cdc23 after arrest in G1, S, G2, or mitosis with cdc10, cdc22, cdc25, or nda3 mutations (Figure 1D). Digestion of DNA with micrococcal nuclease caused complete loss of Cdc23, indicating that Cdc23 that is refractory to detergent extraction is chromatin associated (Figure 1C).
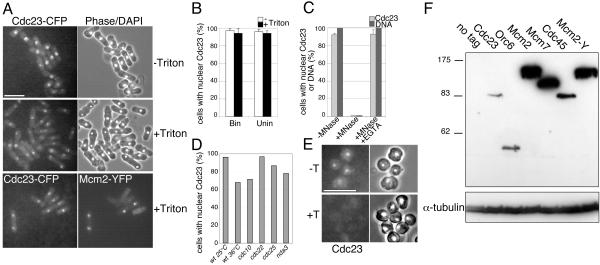
Figure 1.
Cdc23 is chromatin bound throughout the cell cycle. (A) Cdc23 is constitutively located in the nucleus before and after detergent washing. Top and middle panels: an asynchronous log phase culture of strain P1082, permeabilized by zymolyase digestion and either directly fixed (–Triton) or detergent washed and then fixed (+Triton). Bottom panels: strain P1128 (Mcm2-YFP, Cdc23-CFP) permeabilized by zymolyase digestion and detergent washed; Mcm2 is chromatin associated in only binucleate cells (in late M/G1/S), whereas Cdc23 is refractory to detergent extraction in all stages of the cell cycle. Bar, 10 μm. (B) Quantitation of data shown in A. Bin, binucleate (G1/S phase) cells; Unin, uninucleate (G2 phase) cells. (C) Digestion of DNA with micrococcal nuclease releases Cdc23, implying that Cdc23 that is refractory to detergent extraction is bound to chromatin. Cells prepared as in A were either detergent washed only (–MNase), treated with detergent and micrococcal nuclease (+MNase), or treated with detergent and micrococcal nuclease in the presence of EGTA (+MNase, +EGTA). Graphs show the percentage of cells with nuclear Cdc23 or DNA. (D) Cdc23 is chromatin associated in cells arrested at different cell cycle stages. Strains mutant in cdc10 (P1362), cdc22 (P1358), cdc25 (1357), or nda3 (1360) genes were shifted to the restrictive temperature (20°C for nda3, otherwise 36°C) for 3 h, and chromatin binding of Cdc23 was assessed by detergent washing as in A. The wild-type control strain used was P1082. (E) Cdc23 is not chromatin associated in cells arrested in G1 by nitrogen starvation. Log phase cells of strain P1122 was transferred to EMM-nitrogen medium for 16 h at 25°C after which cells were processed as in A and fixed either without (–T) or after detergent washing (+T). Bar, 10 μm. (F) Comparison of Cdc23-CFP levels with other CFP-tagged replication proteins by Western blotting. Strains used were P1122 (Cdc23), P1134 (Orc6), P1051 (Mcm2), and P1054 (Mcm7). Also shown are yellow fluorescent protein (YFP)-tagged Cdc45 (P1083) and Mcm2 (“Mcm2-Y,” P1046) and a wild-type strain (“no tag”). α-Tubulin is shown as a loading control. From quantitative analysis of Western blots with diluted Orc6, Mcm2, and Mcm7 samples, the relative abundance of the proteins is estimated at 1:2:15:15 for Cdc23:Orc6:Mcm2:Mcm7 (unpublished data).
In contrast to the situation with cycling cells, we observed that chromatin association of Cdc23 is altered in stationary phase cells. After arrest in G1 by nitrogen starvation, Cdc23 levels are lower (unpublished data), but the protein is clearly nuclearly localized in cells directly fixed by methanol/acetone (Figure 1E, –T). In contrast to the situation with log phase cells, detergent washing can extract this nuclear Cdc23 (Figure 1E, +T). Taken together these results show that Cdc23 is chromatin associated throughout the cell cycle in log phase cells, but it is displaced from or less tightly associated with chromatin in G1-arrested, quiescent cells.
Mcm10 is present at about two molecules per origin in Xenopus (Wohlschlegel et al., 2002), but there are potentially many copies of the protein per origin in S. cerevisiae, because Mcm10 is similar in abundance to Mcm2–7 and 40–60 times more abundant than ORC (Kawasaki et al., 2000). Given this wide range in abundance we compared Cdc23 levels with other tagged proteins by Western blotting. This showed that in contrast to the situation in budding yeast, Cdc23 is ∼10–20-fold less abundant than Mcm2 and Mcm7 and comparable in abundance to Orc6 (Figure 1F).
Cdc23 Chromatin Reassociation Is Independent of Mcm4
Given that the chromatin association of vertebrate Mcm10 requires the prior chromatin binding of Mcm2–7 (Wohlschlegel et al., 2002), we considered the possibility that Cdc23 might also be loaded in an Mcm2–7-dependent step, but remain on chromatin after completion of DNA replication, thus obscuring any stage-specific association. To investigate this possibility, cells were arrested in G1 by nitrogen starvation, when Cdc23 is detergent extractable, and we followed the reassociation of Cdc23 when cells are refed and carry out DNA replication (Figure 2A). Wild-type cells show an increase in Cdc23 that is not detergent extractable in advance of DNA replication (Figure 2, B–D). To determine the relevance of Mcm2–7 protein in this reassociation, we used a tight allele of mcm4 where a conventional temperature-sensitive allele is N-terminally tagged with a domain that is ubiquitylated and degraded at 37°C (Dohman et al., 1994; Lindner et al., 2002). In contrast to the situation in the single temperature-sensitive mutant, Mcm4 protein is rapidly degraded at the restrictive temperature in this double (mcm4tstd) mutant, and a tighter block to DNA replication is achieved (Lindner et al., 2002). On refeeding the degron mutant at the restrictive temperature, DNA replication is blocked, but the reassociation of Cdc23 with chromatin shows timing similar to that of the wild-type strain (Figure 2, B–D). Thus although Cdc23 reassociates with chromatin before S phase, this step is independent of Mcm4 and presumably pre-RC formation.
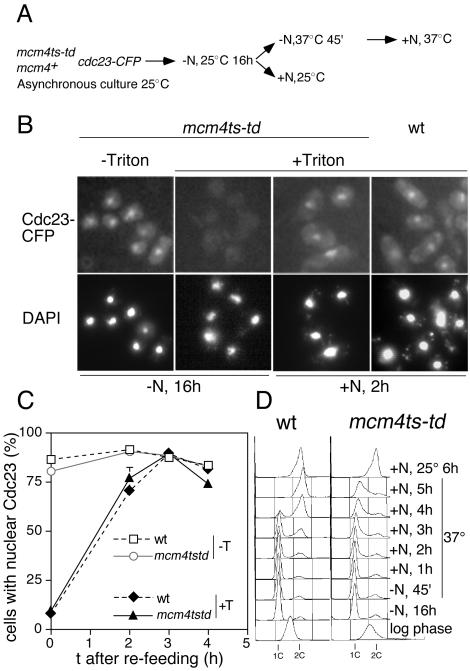
Figure 2.
