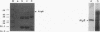Abstract
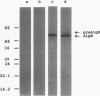
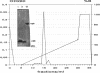
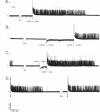
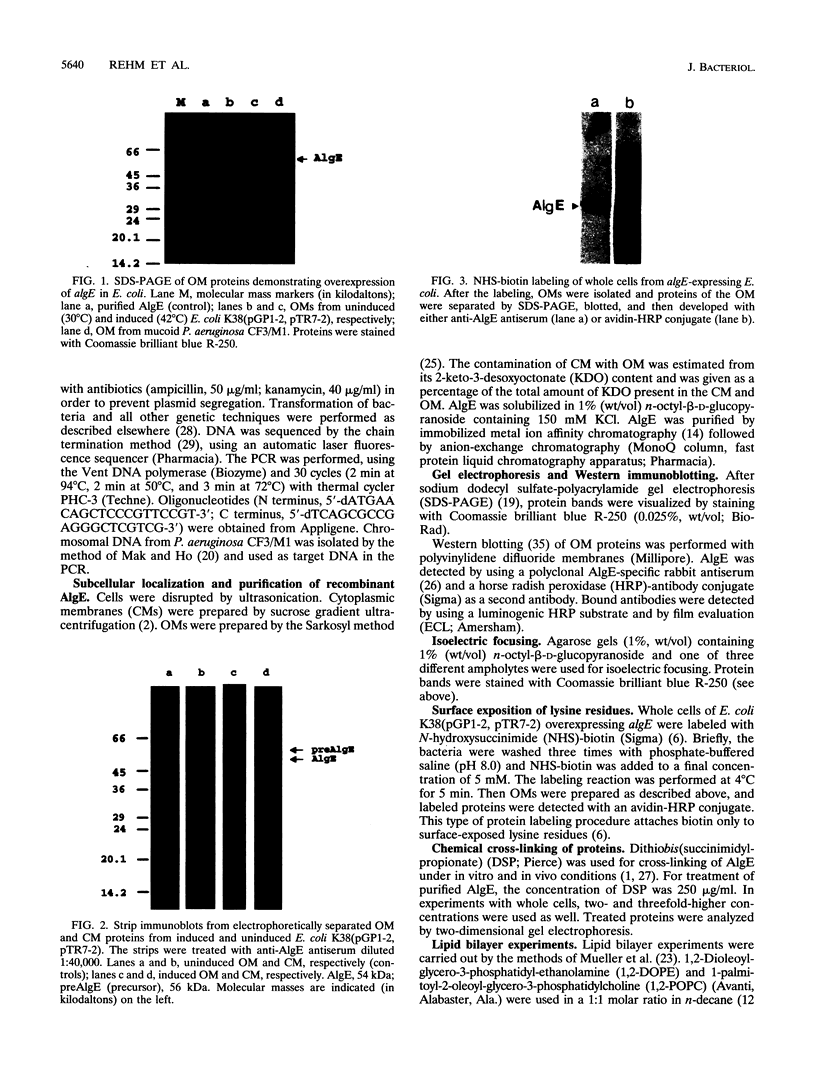
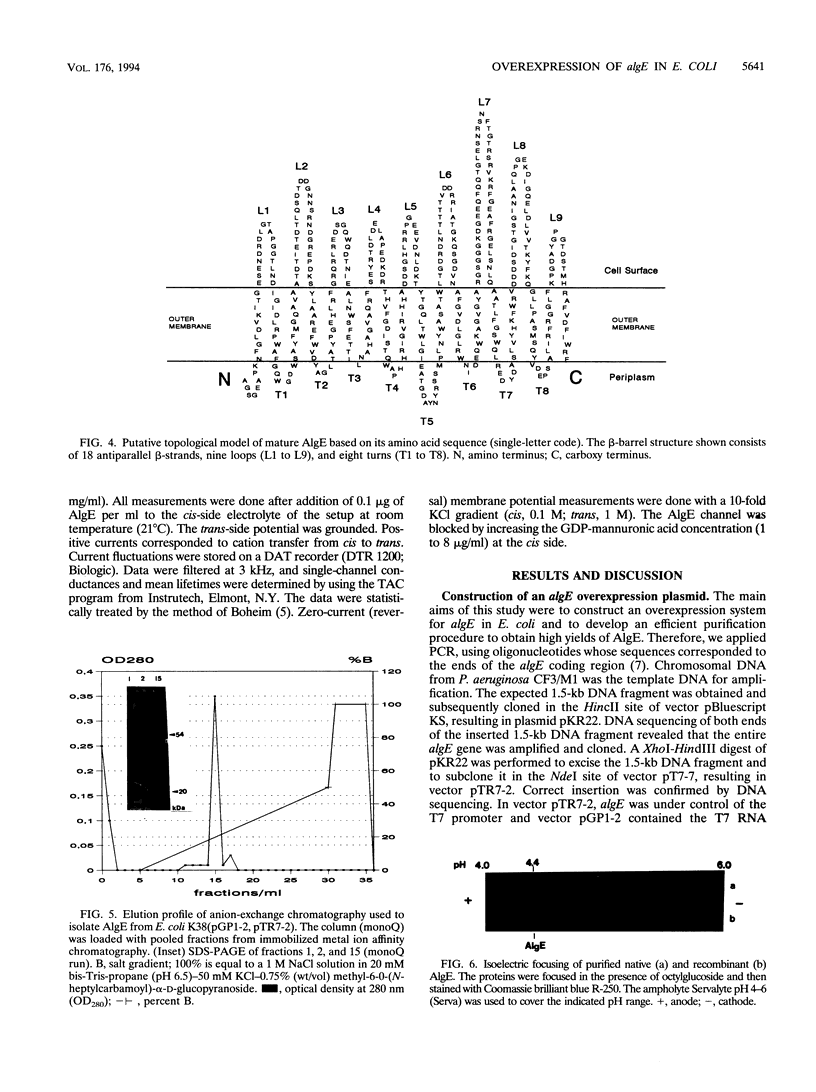
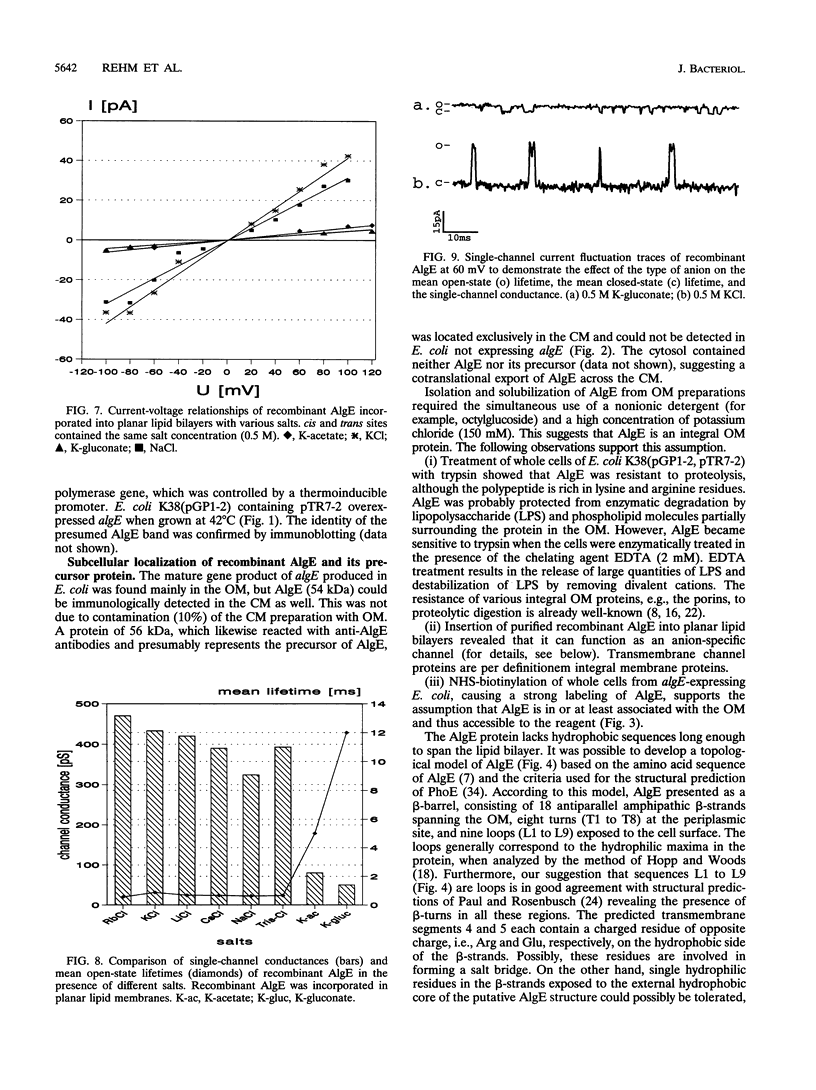
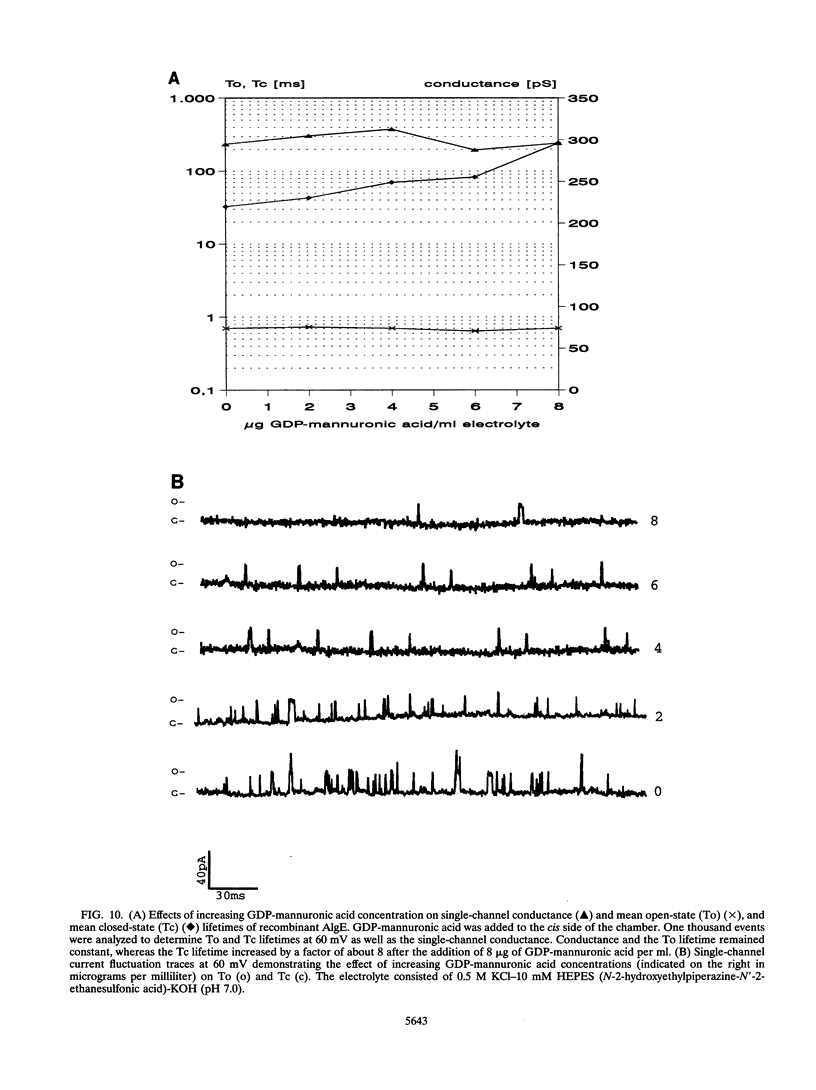
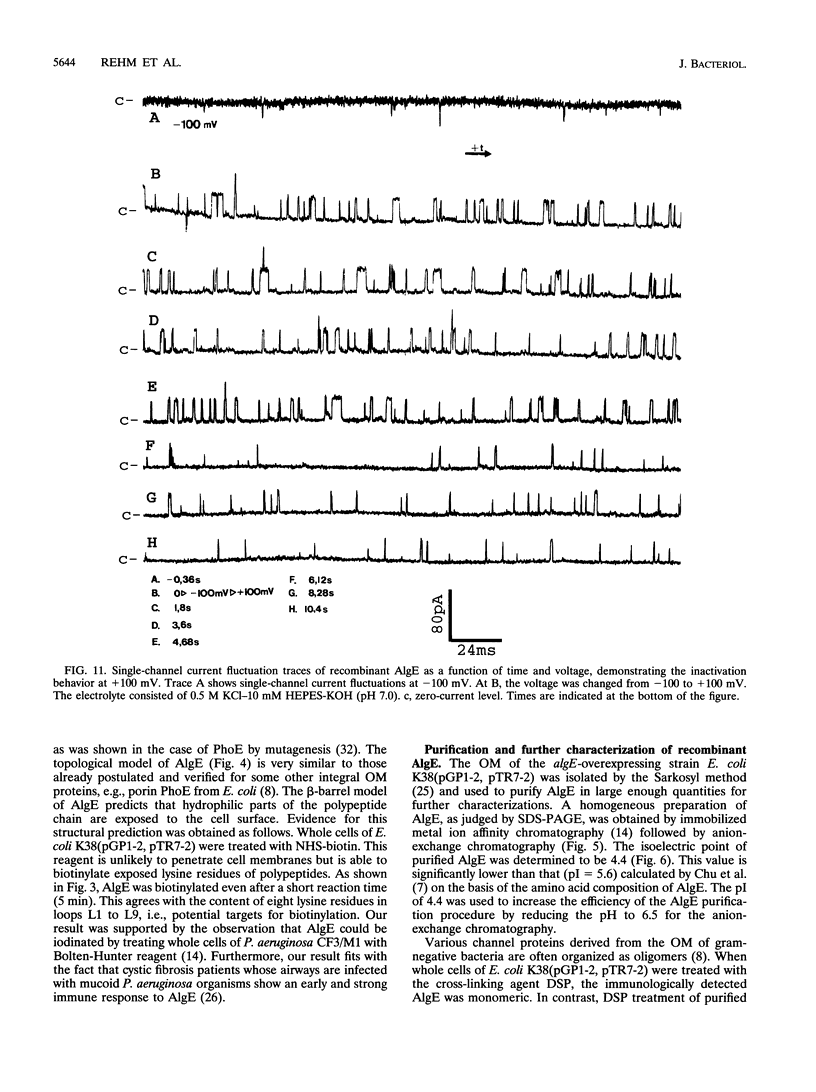
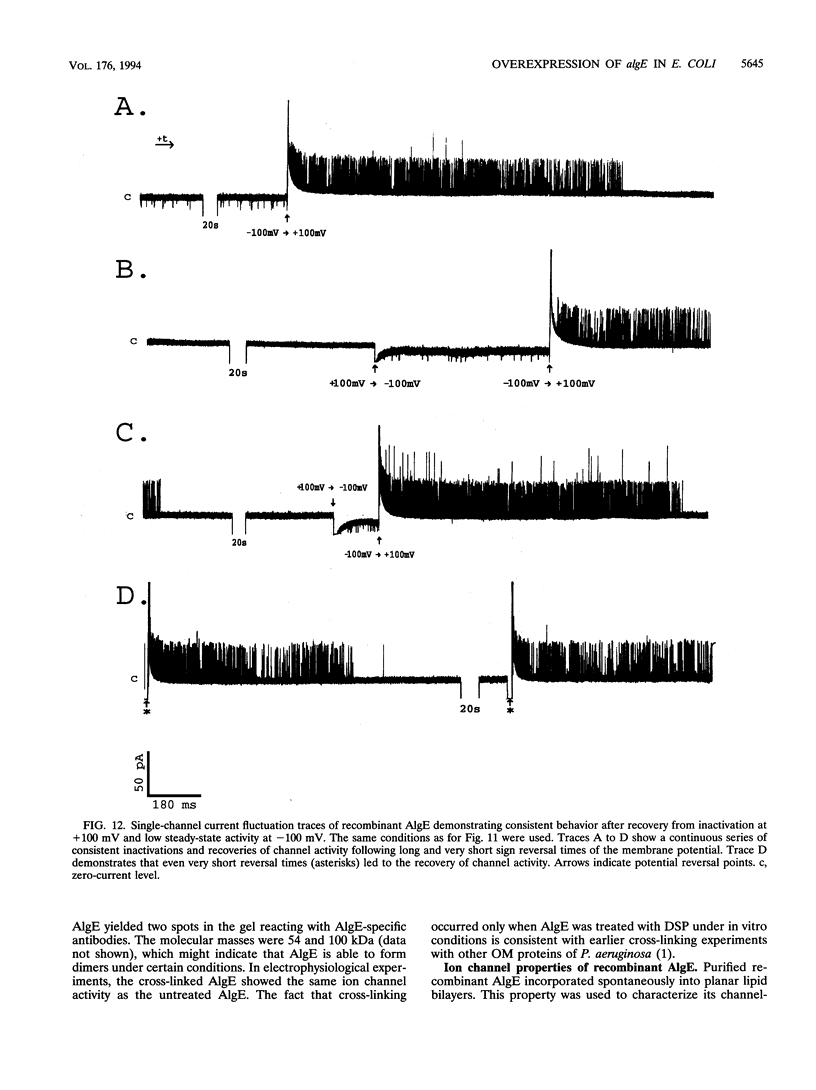
Alginate-producing (mucoid) strains of Pseudomonas aeruginosa possess a 54-kDa outer membrane (OM) protein (AlgE) which is missing in nonmucoid bacteria. The coding region of the algE gene from mucoid P. aeruginosa CF3/M1 was subcloned in the expression vector pT7-7 and expressed in Escherichia coli. The level of expression of recombinant AlgE was seven times higher than that of the native protein in P. aeruginosa. Recombinant AlgE was found mainly in the OM. A putative precursor protein (56 kDa) of AlgE could be immunologically detected in the cytoplasmic membrane (CM). Surface exposition of AlgE in the OM of E. coli was indicated by labeling lysine residues with N-hydroxysuccinimide-biotin. Secondary-structure analysis suggested that AlgE is anchored in the OM by 18 membrane-spanning beta-strands, probably forming a beta-barrel. Recombinant AlgE was purified, and isoelectric focusing revealed a pI of 4.4. Recombinant AlgE was spontaneously