Abstract
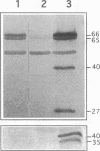
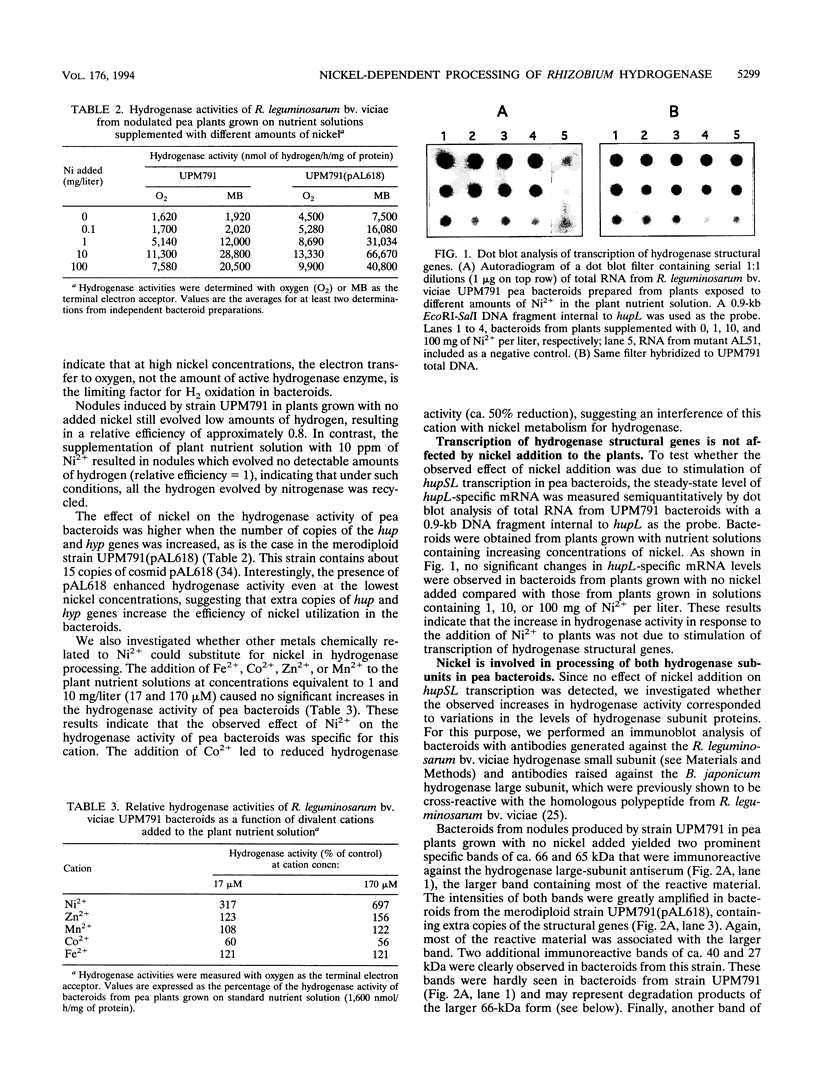
Rhizobium leguminosarum bv. viciae UPM791 induces the synthesis of an [NiFe] hydrogenase in pea (Pisum sativum L.) bacteroids which oxidizes the H2 generated by the nitrogenase complex inside the root nodules. The synthesis of this hydrogenase requires the genes for the small and large hydrogenase subunits (hupS and hupL, respectively) and 15 accessory genes clustered in a complex locus in the symbiotic plasmid. We show here that the bacteroid hydrogenase activity is limited by the availability of nickel to pea plants. Addition of Ni2+ to plant nutrient solutions (up to 10 mg/liter) resulted in sharp increases (up to 15-fold) in hydrogenase activity. This effect was not detected when other divalent cations (Zn2+, Co2+, Fe2+, and Mn2+) were added at the same concentrations. Determinations of the steady-state levels of hupSL-specific mRNA indicated that this increase in hydrogenase activity was not due to stimulation of transcription of structural genes. Immunoblot analysis with antibodies raised against the large and small subunits of the hydrogenase enzyme demonstrated that in the low-nickel situation, both subunits are mainly present in slow-migrating, unprocessed forms. Supplementation of the plant nutrient solution with increasing nickel concentrations caused the conversion of the slow-migrating forms of both subunits into fast-moving, mature forms. This nickel-dependent maturation process of the hydrogenase subunits is mediated by accessory gene products, since bacteroids from H2 uptake-deficient mutants carrying Tn5 insertions in hupG and hupK and in hypB and hypE accumulated the immature forms of both hydrogenase subunits even in the presence of high nickel levels.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bressan G. M., Stanley K. K. pUEX, a bacterial expression vector related to pEX with universal host specificity. Nucleic Acids Res. 1987 Dec 10;15(23):10056–10056. doi: 10.1093/nar/15.23.10056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle C. M., Arp D. J. Nickel affects expression of the nickel-containing hydrogenase of Alcaligenes latus. J Bacteriol. 1988 Sep;170(9):3891–3896. doi: 10.1128/jb.170.9.3891-3896.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans H. J., Harker A. R., Papen H., Russell S. A., Hanus F. J., Zuber M. Physiology, biochemistry, and genetics of the uptake hydrogenase in rhizobia. Annu Rev Microbiol. 1987;41:335–361. doi: 10.1146/annurev.mi.41.100187.002003. [DOI] [PubMed] [Google Scholar]
- Francis K., Patel P., Wendt J. C., Shanmugam K. T. Purification and characterization of two forms of hydrogenase isoenzyme 1 from Escherichia coli. J Bacteriol. 1990 Oct;172(10):5750–5757. doi: 10.1128/jb.172.10.5750-5757.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedrich B., Heine E., Finck A., Friedrich C. G. Nickel requirement for active hydrogenase formation in Alcaligenes eutrophus. J Bacteriol. 1981 Mar;145(3):1144–1149. doi: 10.1128/jb.145.3.1144-1149.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedrich B., Schwartz E. Molecular biology of hydrogen utilization in aerobic chemolithotrophs. Annu Rev Microbiol. 1993;47:351–383. doi: 10.1146/annurev.mi.47.100193.002031. [DOI] [PubMed] [Google Scholar]