Cdc23 is displaced from chromatin after nitrogen starvation, but re-association with chromatin on re-entry to the cell cycle does not require pre-RC formation. (A) Experimental procedure; strains used were P1266 (degron mcm4tstd) and P1122 (mcm4+). (B) Nuclear localization (–Triton) and chromatin association (+Triton) of Cdc23 after nitrogen starvation (–N, 16 h) and re-entry to the cell cycle (+N, 2 h) in wild-type (wt) and mcm4 degron strains (mcm4ts-td). Only data for the mcm4ts-td strain are shown after nitrogen starvation (–N, 16 h), but the mcm4+ strain showed a similar result. (C) Quantitation of data from the experiment shown in B. (D) Flow cytometry, showing that S phase is blocked in the degron mcm4tstd strain and timing of S phase in the wild-type strain at 37°C.
Using Mcm2–7 and Cdc45 Chromatin Binding to Monitor Sequential Steps in Replication Activation in Single Cells
To investigate the function of Cdc23 in DNA replication we first explored whether Cdc45 chromatin association could be used to monitor a late step in replication activation in fission yeast cells, after the initial binding of Mcm2–7 proteins. Cdc45 loading is critical for replication control because it is the last step between origin unwinding and DNA synthesis and is rate-limiting for replication in Xenopus (Edwards et al., 2002). Like Mcm2–7 proteins, Cdc45 (also known as Sna41) is constitutively nuclear during the Schizosaccharomyces pombe cell cycle (Miyake and Yamashita, 1998), but a proportion of binucleate cells (in G1/S phase) is resistant to Cdc45 extraction (Figure 3A). This retained protein is solubilized by micrococcal nuclease digestion (unpublished results), implying that Cdc45 is bound to chromatin in these cells. Analysis of a strain containing Cdc45-YFP, Mcm7-CFP, and α-tubulin-GFP showed that Mcm7 chromatin association occurs first during mid-anaphase, as found for Mcm4 (Kearsey et al., 2000), but Cdc45 chromatin association is only seen in binucleate cells lacking spindles, i.e., after the completion of anaphase (Figure 3, B and C). Most uninucleate (G2) cells were negative for both Mcm7 and Cdc45, implying that both proteins dissociate from chromatin by the end of S phase.
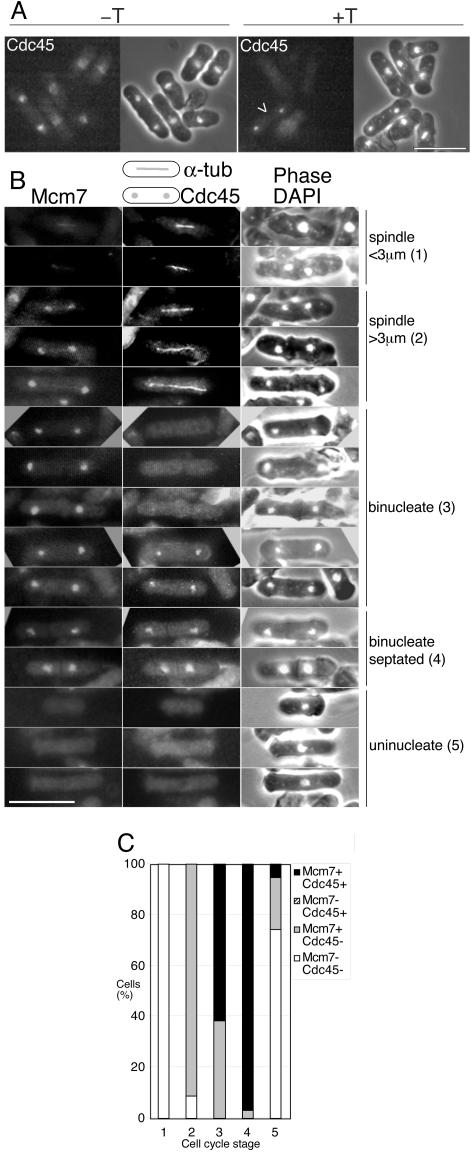
Figure 3.
Sequential chromatin association of Mcm7 and Cdc45 during the fission yeast cell cycle. (A) Cdc45 is chromatin associated only during S phase. An asynchronous culture of strain P1083 was fixed directly (–T) or permeabilized, detergent-washed, and fixed (+T). Cdc45 is constitutively located in the nucleus during the cell cycle in directly fixed cells, confirming a previous result obtained in cells overexpressing Cdc45 (Miyake and Yamashita, 1998), but chromatin association is only seen in some binucleate cells (arrow). Bar, 10 μm. (B) An asynchronous culture of strain P1156, containing Mcm7-CFP, Cdc45-YFP, and α-tubulin-GFP, was processed by detergent extraction to reveal chromatin association of Mcm7 (left panels) and Cdc45 (middle panels). Cells are staged into a cell cycle sequence according to spindle length and nuclear separation: (1) metaphase/early anaphase (spindle <3 μm); (2) mid to late anaphase (spindle >3 μm); (3) G1/S (binucleate, no spindle); (4) G1/S (binucleate, no spindle, septum visible); and (5) G2 (uninucleate). Although GFP (tubulin) fluorescence leaks into the YFP channel in this experiment, tubulin can be unambiguously distinguished from Cdc45 because nuclear fluorescence is not seen in a strain only containing α-tubulin–GFP after detergent extraction and a strain only containing Cdc45-YFP does not show spindle fluorescence. Bar, 10 μm. (C) Quantitation of experiment shown in B, showing percentage of cells positive for Cdc45 or Mcm7 after detergent extraction. Numbers on the x-axis refer to cell cycle stages shown in B. Mcm7 chromatin association occurs from mid-anaphase (stage 2), but Cdc45 chromatin binding is only seen in binucleate cells lacking spindles (stages 3 and 4). Because a proportion of binucleate cells without spindles (stage 3) are positive for Mcm7 and not Cdc45, these may represent G1 cells. Binucleate cells with both Cdc45 and Mcm7 binding are likely to represent cells in S phase.