incorporated into planar lipid bilayers, forming ion channels with a single-channel conductance of 0.76 nS in 1 M KCl and a mean lifetime of 0.7 ms. Single-channel current measurements in the presence of other salts as well as reversal potential measurements in salt gradients revealed that the AlgE channel was strongly anion selective. For chloride ions, a weak binding constant (Km = 0.75 M) was calculated, suggesting that AlgE might constitute an ion channel specific for another particular anion, e.g., polymannuronic acid, which is a precursor of alginate. Consistent with this idea, the open-state probability of the channel decreased when GDP-mannuronic acid was added. The AlgE channel was inactivated when membrane voltages higher than +85 mV were applied. The electrophysiological characteristics of AlgE, including its rectifying properties, are quite different from those of typical porins.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Angus B. L., Hancock R. E. Outer membrane porin proteins F, P, and D1 of Pseudomonas aeruginosa and PhoE of Escherichia coli: chemical cross-linking to reveal native oligomers. J Bacteriol. 1983 Sep;155(3):1042–1051. doi: 10.1128/jb.155.3.1042-1051.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anwar H., Lambert P. A., Brown M. R. Influence of sodium dodecyl sulphate quality on the electrophoretic mobility of the outer membrane proteins of mucoid and non-mucoid Pseudomonas aeruginosa. Biochim Biophys Acta. 1983 Dec 13;761(2):119–125. doi: 10.1016/0304-4165(83)90220-9. [DOI] [PubMed] [Google Scholar]