- Fu C. L., Maier R. J. Identification of a locus within the hydrogenase gene cluster involved in intracellular nickel metabolism in Bradyrhizobium japonicum. Appl Environ Microbiol. 1991 Dec;57(12):3502–3510. doi: 10.1128/aem.57.12.3502-3510.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu C., Maier R. J. A genetic region downstream of the hydrogenase structural genes of Bradyrhizobium japonicum that is required for hydrogenase processing. J Bacteriol. 1993 Jan;175(1):295–298. doi: 10.1128/jb.175.1.295-298.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gollin D. J., Mortenson L. E., Robson R. L. Carboxyl-terminal processing may be essential for production of active NiFe hydrogenase in Azotobacter vinelandii. FEBS Lett. 1992 Sep 14;309(3):371–375. doi: 10.1016/0014-5793(92)80809-u. [DOI] [PubMed] [Google Scholar]
- Hidalgo E., Leyva A., Ruiz-Argüeso T. Nucleotide sequence of the hydrogenase structural genes from Rhizobium leguminosarum. Plant Mol Biol. 1990 Aug;15(2):367–370. doi: 10.1007/BF00036924. [DOI] [PubMed] [Google Scholar]
- Hidalgo E., Palacios J. M., Murillo J., Ruiz-Argüeso T. Nucleotide sequence and characterization of four additional genes of the hydrogenase structural operon from Rhizobium leguminosarum bv. viciae. J Bacteriol. 1992 Jun;174(12):4130–4139. doi: 10.1128/jb.174.12.4130-4139.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holland M. A., Polacco J. C. Urease-null and hydrogenase-null phenotypes of a phylloplane bacterium reveal altered nickel metabolism in two soybean mutants. Plant Physiol. 1992 Mar;98(3):942–948. doi: 10.1104/pp.98.3.942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobi A., Rossmann R., Böck A. The hyp operon gene products are required for the maturation of catalytically active hydrogenase isoenzymes in Escherichia coli. Arch Microbiol. 1992;158(6):444–451. doi: 10.1007/BF00276307. [DOI] [PubMed] [Google Scholar]
- Kim H., Maier R. J. Transcriptional regulation of hydrogenase synthesis by nickel in Bradyrhizobium japonicum. J Biol Chem. 1990 Nov 5;265(31):18729–18732. [PubMed] [Google Scholar]
- Klucas R. V., Hanus F. J., Russell S. A., Evans H. J. Nickel: A micronutrient element for hydrogen-dependent growth of Rhizobium japonicum and for expression of urease activity in soybean leaves. Proc Natl Acad Sci U S A. 1983 Apr;80(8):2253–2257. doi: 10.1073/pnas.80.8.2253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kortlüke C., Friedrich B. Maturation of membrane-bound hydrogenase of Alcaligenes eutrophus H16. J Bacteriol. 1992 Oct;174(19):6290–6293. doi: 10.1128/jb.174.19.6290-6293.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kortlüke C., Horstmann K., Schwartz E., Rohde M., Binsack R., Friedrich B. A gene complex coding for the membrane-bound hydrogenase of Alcaligenes eutrophus H16. J Bacteriol. 1992 Oct;174(19):6277–6289. doi: 10.1128/jb.174.19.6277-6289.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Leyva A., Palacios J. M., Mozo T., Ruiz-Argüeso T. Cloning and characterization of hydrogen uptake genes from Rhizobium leguminosarum. J Bacteriol. 1987 Nov;169(11):4929–4934. doi: 10.1128/jb.169.11.4929-4934.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leyva A., Palacios J. M., Murillo J., Ruiz-Argüeso T. Genetic organization of the hydrogen uptake (hup) cluster from Rhizobium leguminosarum. J Bacteriol. 1990 Mar;172(3):1647–1655. doi: 10.1128/jb.172.3.1647-1655.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maier R. J., Pihl T. D., Stults L., Sray W. Nickel accumulation and storage in Bradyrhizobium japonicum. Appl Environ Microbiol. 1990 Jun;56(6):1905–1911. doi: 10.1128/aem.56.6.1905-1911.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maier T., Jacobi A., Sauter M., Böck A. The product of the hypB gene, which is required for nickel incorporation into hydrogenases, is a novel guanine nucleotide-binding protein. J Bacteriol. 1993 Feb;175(3):630–635. doi: 10.1128/jb.175.3.630-635.