Analysis of a cdc10 mutant, which has been reported to block Mcm6 but not Cdc45 chromatin association (Uchiyama et al., 2001), suggests that Cdc45 might associate with chromatin in a step independent of pre-RC formation, because Cdc10 is necessary for Cdt1 and Cdc18 transcription. To examine this point in more detail we constructed a strain containing Cdc45-YFP, Mcm2-CFP, and a degron mcm4tstd mutation. After shifting this strain to the restrictive temperature, both the associations of Mcm2 and Cdc45 with chromatin that are normally seen in binucleate (late M/G1/S phase) cells are now blocked (Figure 4, A and B). We also showed that as assayed by the in situ chromatin binding assay, Cdc45 chromatin binding is lost in a cdc10 mutant at the restrictive temperature (Figure 4, C and D), which is consistent with a dependence of Cdc45 chromatin loading on Mcm2–7 chromatin binding. In addition, inactivation of either Hsk1 (homologous to S. cerevisiae Cdc7) using a temperature-sensitive allele, or CDK, using a strain with a thiamine-regulatable cdc13+ gene in the background of a triple cyclin B gene deletion, also blocked Cdc45 chromatin association that is normally seen in binucleate (S phase) cells as well as S phase entry (Figure 4, C–F). Relevant to these results is a recent study showing that the Sld3 partner of Cdc45 only binds to chromatin after Hsk1 activation (Nakajima and Masukata, 2002). Taken together these observations indicate that fission yeast Cdc45 has similar properties to homologues in S. cerevisiae and Xenopus, associating with chromatin in a step that occurs after Mcm2–7 chromatin association and that is dependent on pre-RC formation and activation of CDK and Hsk1. Its chromatin association is thus likely to occur around the time of S phase onset.
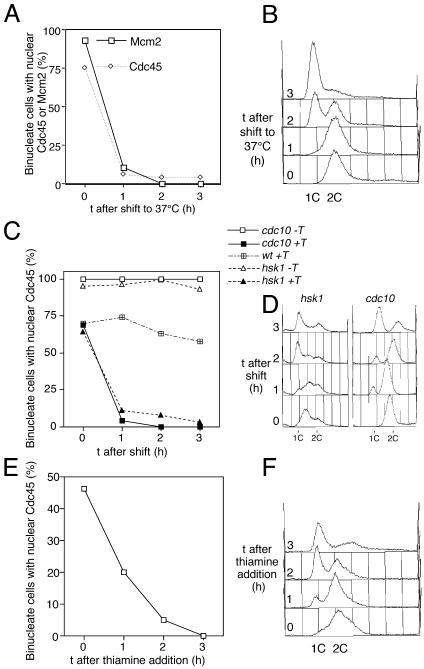
Figure 4.
Chromatin binding of Cdc45 is dependent on pre-RC formation, CDK, and Hsk1. (A) Cdc45 chromatin association is dependent on Mcm4 function. Strain P1162 containing a degron mcm4tstd mutation, Mcm2-CFP, and Cdc45-YFP was shifted to 37°C, and the percentage of binucleate cells with Mcm2 and Cdc45 after detergent extraction was scored. (B) Flow cytometric analysis of experiment shown in A, showing arrest of DNA replication by degron mcm4tstd mutation. (C) Cdc45 chromatin association is dependent on Cdc10 and Hsk1 function. Strains containing YFP-tagged Cdc45 and temperature-sensitive cdc10 (P1166) or hsk1 (P1184) alleles were grown to log phase at 25°C and then shifted to the restrictive temperature (36°C for cdc10, 30°C for hsk1), and the chromatin binding of Cdc45 in binucleate cells was monitored using the in situ chromatin binding assay. The wild-type control strain (shifted to 36°C) was P1083. +T, cells were examined after detergent extraction; –T, cells were fixed directly. (D) Flow cytometric analysis of experiment shown in C. (E) Cdc45 chromatin association is dependent on CDK. Strain P1276, containing a triple cyclin B deletion and an nmt1-regulatable cdc13+ gene was transferred to thiamine-containing medium at t = 0 to inactivate CDK, and chromatin association of Cdc45 was monitored. This block to S phase entry (shown in flow cytometric analysis in F) also prevents Cdc45 chromatin association. In A, C, and E the percentage of binucleate cells with chromatin bound Cdc45 is shown as a percentage of total binucleate cells.
Cdc23 Functions after Mcm2–7 Chromatin Association and Is Required for Cdc45 Chromatin Binding
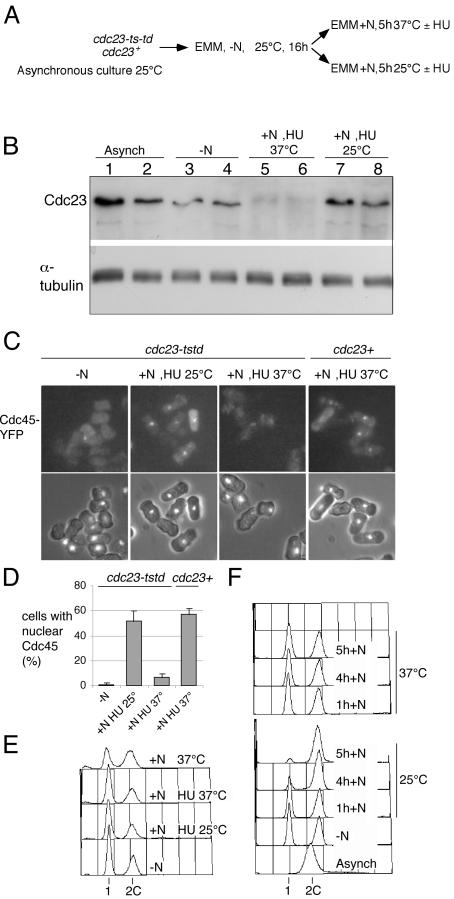
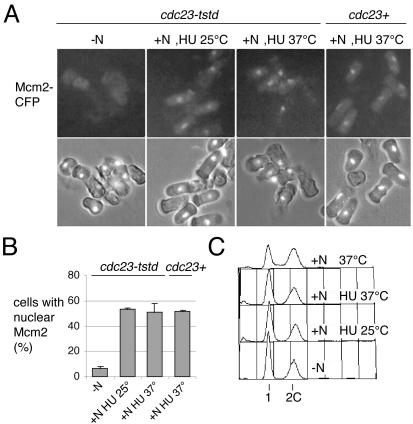
To investigate the effect of Cdc23 inactivation on the sequential chromatin association of Mcm2 and Cdc45, we constructed strains expressing fluorescently tagged versions of these proteins and a degron cdc23 allele. As with mcm4, to make this allele as tight as possible, we made a double mutant (cdc23tstd) where the degron is combined with a temperature-sensitive allele (cdc23-IE2; Grallert and Nurse, 1997). In cycling cells, this degron version of Cdc23 is not efficiently degraded, but if cells are arrested in G1 by nitrogen starvation and then refed 37°C, efficient proteolysis is observed and DNA replication is blocked (Figure 5, B and F). Degron cdc23 strains containing Cdc45-YFP or Mcm2-CFP were thus arrested in G1, when both proteins are not chromatin associated, and refed at either the permissive or restrictive temperature, allowing the reassociation of these proteins with chromatin to be monitored. HU was added to the cultures to prevent displacement of Mcm2 or Cdc45 from chromatin on replication completion at the permissive temperature. On refeeding at 25°C, chromatin association of both proteins is observed (Figures 5, C and D, and 6, A and B). However, at 37°C when Cdc23 is inactivated and degraded, chromatin association of Cdc45 is not detected (Figure 5, C and D), although Mcm2 chromatin association occurs normally (Figure 6, A and B). This indicates that inactivation of Cdc23 does not affect Mcm2 binding and presumably the early step of pre-RC formation, but affects a later one involving association of Cdc45 with chromatin.