- Benz R., Hancock R. E. Properties of the large ion-permeable pores formed from protein F of Pseudomonas aeruginosa in lipid bilayer membranes. Biochim Biophys Acta. 1981 Aug 20;646(2):298–308. doi: 10.1016/0005-2736(81)90336-9. [DOI] [PubMed] [Google Scholar]
- Benz R. Structure and function of porins from gram-negative bacteria. Annu Rev Microbiol. 1988;42:359–393. doi: 10.1146/annurev.mi.42.100188.002043. [DOI] [PubMed] [Google Scholar]
- Boheim G. Statistical analysis of alamethicin channels in black lipid membranes. J Membr Biol. 1974;19(3):277–303. doi: 10.1007/BF01869983. [DOI] [PubMed] [Google Scholar]
- Bradburne J. A., Godfrey P., Choi J. H., Mathis J. N. In vivo labeling of Escherichia coli cell envelope proteins with N-hydroxysuccinimide esters of biotin. Appl Environ Microbiol. 1993 Mar;59(3):663–668. doi: 10.1128/aem.59.3.663-668.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu L., May T. B., Chakrabarty A. M., Misra T. K. Nucleotide sequence and expression of the algE gene involved in alginate biosynthesis by Pseudomonas aeruginosa. Gene. 1991 Oct 30;107(1):1–10. doi: 10.1016/0378-1119(91)90290-r. [DOI] [PubMed] [Google Scholar]
