Abstract
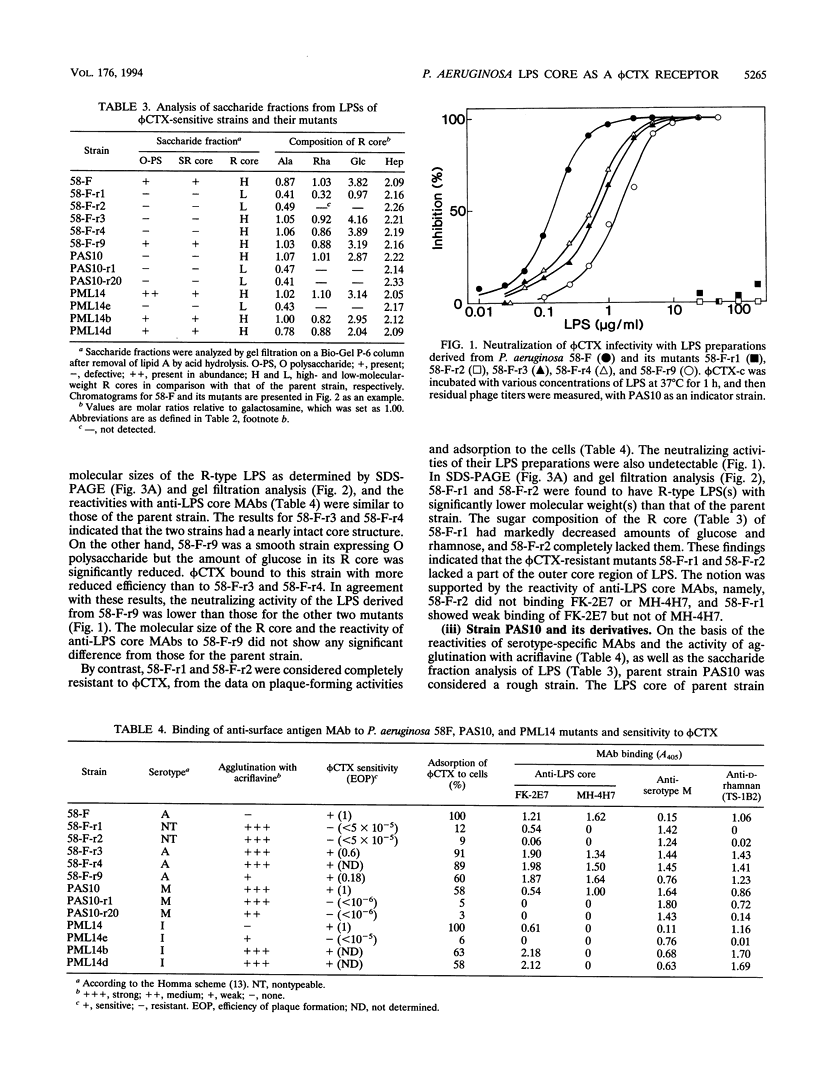
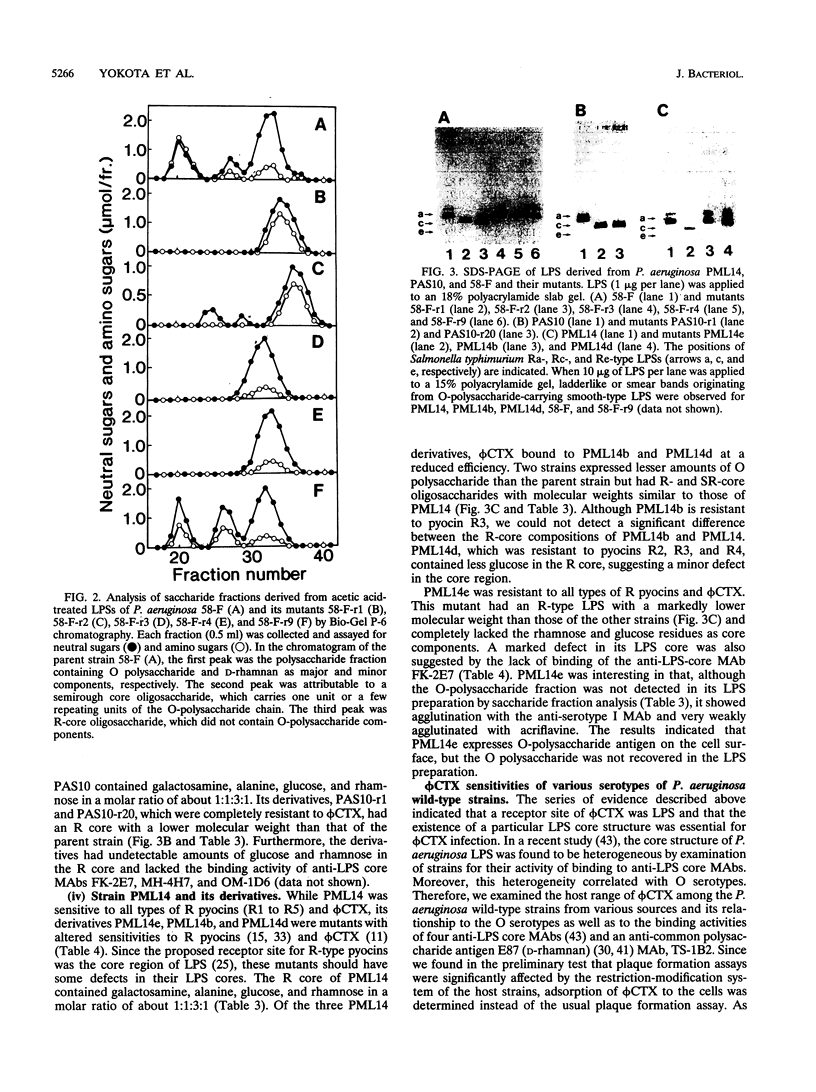
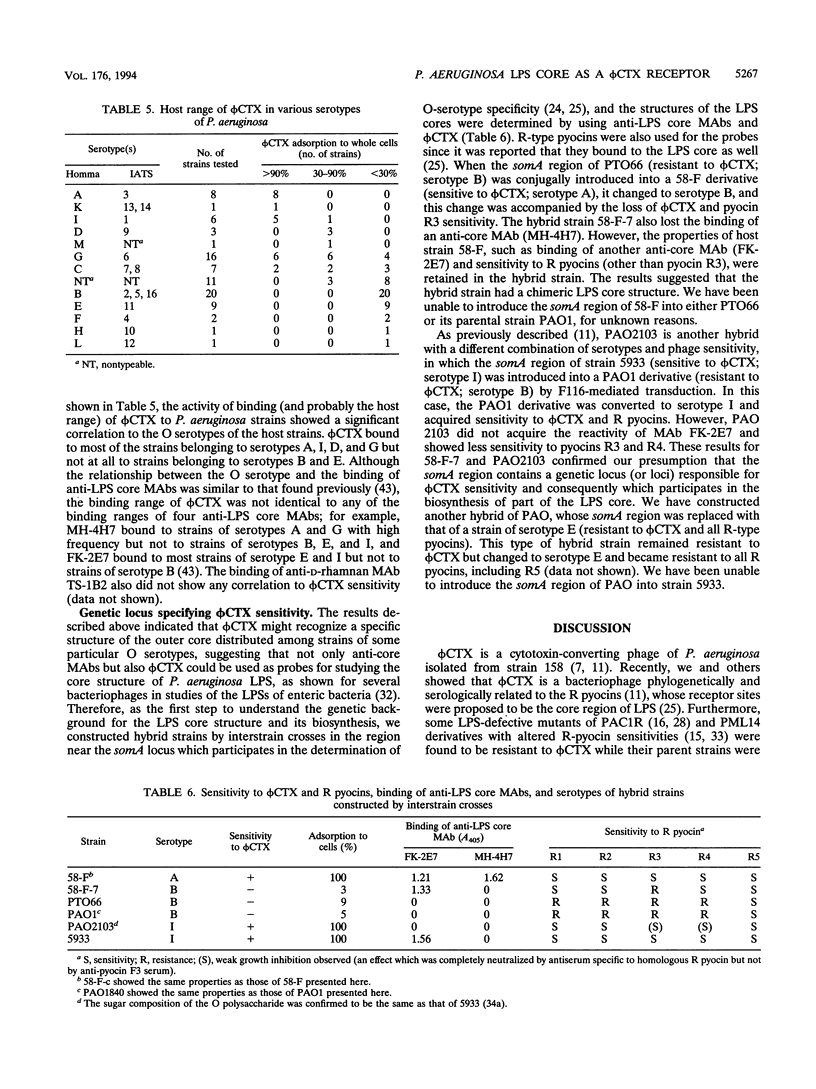
A temperate phage, phi CTX, is a cytotoxin-converting phage of Pseudomonas aeruginosa. In this study, we characterized the lipopolysaccharide (LPS) structures of phi CTX-resistant mutants derived from phi CTX-sensitive strains. phi CTX infectivity was neutralized by LPS preparations derived from sensitive strains but not by those from resistant strains. phi CTX-resistant mutants had lower-molecular-weight rough (R)-type LPS than the parental strains and lacked the reactivity of some anti-LPS core monoclonal antibodies. Some LPS core components were lacking or significantly decreased in the resistant mutants. These results suggested that a receptor site of the cytotoxin-converting phage phi CTX was the LPS core region and that especially L-rhamnose and D-glucose residues in the outer core were involved in phage binding. The host range of phi CTX was nearly O-serotype dependent, probably because of the diversity of the LPS core structure among P. aeruginosa strains. phi CTX bound to most strains of Homma serotypes A, G, and I but not to strains of serotypes B and E. Furthermore, we found that a genetic locus specifying phi CTX sensitivity (and consequently participating in the biosynthesis of part of the LPS core) existed in or near the locus participating in the determination of O-serotype specificity (somA), which has been mapped between leu-10 and eda-9001. phi CTX, as well as anti-LPS core monoclonal antibodies, will be a good tool for structural characterization of the P. aeruginosa LPS core region.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Brown P. R., Clarke P. H. Amino acid substitution in an amidase produced by an acetanilide-utilizing mutant of Pseudomonas aeruginosa. J Gen Microbiol. 1972 Apr;70(2):287–288. doi: 10.1099/00221287-70-2-287. [DOI] [PubMed] [Google Scholar]
- Früh R., Watson J. M., Haas D. Construction of recombination-deficient strains of Pseudomonas aeruginosa. Mol Gen Genet. 1983;191(2):334–337. doi: 10.1007/BF00334835. [DOI] [PubMed] [Google Scholar]