Figure 5.
Inactivation of Cdc23 prevents Cdc45 chromatin association during S phase. (A) Experimental procedure. Strains used were P1100 (cdc23tstd) and P1083 (wt), both of which contain Cdc45-YFP. HU was added to cultures after refeeding to block cells in S phase and thus prevent displacement of Cdc45 at the permissive temperature. (B) Levels of Cdc23 protein during the experiment. Even lanes: protein levels for experiment shown in C; odd lanes: protein levels for the Mcm2 experiment shown in Figure 6. α-Tubulin is shown as a loading control. (C) Cdc45-YFP fluorescence after detergent extraction. For further details see text. (D) Quantitation of data shown in C. (E) Flow cytometric analysis of cdc23tstd cells shown in C; also shown is –HU control. (F) Flow cytometric analysis of degron cdc23tstd cells released from nitrogen starvation in absence of HU, showing block to S phase at 37°C and execution of DNA replication at 25°C.
Figure 6.
Inactivation of Cdc23 has no effect on Mcm2 chromatin association. Experimental procedure is shown in Figure 5A; strains used were P1098 (cdc23tstd) and P1051 (wt), both of which contain Mcm2-CFP. As before, HU was added to cultures after refeeding to block cells in S phase and thus prevent displacement of Mcm2 at the permissive temperature. (A) Mcm2-CFP fluorescence after detergent extraction. For further details see text. (B) Quantitation of data shown in A. (C) Flow cytometric analysis of cdc23tstd cells shown in A; also shown is –HU control.
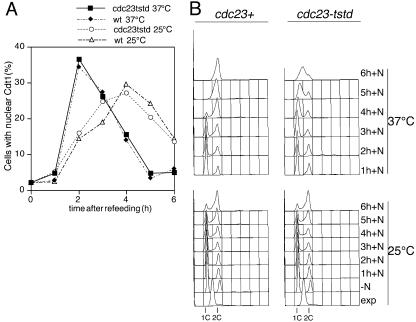
Because Cdc23 functions after Mcm2 chromatin binding but before association of Cdc45, one possibility is that it promotes displacement of Cdt1 from pre-RCs, because Cdt1 is required for Mcm2–7 chromatin association but is not needed for DNA replication after initiation (Nishitani et al., 2000). However, we find that Cdt1 does not persist in cells when Cdc23 is inactivated compared with wild-type cells (Figure 7), indicating that the block to Cdc45 chromatin binding appears to be independent of Cdt1.
Figure 7.
Cdc23 inactivation does not affect Cdt1. Strains expressing Cdt1-CFP and either wild-type (P1214) and or degron cdc23 alleles (nmt-cdc23tstd, P1226) were grown at 25°C to log phase and then transferred to medium lacking nitrogen for 16 h at 25°C to arrest cells in G1. Thiamine was added after 12 h of nitrogen starvation to reduce expression of Cdc23 in the degron strain. At 16 h the cultures were split, refed with nitrogen, and incubated at either 25°C or 37°C and analyzed, at the times indicated, by flow cytometry and fluorescence microscopy to detect nuclear Cdt1. (A) Analysis of percentage of cells showing nuclear Cdt1 after refeeding. Data shown are for directly fixed cells but similar results were obtained for cells that were detergent extracted before fixation. (B) Flow cytometric analysis for data shown in A, showing arrest of DNA replication in the degron cdc23 strain at 37°C.
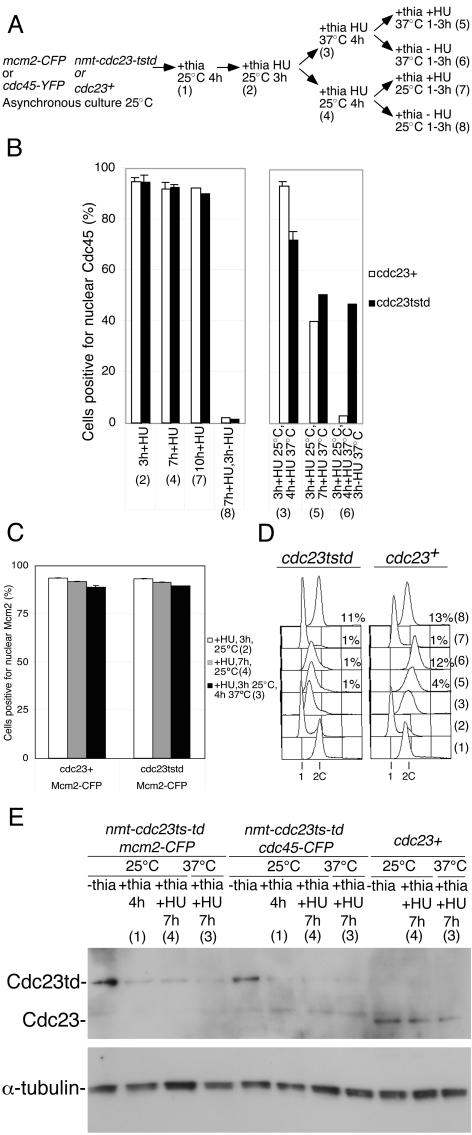
Inactivation of Cdc23 during S Phase Prevents Completion of DNA Replication
In Xenopus, although Mcm10 depletion prevents Cdc45 chromatin association during initiation, it has not been possible to determine whether Mcm10 is also required to maintain Cdc45 chromatin binding after S phase onset. To address this point we were unable to use the cdc23tstd allele, because degradation of Cdc23 in this strain is rather inefficient unless cells are nitrogen starved. We therefore made an improved mutant where transcription of the cdc23tstd allele is from an attenuated version of the nmt1 promoter (nmt-cdc23tstd), which can be repressed with thiamine. These cells were arrested in S phase by adding HU to the medium, which blocks cells with chromatin-associated Cdc45 and Mcm2, and both thiamine addition and a temperature shift to 37°C were used to eliminate Cdc23 activity (Figure 8, A and E). After 4 h at 37°C there is no effect on Mcm2 chromatin association (Figure 8C), although there is a small but reproducible effect on the proportion of cells with chromatin-associated Cdc45 (Figure 8B). When cells were released from the HU block but maintained at 37°C, a significant proportion of cells maintained Cdc45 chromatin binding and nuclear division did not take place, whereas at 25°C or in the wild-type strain at either temperature, DNA synthesis was completed as assessed by loss of Cdc45 chromatin binding and the onset of nuclear division (Figure 8, B and D). The inability of cdc23 mutant cells to complete S phase when released from the HU block implies that Cdc23 is required for the elongation or termination steps of replication and concurs with earlier studies (Nasmyth and Nurse, 1981; Kawasaki et al., 2000). However, inactivation of Cdc23 has a more dramatic effect on the establishment rather than the maintenance of Cdc45 chromatin binding as assessed by detergent extraction. We have been unable to examine the effect of Cdc23 inactivation on maintenance of Cdc45 chromatin association in the absence of HU because of problems in inactivating Cdc23 quickly in nonarrested cells.