- Cowan S. W., Schirmer T., Rummel G., Steiert M., Ghosh R., Pauptit R. A., Jansonius J. N., Rosenbusch J. P. Crystal structures explain functional properties of two E. coli porins. Nature. 1992 Aug 27;358(6389):727–733. doi: 10.1038/358727a0. [DOI] [PubMed] [Google Scholar]
- Franklin M. J., Chitnis C. E., Gacesa P., Sonesson A., White D. C., Ohman D. E. Pseudomonas aeruginosa AlgG is a polymer level alginate C5-mannuronan epimerase. J Bacteriol. 1994 Apr;176(7):1821–1830. doi: 10.1128/jb.176.7.1821-1830.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franklin M. J., Ohman D. E. Identification of algF in the alginate biosynthetic gene cluster of Pseudomonas aeruginosa which is required for alginate acetylation. J Bacteriol. 1993 Aug;175(16):5057–5065. doi: 10.1128/jb.175.16.5057-5065.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frosch M., Müller D., Bousset K., Müller A. Conserved outer membrane protein of Neisseria meningitidis involved in capsule expression. Infect Immun. 1992 Mar;60(3):798–803. doi: 10.1128/iai.60.3.798-803.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Govan J. R., Harris G. S. Pseudomonas aeruginosa and cystic fibrosis: unusual bacterial adaptation and pathogenesis. Microbiol Sci. 1986 Oct;3(10):302–308. [PubMed] [Google Scholar]
