Abstract
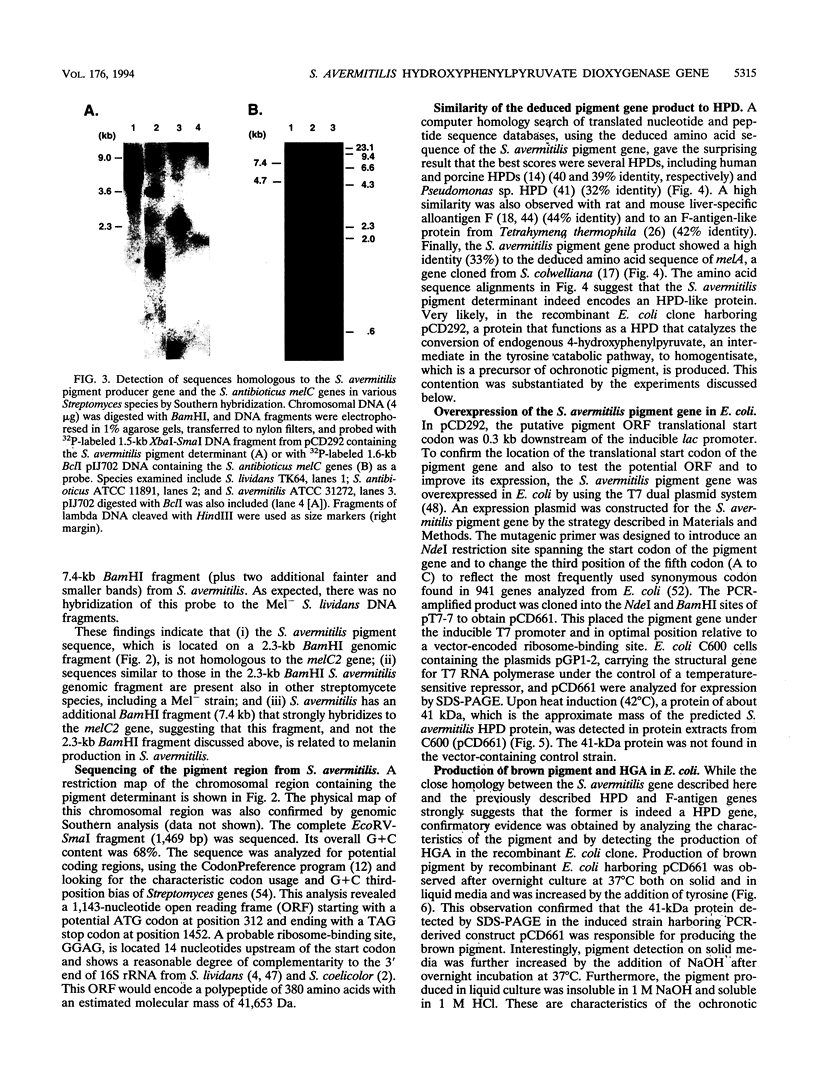
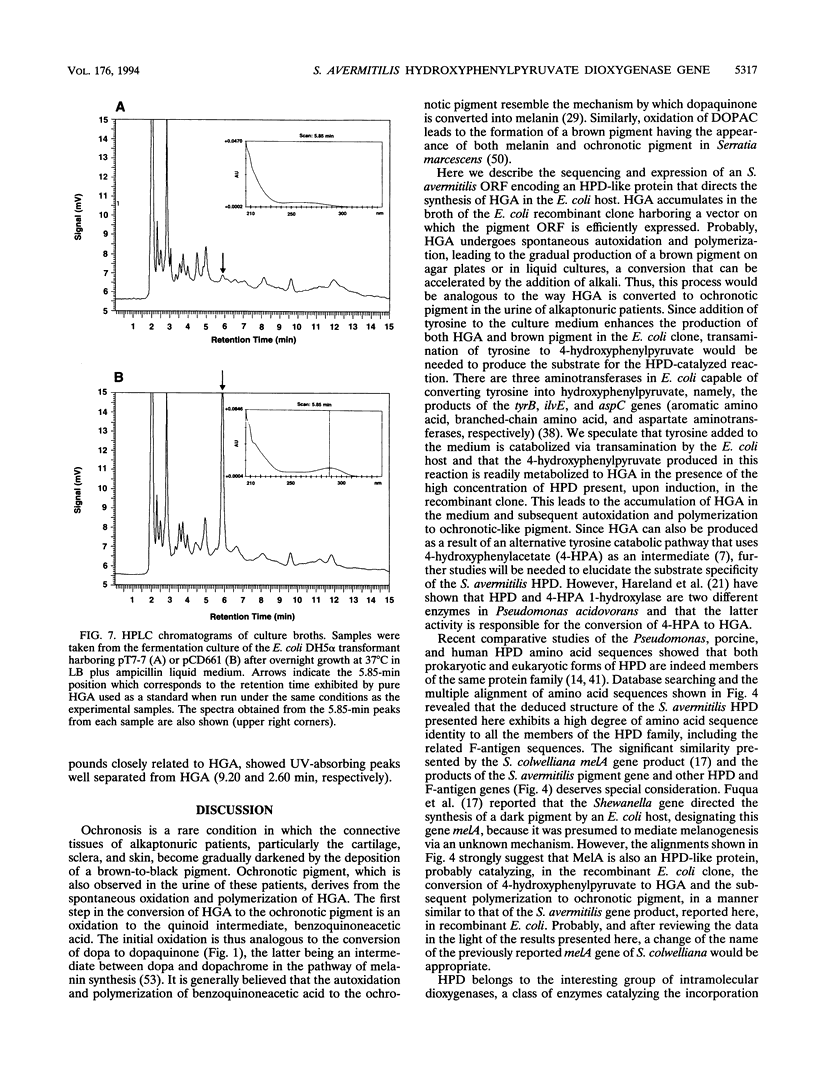
A 1.5-kb genomic fragment isolated from Streptomyces avermitilis that directs the synthesis of a brown pigment in Escherichia coli was characterized. Since pigment production in recombinant E. coli was enhanced by the addition of tyrosine to the medium, it had been inferred that the cloned DNA might be associated with melanin biosynthesis. Hybridization studies, however, showed that the pigment gene isolated from S. avermitilis was unrelated to the Streptomyces antibioticus melC2 determinant, which is the prototype of melanin genes in Streptomyces spp. Sequence analysis of the 1.5-kb DNA that caused pigment production revealed a single open reading frame encoding a protein of 41.6 kDa (380 amino acids) that resembled several prokaryotic and eukaryotic 4-hydroxyphenylpyruvate dioxygenases (HPDs). When this open reading frame was overexpressed in E. coli, a protein of about 41 kDa was detected. This E. coli clone produced homogentisic acid (HGA), which is the expected product of the oxidation of 4-hydroxyphenylpyruvate catalyzed by an HPD, and also a brown pigment with characteristics similar to the pigment observed in the urine of alkaptonuric patients. Alkaptonuria is a genetic disease in which inability to metabolize HGA leads to increasing concentrations of this acid in urine, followed by oxidation and polymerization of HGA to an ochronotic pigment. Similarly, the production of ochronotic-like pigment in the recombinant E. coli clone overexpressing the S. avermitilis gene encoding HPD is likely to be due to the spontaneous oxidation and polymerization of the HGA accumulated in the medium by this clone.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Akesson B., Forslind K., Wollheim F. Analysis of homogentisic acid in body fluids by high-performance liquid chromatography. J Chromatogr. 1987 Jan 23;413:233–236. doi: 10.1016/0378-4347(87)80231-1. [DOI] [PubMed] [Google Scholar]
- Baylis H. A., Bibb M. J. The nucleotide sequence of a 16S rRNA gene from Streptomyces coelicolor A3(2) Nucleic Acids Res. 1987 Sep 11;15(17):7176–7176. doi: 10.1093/nar/15.17.7176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernan V., Filpula D., Herber W., Bibb M., Katz E. The nucleotide sequence of the tyrosinase gene from Streptomyces antibioticus and characterization of the gene product. Gene. 1985;37(1-3):101–110. doi: 10.1016/0378-1119(85)90262-8. [DOI] [PubMed] [Google Scholar]
- Bibb M. J., Cohen S. N. Gene expression in Streptomyces: construction and application of promoter-probe plasmid vectors in Streptomyces lividans. Mol Gen Genet. 1982;187(2):265–277. doi: 10.1007/BF00331128. [DOI] [PubMed] [Google Scholar]