Figure 8.
Inactivation of Cdc23 during an S phase arrest blocks S phase completion (A) Experimental procedure. Strains used were P1051 (mcm2-CFP); P1220 (mcm2-CFP, nmt-cdc23tstd); P1083 (cdc45-YFP), and P1221 (cdc45-YFP, nmt-cdc23tstd). Numbers in parentheses indicate experimental stages, which are referred to in other parts of the figure. (B) Quantitation of chromatin binding data for Cdc45 for the stages in the experiment shown in A. Left panel: data for cells at 25°C; right panel: data for where cells were shifted to 37°C. (C) Quantitation of chromatin binding data for Mcm2 for the stages in the experiment shown in A. The Mcm2 experiments were terminated at stages 3 and 4. (D) Flow cytometry analysis for data shown in B. In this experiment the peaks drift to the right due to cell elongation at 37°C. The percentages of binucleate cells at stages (5–8) are also shown. (E) Western analysis of Cdc23 levels during experiment; α-tubulin is shown as a loading control.
DISCUSSION
In this article we have extended the use of fluorescently tagged replication proteins to study the function of replication factors in fission yeast. Cells containing tagged Mcm2–7 and Cdc45 allow two steps leading to S phase to be monitored in single cells, one corresponding to pre-RC formation and the other occurring around DNA replication onset. Cdc45 chromatin association should provide a useful cytological method to distinguish cells in late mitosis/G1 from those in S phase and offers an alternative to methods that cannot be applied to single cells and require synchronization of cell populations.
Using this approach, we have shown that Cdc23 functions after Mcm2 chromatin binding, implying that it is not needed for pre-RC formation, but is necessary for the chromatin association of Cdc45 during replication initiation. This function is conserved between vertebrates and fission yeast, because comparable findings for Mcm10 function have been reported using a soluble in vitro system for DNA replication derived from Xenopus eggs (Wohlschlegel et al., 2002). However, in S. cerevisiae, inactivation of Mcm10 leads to loss of chromatin-associated Mcm2 in G1-arrested cells, leading to the conclusion that Mcm10 is necessary for the earlier step of pre-RC formation (Homesley et al., 2000). Further work will be required to determine whether this represents an Mcm10 function not conserved in fission yeast or Xenopus or is a result of differences in experimental design.
What precisely is the biochemical role of Cdc23/Mcm10 in stimulating Cdc45 chromatin binding? One possibility is that it acts as a molecular tether between Cdc45 and other components of the pre-RC, to allow Cdc45 to associate with chromatin. Once loaded, Cdc45 could carry out origin unwinding (Walter and Newport, 2000) and subsequent assembly of RPA, polymerase α, and polymerase ε during initiation (Mimura and Takisawa, 1998; Mimura et al., 2000; Uchiyama et al., 2001). Cdc45 is required for elongation of replication forks (Tercero et al., 2000), and Cdc23/Mcm10 could be essential for elongation by maintaining Cdc45 chromatin association during DNA synthesis. However, we find that if cells are arrested in S phase with HU after the Cdc45 chromatin-binding step, and then Cdc23 is inactivated, most cells retain Cdc45 chromatin association. The possibility that incomplete inactivation of Cdc23 is the explanation for a modest reduction in chromatin bound Cdc45 is not supported by the observation that when cells are released from the HU block in this experiment, they fail to complete S phase. This implies that the cells that retain chromatin associated Cdc45 are incapable of completing DNA replication in the absence of Cdc23 function. One interpretation of these results is that Cdc23/Mcm10 is not simply a tether for Cdc45, but affects Cdc45 chromatin association indirectly. For instance, Cdc23/Mcm10 could catalyze a step after pre-RC formation that is needed for both initiation and elongation. Cdc45 could bind as a consequence of this function at initiation, but, once bound, maintenance of its chromatin association would not be so dependent on Cdc23/Mcm10's function during elongation. While this article was under review, Lee et al. (2003) reported that the in vitro phosphorylation of Mcm2 and Mcm4 by Hsk1 is stimulated by Cdc23. If the critical event for Cdc45 chromatin binding is this Mcm2,4 phosphorylation event, this would explain the dependence of Cdc45 chromatin binding on Mcm4, Hsk1, and Cdc23 reported here.
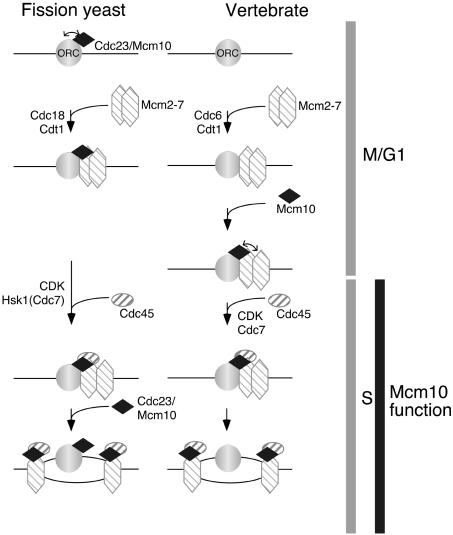
In spite of the common Cdc23/Mcm10 function between fission yeast and vertebrates, the periodicity of Mcm10 chromatin association during the vertebrate cell cycle contrasts with the constitutive binding of Cdc23 in fission yeast cells. The possible protein interactions that are important for Cdc23/Mcm10 chromatin association are shown in the model in Figure 9. Chromatin association of fission yeast Cdc23 is shown to occur via ORC and, although direct evidence is lacking, this interaction is plausible based on interactions with ORC subunits in fission yeast (Hart et al., 2002) and humans (Izumi et al., 2000) as well as enrichment at origin sequences in S. cerevisiae (Homesley et al., 2000). To explain the elongation requirement for Cdc23, Cdc23 is shown departing with the replication forks in association with the putative Mcm2–7 helicase, allowing ORC to bind free Cdc23. Interaction between Cdc23 and Mcm2–7 proteins is suggested by a number of studies (Merchant et al., 1997; Homesley et al., 2000; Izumi et al., 2000; Liang and Forsburg, 2001; Hart et al., 2002), although from a consideration of the relative levels of these proteins in fission yeast only a small proportion of total Mcm2–7 can be associated in a complex with Cdc23. An alternative explanation for the function of Cdc23 during elongation that does not require its participation at the replication fork is that its presence at ORC could facilitate the passive replication of unfired origins, as suggested by pausing of forks at origins in a budding yeast mcm10 mutant (Homesley et al., 2000).