- Grabert E., Wingender J., Winkler U. K. An outer membrane protein characteristic of mucoid strains of Pseudomonas aeruginosa. FEMS Microbiol Lett. 1990 Mar 1;56(1-2):83–87. doi: 10.1111/j.1574-6968.1990.tb04127.x. [DOI] [PubMed] [Google Scholar]
- Hancock R. E., Benz R. Demonstration and chemical modification of a specific phosphate binding site in the phosphate-starvation-inducible outer membrane porin protein P of Pseudomonas aeruginosa. Biochim Biophys Acta. 1986 Sep 11;860(3):699–707. doi: 10.1016/0005-2736(86)90569-9. [DOI] [PubMed] [Google Scholar]
- Hopp T. P., Woods K. R. Prediction of protein antigenic determinants from amino acid sequences. Proc Natl Acad Sci U S A. 1981 Jun;78(6):3824–3828. doi: 10.1073/pnas.78.6.3824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lugtenberg B., Meijers J., Peters R., van der Hoek P., van Alphen L. Electrophoretic resolution of the "major outer membrane protein" of Escherichia coli K12 into four bands. FEBS Lett. 1975 Oct 15;58(1):254–258. doi: 10.1016/0014-5793(75)80272-9. [DOI] [PubMed] [Google Scholar]
- MUELLER P., RUDIN D. O., TIEN H. T., WESCOTT W. C. Reconstitution of cell membrane structure in vitro and its transformation into an excitable system. Nature. 1962 Jun 9;194:979–980. doi: 10.1038/194979a0. [DOI] [PubMed] [Google Scholar]