- Goldberg J. B., Hatano K., Meluleni G. S., Pier G. B. Cloning and surface expression of Pseudomonas aeruginosa O antigen in Escherichia coli. Proc Natl Acad Sci U S A. 1992 Nov 15;89(22):10716–10720. doi: 10.1073/pnas.89.22.10716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HABS I. Untersuchungen über die O-Antigene von Pseudomonas aeruginosa. Z Hyg Infektionskr. 1957;144(3):218–228. [PubMed] [Google Scholar]
- Hayashi T., Baba T., Matsumoto H., Terawaki Y. Phage-conversion of cytotoxin production in Pseudomonas aeruginosa. Mol Microbiol. 1990 Oct;4(10):1703–1709. doi: 10.1111/j.1365-2958.1990.tb00547.x. [DOI] [PubMed] [Google Scholar]
- Hayashi T., Kamio Y., Hishinuma F., Usami Y., Titani K., Terawaki Y. Pseudomonas aeruginosa cytotoxin: the nucleotide sequence of the gene and the mechanism of activation of the protoxin. Mol Microbiol. 1989 Jul;3(7):861–868. doi: 10.1111/j.1365-2958.1989.tb00235.x. [DOI] [PubMed] [Google Scholar]
- Hayashi T., Kamio Y., Terawaki Y. Purification and characterization of cytotoxin from the crude extract of Pseudomonas aeruginosa. Microb Pathog. 1989 Feb;6(2):103–112. doi: 10.1016/0882-4010(89)90013-2. [DOI] [PubMed] [Google Scholar]
- Hayashi T., Matsumoto H., Ohnishi M., Terawaki Y. Molecular analysis of a cytotoxin-converting phage, phi CTX, of Pseudomonas aeruginosa: structure of the attP-cos-ctx region and integration into the serine tRNA gene. Mol Microbiol. 1993 Mar;7(5):657–667. doi: 10.1111/j.1365-2958.1993.tb01157.x. [DOI] [PubMed] [Google Scholar]
- Kageyama M., Shinomiya T., Aihara Y., Kobayashi M. Characterization of a bacteriophage related to R-type pyocins. J Virol. 1979 Dec;32(3):951–957. doi: 10.1128/jvi.32.3.951-957.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koval S. F., Meadow P. M. The isolation and characterization of lipopolysaccharide-defective mutants of Pseudomonas aeruginosa PAC1. J Gen Microbiol. 1977 Feb;98(2):387–398. doi: 10.1099/00221287-98-2-387. [DOI] [PubMed] [Google Scholar]
- Kropinski A. M., Chan L. C., Milazzo F. H. The extraction and analysis of lipopolysaccharides from Pseudomonas aeruginosa strain PAO, and three rough mutants. Can J Microbiol. 1979 Mar;25(3):390–398. doi: 10.1139/m79-060. [DOI] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lightfoot J., Lam J. S. Chromosomal mapping, expression and synthesis of lipopolysaccharide in Pseudomonas aeruginosa: a role for guanosine diphospho (GDP)-D-mannose. Mol Microbiol. 1993 May;8(4):771–782. doi: 10.1111/j.1365-2958.1993.tb01620.x. [DOI] [PubMed] [Google Scholar]