Figure 9.
Model showing possible basis of Cdc23/Mcm10 chromatin association in fission yeast and vertebrate cell cycles. Arrows indicate hypothetical protein-protein interactions that may be important for establishing chromatin association of Cdc23/Mcm10. For details see text.
In vertebrates, the main difference compared with fission yeast is that Mcm10 only binds after Mcm2–7 chromatin association, perhaps as interaction with these proteins rather than ORC is important for Mcm10 chromatin binding (Figure 9). This is consistent with the observation that during S phase, Mcm10 disassociates along with Mcm2–7 proteins from chromatin (Wohlschlegel et al., 2002). Comparison of Mcm10 sequences reveals that metazoan proteins have a C-terminal extension not found in yeasts (Izumi et al., 2000), which contains a conserved domain, and it will be of interest to determine whether this is relevant to the distinct chromatin binding properties of vertebrate Mcm10.
These nonconserved patterns of Cdc23/Mcm10 chromatin association during the cell cycle comparing yeasts and vertebrates are intriguingly similar to those seen with Cdc7. S. cerevisiae Cdc7 is also bound to chromatin throughout the cell cycle (Weinreich and Stillman, 1999), and the chromatin interaction of the Dbf4 regulatory subunit of Cdc7 is dependent on ORC (Pasero et al., 1999; Duncker et al., 2002). In contrast, Xenopus Cdc7 requires prior binding of Mcm2–7 proteins for its chromatin interaction (Jares and Blow, 2000). If chromatin associations of Cdc7 and Mcm10, which both function around initiation, are solely dependent on Mcm2–7 proteins in vertebrates, but dependent on ORC in yeasts, this could reflect a dispensability of ORC for initiation in vertebrates once Mcm2–7 chromatin binding has occurred. This is relevant to the consideration of models suggesting that Mcm2–7 complexes in vertebrates may become distributed over a large region of DNA after loading at ORC, before initiation, (Ritzi et al., 1998; Edwards et al., 2002), thus potentially allowing initiation away from ORC.
In this work we have established that quiescent fission yeast cells must reestablish Cdc23 chromatin binding as a requirement for DNA replication, and it will be of interest to establish whether this possible coupling between growth and DNA replication has any regulatory significance. We have shown that this event is independent of Mcm2–7 chromatin association and thus does not seem to be related to the discrete binding of Mcm10 that occurs in vertebrate cell cycles after pre-RC formation. Growing fission yeast cells differ from vertebrate cells in that Mcm10 chromatin binding does not have to be re-established after mitosis, and if this step is rate limiting, it is possible that vertebrates thus have an additional regulatory step in G1 to control DNA replication that is not present in yeast. There are precedents for differences in replication control comparing eukaryotes. Yeasts lack vertebrate replication controls involving geminin (Wohlschlegel et al., 2000; Tada et al., 2001) and destabilization of ORC1 chromatin association after replication initiation (Mendez et al., 2002; Sun et al., 2002), perhaps because unicellular organisms with small genomes can tolerate a lower fidelity of replication control.
Acknowledgments
The authors thank Karim Labib for anti-GFP antibody and Robin Allshire, Beata Grallert, Angus Lamond, Janet Leatherwood, Hisao Masai, Alison Pidoux, and Paul Nurse's lab for strains and plasmids. The authors also thank Shao-Win Wang and Karim Labib for comments on the manuscript and Lynne Larkman for technical assistance. This work was supported by grants from Cancer Research UK and the Association for International Cancer Research.
Article published online ahead of print. Mol. Biol. Cell 10.1091/mbc.E03–02–0090. Article and publication date are available at www.molbiolcell.org/cgi/doi/10.1091/mbc.E03-02-0090.
Abbreviations used: CDK, cyclin dependent kinase; CFP, cyan fluorescent protein; GFP, green fluorescent protein; HU, hydroxyurea; Mcm, minichromosome maintenance; ORC, origin recognition complex; pre-RC, pre-replicative complex; YFP, yellow fluorescent protein
References
- Aparicio, O.M., Weinstein, D.M., and Bell, S.P. (1997). Components and dynamics of DNA replication complexes in S. cerevisiae: redistribution of MCM proteins and Cdc45p during S phase. Cell 91, 59–69. [DOI] [PubMed] [Google Scholar]
- Aves, S.J., Tongue, N., Foster, A.J., and Hart, E.A. (1998). The essential Schizosaccharomyces pombe cdc23 DNA replication gene shares structural and functional homology with the Saccharomyces cerevisiae DNA43 (MCM10) gene. Curr. Genet. 34, 164–171. [DOI] [PubMed] [Google Scholar]
- Basi, G., Schmid, E., and Maundrell, K. (1993). TATA box mutations in the Schizosaccharomyces pombe nmt1 promoter affect transcription efficiency but not the transcription start point or thiamine repressibility. Gene 123, 131–136. [DOI] [PubMed] [Google Scholar]
- Bell, S.P., and Dutta, A. (2002). DNA replication in eukaryotic cells. Annu. Rev Biochem. 71, 333–374. [DOI] [PubMed] [Google Scholar]
- Blow, J.J., and Hodgson, B. (2002). Replication licensing— defining the proliferation control. Trends. Cell Biol. 12, 72–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diffley, J.F.X., and Labib, K. (2002). The chromosome replication cycle. J. Cell Sci. 115, 869–872. [DOI] [PubMed] [Google Scholar]
- Ding, R., and Smith, G.R. (1998). Global control of meiotic recombination genes by Schizosaccharomyces pombe rec16 (rep1). Mol. Gen. Genet. 258, 663–670. [DOI] [PubMed] [Google Scholar]
- Dohman, R.J., Wu, P., and Varshavsky, A. (1994). Heat-inducible degron: a method for constructing temperature-sensitive mutants. Science 263, 1273–1276. [DOI] [PubMed] [Google Scholar]