- Mak Y. M., Ho K. K. An improved method for the isolation of chromosomal DNA from various bacteria and cyanobacteria. Nucleic Acids Res. 1992 Aug 11;20(15):4101–4102. doi: 10.1093/nar/20.15.4101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- May T. B., Shinabarger D., Maharaj R., Kato J., Chu L., DeVault J. D., Roychoudhury S., Zielinski N. A., Berry A., Rothmel R. K. Alginate synthesis by Pseudomonas aeruginosa: a key pathogenic factor in chronic pulmonary infections of cystic fibrosis patients. Clin Microbiol Rev. 1991 Apr;4(2):191–206. doi: 10.1128/cmr.4.2.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morona R., Tommassen J., Henning U. Demonstration of a bacteriophage receptor site on the Escherichia coli K12 outer-membrane protein OmpC by the use of a protease. Eur J Biochem. 1985 Jul 1;150(1):161–169. doi: 10.1111/j.1432-1033.1985.tb09002.x. [DOI] [PubMed] [Google Scholar]
- Paul C., Rosenbusch J. P. Folding patterns of porin and bacteriorhodopsin. EMBO J. 1985 Jun;4(6):1593–1597. doi: 10.1002/j.1460-2075.1985.tb03822.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rehm B. H., Grabert E., Hein J., Winkler U. K. Antibody response of rabbits and cystic fibrosis patients to an alginate-specific outer membrane protein of a mucoid strain of Pseudomonas aeruginosa. Microb Pathog. 1994 Jan;16(1):43–51. doi: 10.1006/mpat.1994.1004. [DOI] [PubMed] [Google Scholar]