- Lindberg A. A. Studies of a receptor for felix O-1 phage in Salmonella minnesota. J Gen Microbiol. 1967 Aug;48(2):225–233. doi: 10.1099/00221287-48-2-225. [DOI] [PubMed] [Google Scholar]
- Ratnaningsih E., Dharmsthiti S., Krishnapillai V., Morgan A., Sinclair M., Holloway B. W. A combined physical and genetic map of Pseudomonas aeruginosa PAO. J Gen Microbiol. 1990 Dec;136(12):2351–2357. doi: 10.1099/00221287-136-12-2351. [DOI] [PubMed] [Google Scholar]
- Roehl R. A., Feary T. W., Phibbs P. V., Jr Clustering of mutations affecting central pathway enzymes of carbohydrate catabolism in Pseudomonas aeruginosa. J Bacteriol. 1983 Dec;156(3):1123–1129. doi: 10.1128/jb.156.3.1123-1129.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowe P. S., Meadow P. M. Structure of the Core oligosaccharide from the lipopolysaccharide of Pseudomonas aeruginosa PAC1R and its defective mutants. Eur J Biochem. 1983 May 2;132(2):329–337. doi: 10.1111/j.1432-1033.1983.tb07366.x. [DOI] [PubMed] [Google Scholar]
- Royle P. L., Matsumoto H., Holloway B. W. Genetic circularity of the Pseudomonas aeruginosa PAO chromosome. J Bacteriol. 1981 Jan;145(1):145–155. doi: 10.1128/jb.145.1.145-155.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawada S., Kawamura T., Masuho Y., Tomibe K. A new common polysaccharide antigen of strains of Pseudomonas aeruginosa detected with a monoclonal antibody. J Infect Dis. 1985 Dec;152(6):1290–1299. doi: 10.1093/infdis/152.6.1290. [DOI] [PubMed] [Google Scholar]
- Scharmann W. Formation and isolation of leucocidin from Pseudomonas aeruginosa. J Gen Microbiol. 1976 Apr;93(2):283–291. doi: 10.1099/00221287-93-2-283. [DOI] [PubMed] [Google Scholar]