- Donovan, S., J, Harwood, Drury, LS, and Diffley, J.F.X. (1997). Cdc6p-dependent loading of Mcm proteins onto pre-replicative chromatin in budding yeast. Proc. Natl. Acad. Sci. USA 94, 5611–5616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumas, L.B., Lussky, J.P., McFarland, E.J., and Shampay, J. (1982). New temperature-sensitive mutants of Saccharomyces cerevisiae affecting DNA replication. Mol. Gen. Genet. 187, 42–46. [DOI] [PubMed] [Google Scholar]
- Duncker, B.P., Shimada, K., Tsai Pflugfelder, M., Pasero, P., and Gasser, S.M. (2002). An N-terminal domain of Dbf4p mediates interaction with both origin recognition complex (ORC) and Rad53p and can deregulate late origin firing. Proc. Natl. Acad. Sci. Sci. USA 99, 16087–16092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards, M.C. et al. (2002) MCM2–7 Complexes bind chromatin in a distributed pattern surrounding the origin recognition complex in Xenopus egg extracts. J. Biol. Chem. 277, 33049–57. [DOI] [PubMed] [Google Scholar]
- Fisher, D.L., and Nurse, P. (1995). A single fission yeast mitotic cyclin B-p34cdc2 kinase promotes both S-phase and mitosis in the absence of G1-cyclins. EMBO J. 15, 850–860. [PMC free article] [PubMed] [Google Scholar]
- Grallert, B., Kearsey, S.E., Lenhard, M., Carlson, C.R., Nurse, P., Boye, E., and Labib, K. (2000). A fission yeast general translation factor reveals links between protein synthesis and cell cycle controls. J. Cell Sci. 113, 1447–1458. [DOI] [PubMed] [Google Scholar]
- Grallert, B., and Nurse, P. (1997). An approach to identify functional homologues and suppressors of genes in fission yeast. Curr. Genet. 32, 27–31. [DOI] [PubMed] [Google Scholar]
- Hart, E.A., Bryant, John A., Moore, K., and Aves, S.J. (2002). Fission yeast Cdc23 interactions with DNA replication initiation proteins. Curr. Genet. 41, 342–348. [DOI] [PubMed] [Google Scholar]
- Homesley, L., Lei, M., Kawasaki, Y., Sawyer, S., Christensen, T., and Tye, B.K. (2000). Mcm10 and the MCM2–7 complex interact to initiate DNA synthesis and to release replication factors from origins. Genes Dev. 14, 913–926. [PMC free article] [PubMed] [Google Scholar]
- Ishimi, Y. (1997). A DNA helicase activity is associated with an MCM4, 6 and -7 protein complex. J. Biol. Chem. 272, 24508–24513. [DOI] [PubMed] [Google Scholar]
- Izumi, M., Yanagi, K., Mizuno, T., Yokoi, M., Kawasaki, Y., Moon, K.Y., Hurwitz, J., Yatagai, F., and Hanaoka, F. (2000). The human homolog of Saccharomyces cerevisiae Mcm10 interacts with replication factors and dissociates from nuclease-resistant nuclear structures in G(2) phase. Nucleic Acids Res. 28, 4769–4777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izumi, M., Yatagai, F., and Hanaoka, F. (2001). Cell cycle-dependent proteolysis and phosphorylation of human Mcm10. J. Biol. Chem. 276, 48526–48531. [DOI] [PubMed] [Google Scholar]
- Jares, P., and Blow, J.J. (2000). Xenopus cdc7 function is dependent on licensing but not on XORC, XCdc6, or CDK activity and is required for XCdc45 loading. Genes Dev. 14, 1528–1540. [PMC free article] [PubMed] [Google Scholar]
- Kamimura, Y., Tak, Y.S., Sugino, A., and Araki, H. (2001). Sld3, which interacts with Cdc45 (Sld4), functions for chromosomal DNA replication in Saccharomyces cerevisiae. EMBO J. 20, 2097–2107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawasaki, Y., Hiraga, S., and Sugino, A. (2000). Interactions between Mcm10p and other replication factors are required for proper initiation and elongation of chromosomal DNA replication in Saccharomyces cerevisiae. Genes Cells 5, 975–989. [DOI] [PubMed] [Google Scholar]
- Kearsey, S.E., Montgomery, S. Labib, K., and Lindner, K. (2000) Chromatin binding of the fission yeast replication factor mcm4 occurs during anaphase and requires ORC and cdc18. EMBO J. 19, 1681–1690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, J.K., Seo, Y.S., and Hurwitz, J. (2003). The Cdc23 (Mcm10) protein is required for the phosphorylation of minichromosome maintenance complex by the Dfp1-Hsk1 kinase. Proc. Natl. Acad. Sci. USA 100, 2334–2339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang, D.T., and Forsburg, S.L. (2001). Characterization of Schizosaccharomyces pombe mcm7+ and cdc23+ (MCM10) and interactions with replication checkpoints. Genetics 159, 471–486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindner, K., Gregan, J., Montgomery, S., and Kearsey, S. (2002). Essential role of MCM proteins in pre-meiotic DNA replication. Mol. Biol. Cell 13, 435–444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lygerou, Z., and Nurse, P. (1999). The fission yeast origin recognition complex is constitutively associated with chromatin and is differentially modified through the cell cycle. J. Cell Sci. 112, 3703–3712. [DOI] [PubMed] [Google Scholar]
- Maine, G.T., Sinha, P., and Tye, B.K. (1984). Mutants of S. cerevisiae defective in the maintenance of minichromosomes. Genetics 106, 365–385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maiorano, D., Moreau, J., and Mechali, M. (2000). XCDT1 is required for the assembly of pre-replicative complexes in Xenopus laevis. Nature 404, 622–625. [DOI] [PubMed] [Google Scholar]
- Masai, H., and Arai, K.I. (2002). Cdc7 kinase complex: a key regulator in the initiation of DNA replication. J. Cell Physiol. 190, 287–296. [DOI] [PubMed] [Google Scholar]
- McGarry, T.J., and Kirschner, M.W. (1998). Geminin, an inhibitor of DNA replication, is degraded during mitosis. Cell 93, 1043–1053. [DOI] [PubMed] [Google Scholar]
- Mendez, J., Zou Yang, X.H., Kim, S.Y., Hidaka, M., Tansey, W.P., and Stillman, B. (2002). Human origin recognition complex large subunit is degraded by ubiquitin-mediated proteolysis after initiation of DNA replication. Mol. Cell 9, 481–491. [DOI] [PubMed] [Google Scholar]
- Merchant, A.M., Kawasaki, Y., Chen, Y., Lei, M., and Tye, B.K. (1997). A lesion in the DNA replication initiation factor MCM10 induces pausing of replication forks through chromosomal origins in S. cerevisiae. Mol. Cell. Biol. 17, 3261–3271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mimura, S., Masuda, T., Matsui, T., and Takisawa, H. (2000). Central role for cdc45 in establishing an initiation complex of DNA replication in Xenopus egg extracts. Genes Cells 5, 439–52. [DOI] [PubMed] [Google Scholar]