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menon A. L., Robson R. L. In vivo and in vitro nickel-dependent processing of the [NiFe] hydrogenase in Azotobacter vinelandii. J Bacteriol. 1994 Jan;176(2):291–295. doi: 10.1128/jb.176.2.291-295.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menon N. K., Robbins J., Der Vartanian M., Patil D., Peck H. D., Jr, Menon A. L., Robson R. L., Przybyla A. E. Carboxy-terminal processing of the large subunit of [NiFe] hydrogenases. FEBS Lett. 1993 Sep 27;331(1-2):91–95. doi: 10.1016/0014-5793(93)80303-c. [DOI] [PubMed] [Google Scholar]
- Menon N. K., Robbins J., Wendt J. C., Shanmugam K. T., Przybyla A. E. Mutational analysis and characterization of the Escherichia coli hya operon, which encodes [NiFe] hydrogenase 1. J Bacteriol. 1991 Aug;173(15):4851–4861. doi: 10.1128/jb.173.15.4851-4861.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moshiri F., Maier R. J. Conformational changes in the membrane-bound hydrogenase of Bradyrhizobium japonicum. Evidence that the redox state of the enzyme affects its accessibility to protease and membrane-impermeant reagents. J Biol Chem. 1988 Nov 25;263(33):17809–17816. [PubMed] [Google Scholar]
- Palacios J. M., Murillo J., Leyva A., Ditta G., Ruiz-Argüeso T. Differential expression of hydrogen uptake (hup) genes in vegetative and symbiotic cells of Rhizobium leguminosarum. Mol Gen Genet. 1990 May;221(3):363–370. doi: 10.1007/BF00259401. [DOI] [PubMed] [Google Scholar]
- Partridge C. D., Yates M. G. Effect of chelating agents on hydrogenase in Azotobacter chroococcum. Evidence that nickel is required for hydrogenase synthesis. Biochem J. 1982 Apr 15;204(1):339–344. doi: 10.1042/bj2040339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips D. A., Kapulnik Y., Bedmar E. J., Joseph C. M. Development and Partial Characterization of Nearly Isogenic Pea Lines (Pisum sativum L.) that Alter Uptake Hydrogenase Activity in Symbiotic Rhizobium. Plant Physiol. 1990 Apr;92(4):983–989. doi: 10.1104/pp.92.4.983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Przybyla A. E., Robbins J., Menon N., Peck H. D., Jr Structure-function relationships among the nickel-containing hydrogenases. FEMS Microbiol Rev. 1992 Feb;8(2):109–135. doi: 10.1111/j.1574-6968.1992.tb04960.x. [DOI] [PubMed] [Google Scholar]
- Rey L., Hidalgo E., Palacios J., Ruiz-Argüeso T. Nucleotide sequence and organization of an H2-uptake gene cluster from Rhizobium leguminosarum bv. viciae containing a rubredoxin-like gene and four additional open reading frames. J Mol Biol. 1992 Dec 5;228(3):998–1002. doi: 10.1016/0022-2836(92)90886-o. [DOI] [PubMed] [Google Scholar]
- Rey L., Murillo J., Hernando Y., Hidalgo E., Cabrera E., Imperial J., Ruiz-Argüeso T. Molecular analysis of a microaerobically induced operon required for hydrogenase synthesis in Rhizobium leguminosarum biovar viciae. Mol Microbiol. 1993 May;8(3):471–481. doi: 10.1111/j.1365-2958.1993.tb01591.x. [DOI] [PubMed] [Google Scholar]
- Smith P. K., Krohn R. I., Hermanson G. T., Mallia A. K., Gartner F. H., Provenzano M. D., Fujimoto E. K., Goeke N. M., Olson B. J., Klenk D. C. Measurement of protein using bicinchoninic acid. Anal Biochem. 1985 Oct;150(1):76–85. doi: 10.1016/0003-2697(85)90442-7. [DOI] [PubMed] [Google Scholar]
- Stults L. W., Mallick S., Maier R. J. Nickel uptake in Bradyrhizobium japonicum. J Bacteriol. 1987 Apr;169(4):1398–1402. doi: 10.1128/jb.169.4.1398-1402.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vignais P. M., Toussaint B. Molecular biology of membrane-bound H2 uptake hydrogenases. Arch Microbiol. 1994;161(1):1–10. doi: 10.1007/BF00248887. [DOI] [PubMed] [Google Scholar]