- Schnaitman C. A., Klena J. D. Genetics of lipopolysaccharide biosynthesis in enteric bacteria. Microbiol Rev. 1993 Sep;57(3):655–682. doi: 10.1128/mr.57.3.655-682.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinomiya T., Shiga S. Bactericidal activity of the tail of Pseudomonas aeruginosa bacteriophage PS17. J Virol. 1979 Dec;32(3):958–967. doi: 10.1128/jvi.32.3.958-967.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinomiya T., Shiga S., Kikuchi A., Kageyama M. Genetic determinant of pyocin R2 in Pseudomonas aeruginosa PAO. II. Physical characterization of pyocin R2 genes using R-prime plasmids constructed from R68.45. Mol Gen Genet. 1983;189(3):382–389. doi: 10.1007/BF00325899. [DOI] [PubMed] [Google Scholar]
- Tsai C. M., Frasch C. E. A sensitive silver stain for detecting lipopolysaccharides in polyacrylamide gels. Anal Biochem. 1982 Jan 1;119(1):115–119. doi: 10.1016/0003-2697(82)90673-x. [DOI] [PubMed] [Google Scholar]
- Tsuji A., Kinoshita T., Hoshino M. Microdetermination of hexosamines. Chem Pharm Bull (Tokyo) 1969 Jan;17(1):217–218. doi: 10.1248/cpb.17.217. [DOI] [PubMed] [Google Scholar]
- VERDER E., EVANS J. A proposed antigenic schema for the identification of strains of Pseudomonas aeruginosa. J Infect Dis. 1961 Sep-Oct;109:183–193. doi: 10.1093/infdis/109.2.183. [DOI] [PubMed] [Google Scholar]
- Wright B. G., Rebers P. A. Procedure for determining heptose and hexose in lipopolysaccharides. Modification of the cysteine-sulfuric acid method. Anal Biochem. 1972 Oct;49(2):307–319. doi: 10.1016/0003-2697(72)90433-2. [DOI] [PubMed] [Google Scholar]
- Yokota S., Kaya S., Araki Y., Ito E., Kawamura T., Sawada S. Occurrence of D-rhamnan as the common antigen reactive against monoclonal antibody E87 in Pseudomonas aeruginosa IFO 3080 and other strains. J Bacteriol. 1990 Oct;172(10):6162–6164. doi: 10.1128/jb.172.10.6162-6164.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokota S., Ochi H., Ohtsuka H., Kato M., Noguchi H. Heterogeneity of the L-rhamnose residue in the outer core of Pseudomonas aeruginosa lipopolysaccharide, characterized by using human monoclonal antibodies. Infect Immun. 1989 Jun;57(6):1691–1696. doi: 10.1128/iai.57.6.1691-1696.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokota S., Terashima M., Chiba J., Noguchi H. Variable cross-reactivity of Pseudomonas aeruginosa lipopolysaccharide-code-specific monoclonal antibodies and its possible relationship with serotype. J Gen Microbiol. 1992 Feb;138(2):289–296. doi: 10.1099/00221287-138-2-289. [DOI] [PubMed] [Google Scholar]