- Binnie C., Warren M., Butler M. J. Cloning and heterologous expression in Streptomyces lividans of Streptomyces rimosus genes involved in oxytetracycline biosynthesis. J Bacteriol. 1989 Feb;171(2):887–895. doi: 10.1128/jb.171.2.887-895.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birnboim H. C., Doly J. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 1979 Nov 24;7(6):1513–1523. doi: 10.1093/nar/7.6.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blakley E. R. Microbial conversion of p-hydroxyphenylacetic acid to homogentisic acid. Can J Microbiol. 1972 Aug;18(8):1247–1255. doi: 10.1139/m72-193. [DOI] [PubMed] [Google Scholar]
- Burg R. W., Miller B. M., Baker E. E., Birnbaum J., Currie S. A., Hartman R., Kong Y. L., Monaghan R. L., Olson G., Putter I. Avermectins, new family of potent anthelmintic agents: producing organism and fermentation. Antimicrob Agents Chemother. 1979 Mar;15(3):361–367. doi: 10.1128/aac.15.3.361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper R. A., Skinner M. A. Catabolism of 3- and 4-hydroxyphenylacetate by the 3,4-dihydroxyphenylacetate pathway in Escherichia coli. J Bacteriol. 1980 Jul;143(1):302–306. doi: 10.1128/jb.143.1.302-306.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denoya C. D., Bechhofer D. H., Dubnau D. Translational autoregulation of ermC 23S rRNA methyltransferase expression in Bacillus subtilis. J Bacteriol. 1986 Dec;168(3):1133–1141. doi: 10.1128/jb.168.3.1133-1141.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devereux J., Haeberli P., Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984 Jan 11;12(1 Pt 1):387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duff G. A., Roberts J. E., Foster N. Analysis of the structure of synthetic and natural melanins by solid-phase NMR. Biochemistry. 1988 Sep 6;27(18):7112–7116. doi: 10.1021/bi00418a067. [DOI] [PubMed] [Google Scholar]
- Endo F., Awata H., Tanoue A., Ishiguro M., Eda Y., Titani K., Matsuda I. Primary structure deduced from complementary DNA sequence and expression in cultured cells of mammalian 4-hydroxyphenylpyruvic acid dioxygenase. Evidence that the enzyme is a homodimer of identical subunits homologous to rat liver-specific alloantigen F. J Biol Chem. 1992 Dec 5;267(34):24235–24240. [PubMed] [Google Scholar]
- Fravi G., Lindenmann J. Induction by allogeneic extracts of liver-specific precipitating autoantibodies in the mouse. Nature. 1968 Apr 13;218(5137):141–143. doi: 10.1038/218141a0. [DOI] [PubMed] [Google Scholar]
- Fuqua W. C., Coyne V. E., Stein D. C., Lin C. M., Weiner R. M. Characterization of melA: a gene encoding melanin biosynthesis from the marine bacterium Shewanella colwelliana. Gene. 1991 Dec 20;109(1):131–136. doi: 10.1016/0378-1119(91)90598-6. [DOI] [PubMed] [Google Scholar]
- Gershwin M. E., Coppel R. L., Bearer E., Peterson M. G., Sturgess A., Mackay I. R. Molecular cloning of the liver-specific rat F antigen. J Immunol. 1987 Dec 1;139(11):3828–3833. [PubMed] [Google Scholar]