- Reithmeier R. A., Bragg P. D. Cross-linking of the proteins in the outer membrane of Escherichia coli. Biochim Biophys Acta. 1977 Apr 18;466(2):245–256. doi: 10.1016/0005-2736(77)90222-x. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiller N. L., Monday S. R., Boyd C. M., Keen N. T., Ohman D. E. Characterization of the Pseudomonas aeruginosa alginate lyase gene (algL): cloning, sequencing, and expression in Escherichia coli. J Bacteriol. 1993 Aug;175(15):4780–4789. doi: 10.1128/jb.175.15.4780-4789.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindler H., Rosenbusch J. P. Matrix protein from Escherichia coli outer membranes forms voltage-controlled channels in lipid bilayers. Proc Natl Acad Sci U S A. 1978 Aug;75(8):3751–3755. doi: 10.1073/pnas.75.8.3751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabor S., Richardson C. C. A bacteriophage T7 RNA polymerase/promoter system for controlled exclusive expression of specific genes. Proc Natl Acad Sci U S A. 1985 Feb;82(4):1074–1078. doi: 10.1073/pnas.82.4.1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Towbin H., Staehelin T., Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc Natl Acad Sci U S A. 1979 Sep;76(9):4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wicker-Böckelmann U., Wingender J., Winkler U. K. Alginate lyase releases cell-bound lipase from mucoid strains of Pseudomonas aeruginosa. Zentralbl Bakteriol Mikrobiol Hyg A. 1987 Oct;266(3-4):379–389. doi: 10.1016/s0176-6724(87)80218-3. [DOI] [PubMed] [Google Scholar]