- Mimura, S., and Takisawa, H. (1998). Xenopus Cdc45-dependent loading of DNA polymerase alpha onto chromatin under the control of S-phase Cdk. EMBO J. 17, 5699–5707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyake, S., and Yamashita, S. (1998). Identification of sna41 gene, which is the suppressor of nda4 mutation and is involved in DNA replication in Schizosaccharomyces pombe. Genes Cells 3, 157–166. [DOI] [PubMed] [Google Scholar]
- Moreno, S., Klar, A., and Nurse, P. (1991). Molecular genetic analysis of the fission yeast Schizosaccharomyces pombe. Methods Enzymol. 194, 795–823. [DOI] [PubMed] [Google Scholar]
- Nakajima, R., and Masukata, H. (2002) SpSld3 is required for loading and maintenance of SpCdc45 on chromatin in DNA replication in fission yeast. Mol. Biol. Cell 13, 1462–1472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasmyth, K.A., and Nurse, P. (1981). Cell division mutants altered in DNA replication and mitosis in the fission yeast Schizosaccharomyces pombe. Mol. Gen. Genet. 182, 119–124. [DOI] [PubMed] [Google Scholar]
- Nguyen, V.Q., Co, C., and Li, J.J. (2001). Cyclin-dependent kinases prevent DNA re-replication through multiple mechanisms. Nature 411, 1068–1073. [DOI] [PubMed] [Google Scholar]
- Nishitani, H., Lygerou, Z., Nishimoto, T., and Nurse, P. (2000). The Cdt1 protein is required to license DNA for replication in fission yeast. Nature 404, 625–628. [DOI] [PubMed] [Google Scholar]
- Pasero, P., Duncker, B.P., Schwob, E., and Gasser, S.M. (1999). A role for the Cdc7 kinase regulatory subunit Dbf4p in the formation of initiation-competent origins of replication. Genes Dev. 13, 2159–2176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pidoux, A., Uzawa, S., Perry, P., Cande, W., and Allshire, R. (2000). Live analysis of lagging chromosome during anaphase and their effect on spindle elongation rate in fission yeast. J. Cell Sci. 113, 4177–4191. [DOI] [PubMed] [Google Scholar]
- Ritzi, M., Baack, M., Musahl, C., Romanowski, P., Laskey, R.A., and Knippers, R. (1998). Human minichromosome maintenance proteins and human origin recognition complex 2 protein on chromatin. J. Biol. Chem. 273, 24543–24549. [DOI] [PubMed] [Google Scholar]
- Sclafani, R.A. (2000). Cdc7p-Dbf4p becomes famous in the cell cycle. J. Cell Sci. 113, 2111–2117. [DOI] [PubMed] [Google Scholar]
- Solomon, N.A., Wright, M.B., Chang, S., Buckley, A.M., Dumas, L.B., and Gaber, R.F. (1992). Genetic and molecular analysis of DNA43 and DNA52: two new cell-cycle genes in Saccharomyces cerevisiae. Yeast 8, 273–289. [DOI] [PubMed] [Google Scholar]
- Sun, W.H., Coleman, T.R., and DePamphilis, M.L. (2002). Cell cycle-dependent regulation of the association between origin recognition proteins and somatic cell chromatin. EMBO J. 21, 1437–1446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tada, S., Li, A., Maiorano, D., Méchali, M., and Blow, J.J. (2001). Repression of origin assembly in metaphase depends on inhibition of RLF-B/Cdt1 by geminin. Nat. Cell Biol. 3, 107–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeda, T., Ogino, K., Tatebayashi, K., Ikeda, H., Arai, K., and Masai, H. (2001). Regulation of initiation of S phase, replication checkpoint signaling, and maintenance of mitotic chromosome structures during S phase by Hsk1 kinase in the fission yeast. Mol. Biol. Cell 12, 1257–1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takisawa, H., Mimura, S., and Kubota, Y. (2000). Eukaryotic DNA replication: from pre-replication complex to initiation complex. Curr. Opin. Cell Biol. 12, 690–696. [DOI] [PubMed] [Google Scholar]
- Tanaka, H., Tanaka, K., Murakami, H., and Okayama, H. (1999). Fission yeast cdc24 is a replication factor Cand proliferating cell nuclear antigen-interacting factor essential for S-phase completion. Mol. Cell. Biol. 19, 1038–1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka, T., Knapp, D., and Nasmyth, K. (1997). Loading of an Mcm protein onto DNA replication origins is regulated by Cdc6p and CDKs. Cell 90, 649–660. [DOI] [PubMed] [Google Scholar]
- Tercero, J.A., Labib, K., and Diffley, J.F. (2000). DNA synthesis at individual replication forks requires the essential initiation factor Cdc45p. EMBO J. 19, 2082–2093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uchiyama, M., Griffiths, D., Arai, K., and Masai, H. (2001). Essential role of Sna41/Cdc45 in loading of DNA polymerase alpha onto minichromosome maintenance proteins in fission yeast. J. Biol. Chem. 276, 26189–26196. [DOI] [PubMed] [Google Scholar]
- Walter, J., and Newport, J. (2000). Initiation of eukaryotic DNA replication: origin unwinding and sequential chromatin association of Cdc45, RPA, and DNA polymerase alpha. Mol. Cell 5, 617–627. [DOI] [PubMed] [Google Scholar]
- Weinreich, M., and Stillman, B. (1999). Cdc7p-Dbf4p kinase binds to chromatin during S phase and is regulated by both the APC and the RAD53 checkpoint pathway. EMBO J. 18, 5334–5346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wohlschlegel, J.A., Dhar, S.K., Prokhorova, T.A., Dutta, A., and Walter, J.C. (2002). Xenopus Mcm10 binds to origins of DNA replication after Mcm2–7 and stimulates origin binding of Cdc45. Mol. Cell 9, 233–240. [DOI] [PubMed] [Google Scholar]
- Wohlschlegel, J.A., Dwyer, B.T., Dhar, S.K., Cvetic, C., Walter, J.C., and Dutta, A. (2000). Inhibition of eukaryotic DNA replication by geminin binding to Cdt1. Science 290, 2309–2312. [DOI] [PubMed] [Google Scholar]
- Zou, L., and Stillman, B. (2000). Assembly of a complex containing Cdc45p, replication protein A, and Mcm2p at replication origins controlled by S-phase cyclin-dependent kinases and Cdc7p-Dbf4p kinase. Mol. Cell. Biol. 20, 3086–3096. [DOI] [PMC free article] [PubMed] [Google Scholar]