- Hareland W. A., Crawford R. L., Chapman P. J., Dagley S. Metabolic function and properties of 4-hydroxyphenylacetic acid 1-hydroxylase from Pseudomonas acidovorans. J Bacteriol. 1975 Jan;121(1):272–285. doi: 10.1128/jb.121.1.272-285.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Held T., Kutzner H. J. Genetic recombination in Streptomyces michiganensis DSM 40,015 revealed three genes responsible for the formation of melanin. J Basic Microbiol. 1991;31(2):127–134. doi: 10.1002/jobm.3620310211. [DOI] [PubMed] [Google Scholar]
- Henikoff S. Unidirectional digestion with exonuclease III in DNA sequence analysis. Methods Enzymol. 1987;155:156–165. doi: 10.1016/0076-6879(87)55014-5. [DOI] [PubMed] [Google Scholar]
- Hintermann G., Zatchej M., Hütter R. Cloning and expression of the genetically unstable tyrosinase structural gene from Streptomyces glaucescens. Mol Gen Genet. 1985;200(3):422–432. doi: 10.1007/BF00425726. [DOI] [PubMed] [Google Scholar]
- Hummel R., Nørgaard P., Andreasen P. H., Neve S., Skjødt K., Tornehave D., Kristiansen K. Tetrahymena gene encodes a protein that is homologous with the liver-specific F-antigen and associated with membranes of the Golgi apparatus and transport vesicles. J Mol Biol. 1992 Dec 5;228(3):850–861. doi: 10.1016/0022-2836(92)90869-l. [DOI] [PubMed] [Google Scholar]
- Katz E., Thompson C. J., Hopwood D. A. Cloning and expression of the tyrosinase gene from Streptomyces antibioticus in Streptomyces lividans. J Gen Microbiol. 1983 Sep;129(9):2703–2714. doi: 10.1099/00221287-129-9-2703. [DOI] [PubMed] [Google Scholar]
- Lerch K., Ettinger L. Purification and characterization of a tyrosinase from Streptomyces glaucescens. Eur J Biochem. 1972 Dec 18;31(3):427–437. doi: 10.1111/j.1432-1033.1972.tb02549.x. [DOI] [PubMed] [Google Scholar]
- Lindblad B., Lindstedt G., Lindstedt S. The mechanism of enzymic formation of homogentisate from p-hydroxyphenylpyruvate. J Am Chem Soc. 1970 Dec 16;92(25):7446–7449. doi: 10.1021/ja00728a032. [DOI] [PubMed] [Google Scholar]
- Lindstedt S., Odelhög B. 4-Hydroxyphenylpyruvate dioxygenase from human liver. Methods Enzymol. 1987;142:139–142. doi: 10.1016/s0076-6879(87)42021-1. [DOI] [PubMed] [Google Scholar]
- Lindstedt S., Odelhög B., Rundgren M. Purification and some properties of 4-hydroxyphenylpyruvate dioxygenase from Pseudomonas sp. P. J. 874. Biochemistry. 1977 Jul 26;16(15):3369–3377. doi: 10.1021/bi00634a013. [DOI] [PubMed] [Google Scholar]
- Müller G., Ruppert S., Schmid E., Schütz G. Functional analysis of alternatively spliced tyrosinase gene transcripts. EMBO J. 1988 Sep;7(9):2723–2730. doi: 10.1002/j.1460-2075.1988.tb03126.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliveira D. B., Vindlacheruvu S. The phylogenetic distribution of the liver protein F antigen. Comp Biochem Physiol B. 1987;87(1):87–90. doi: 10.1016/0305-0491(87)90474-3. [DOI] [PubMed] [Google Scholar]
- Richardson M. A., Kuhstoss S., Solenberg P., Schaus N. A., Rao R. N. A new shuttle cosmid vector, pKC505, for streptomycetes: its use in the cloning of three different spiramycin-resistance genes from a Streptomyces ambofaciens library. Gene. 1987;61(3):231–241. doi: 10.1016/0378-1119(87)90187-9. [DOI] [PubMed] [Google Scholar]
- Roche P. A., Moorehead T. J., Hamilton G. A. Purification and properties of hog liver 4-hydroxyphenylpyruvate dioxygenase. Arch Biochem Biophys. 1982 Jun;216(1):62–73. doi: 10.1016/0003-9861(82)90188-6. [DOI] [PubMed] [Google Scholar]
- Rüetschi U., Odelhög B., Lindstedt S., Barros-Söderling J., Persson B., Jörnvall H. Characterization of 4-hydroxyphenylpyruvate dioxygenase. Primary structure of the Pseudomonas enzyme. Eur J Biochem. 1992 Apr 15;205(2):459–466. doi: 10.1111/j.1432-1033.1992.tb16800.x. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schofield J. P., Vijayakumar R. K., Oliveira D. B. Sequences of the mouse F protein alleles and identification of a T cell epitope. Eur J Immunol. 1991 May;21(5):1235–1240. doi: 10.1002/eji.1830210521. [DOI] [PubMed] [Google Scholar]
- Skinner D. D., Denoya C. D. A simple DNA polymerase chain reaction method to locate and define orientation of specific sequences in cloned bacterial genomic fragments. Microbios. 1993;75(303):125–129. [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Suzuki Y., Yamada T. The nucleotide sequence of 16S rRNA gene from Streptomyces lividans TK21. Nucleic Acids Res. 1988 Jan 11;16(1):370–370. doi: 10.1093/nar/16.1.370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tibbetts M. W., Hafner E. W., Morgenstern M. R., Skinner D. D., Denoya C. D. Cloning of a DNA fragment involved in pigment production in Streptomyces avermitilis. FEMS Microbiol Lett. 1992 Feb 1;70(1):9–13. doi: 10.1016/0378-1097(92)90555-3. [DOI] [PubMed] [Google Scholar]
- Trias J., Viñas M., Guinea J., Lorèn J. G. Brown pigmentation in Serratia marcescens cultures associated with tyrosine metabolism. Can J Microbiol. 1989 Nov;35(11):1037–1042. doi: 10.1139/m89-172. [DOI] [PubMed] [Google Scholar]
- Wada G. H., Fellman J. H., Fujita T. S., Roth E. S. Purification and properties of avian liver p-hydroxyphenylpyruvate hydroxylase. J Biol Chem. 1975 Sep 10;250(17):6720–6726. [PubMed] [Google Scholar]
- Wada K., Aota S., Tsuchiya R., Ishibashi F., Gojobori T., Ikemura T. Codon usage tabulated from the GenBank genetic sequence data. Nucleic Acids Res. 1990 Apr 25;18 (Suppl):2367–2411. doi: 10.1093/nar/18.suppl.2367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright F., Bibb M. J. Codon usage in the G+C-rich Streptomyces genome. Gene. 1992 Apr 1;113(1):55–65. doi: 10.1016/0378-1119(92)90669-g. [DOI] [PubMed] [Google Scholar]