Abstract
Plasmid pEP2 was found to encode a protein, RepA, which is essential and rate limiting for its replication in Escherichia coli and Corynebacterium pseudotuberculosis. Mutations which altered the rate of synthesis of this protein in E. coli affected the copy number and segregational stability of pEP2 in the two hosts. RepA contains 483 amino acid residues and has the calculated molecular weight of 53,925. It shows 45% amino acid residue identity with open reading frame ORF2 of pSR1, a plasmid isolated from Corynebacterium glutamicum (J. A. C. Archer and A. J. Sinskey, J. Gen. Microbiol. 139:1753-1759, 1993). Plasmid pEP2 was shown to accumulate single-stranded DNA corresponding to the RepA coding strand during its replication in E. coli and C. pseudotuberculosis, suggesting that it may replicate by a rolling circle mechanism. However, RepA has no significant sequence homology with the replication initiator proteins of plasmids known to use this mode of replication.
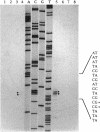
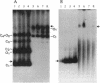
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Aiba H., Adhya S., de Crombrugghe B. Evidence for two functional gal promoters in intact Escherichia coli cells. J Biol Chem. 1981 Nov 25;256(22):11905–11910. [PubMed] [Google Scholar]
- Archer J. A., Sinskey A. J. The DNA sequence and minimal replicon of the Corynebacterium glutamicum plasmid pSR1: evidence of a common ancestry with plasmids from C. diphtheriae. J Gen Microbiol. 1993 Aug;139(8):1753–1759. doi: 10.1099/00221287-139-8-1753. [DOI] [PubMed] [Google Scholar]
- Bonnassie S., Burini J. F., Oreglia J., Trautwetter A., Patte J. C., Sicard A. M. Transfer of plasmid DNA to Brevibacterium lactofermentum by electrotransformation. J Gen Microbiol. 1990 Oct;136(10):2107–2112. doi: 10.1099/00221287-136-10-2107. [DOI] [PubMed] [Google Scholar]
- Bouia A., Bringel F., Frey L., Kammerer B., Belarbi A., Guyonvarch A., Hubert J. C. Structural organization of pLP1, a cryptic plasmid from Lactobacillus plantarum CCM 1904. Plasmid. 1989 Nov;22(3):185–192. doi: 10.1016/0147-619x(89)90001-2. [DOI] [PubMed] [Google Scholar]
- Carleton S., Projan S. J., Highlander S. K., Moghazeh S. M., Novick R. P. Control of pT181 replication II. Mutational analysis. EMBO J. 1984 Oct;3(10):2407–2414. doi: 10.1002/j.1460-2075.1984.tb02147.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catchpole I., Thomas C., Davies A., Dyke K. G. The nucleotide sequence of Staphylococcus aureus plasmid pT48 conferring inducible macrolide-lincosamide-streptogramin B resistance and comparison with similar plasmids expressing constitutive resistance. J Gen Microbiol. 1988 Mar;134(3):697–709. doi: 10.1099/00221287-134-3-697. [DOI] [PubMed] [Google Scholar]
- Geider K., Kornberg A. Conversion of the M13 viral single strand to the double-stranded replicative forms by purified proteins. J Biol Chem. 1974 Jul 10;249(13):3999–4005. [PubMed] [Google Scholar]
- Gerdes K., Larsen J. E., Molin S. Stable inheritance of plasmid R1 requires two different loci. J Bacteriol. 1985 Jan;161(1):292–298. doi: 10.1128/jb.161.1.292-298.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gil D., Bouché J. P. ColE1-type vectors with fully repressible replication. Gene. 1991 Aug 30;105(1):17–22. doi: 10.1016/0378-1119(91)90508-9. [DOI] [PubMed] [Google Scholar]
- Goodrich J. A., Schwartz M. L., McClure W. R. Searching for and predicting the activity of sites for DNA binding proteins: compilation and analysis of the binding sites for Escherichia coli integration host factor (IHF). Nucleic Acids Res. 1990 Sep 11;18(17):4993–5000. doi: 10.1093/nar/18.17.4993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gros M. F., te Riele H., Ehrlich S. D. Replication origin of a single-stranded DNA plasmid pC194. EMBO J. 1989 Sep;8(9):2711–2716. doi: 10.1002/j.1460-2075.1989.tb08412.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gros M. F., te Riele H., Ehrlich S. D. Rolling circle replication of single-stranded DNA plasmid pC194. EMBO J. 1987 Dec 1;6(12):3863–3869. doi: 10.1002/j.1460-2075.1987.tb02724.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruss A. D., Ross H. F., Novick R. P. Functional analysis of a palindromic sequence required for normal replication of several staphylococcal plasmids. Proc Natl Acad Sci U S A. 1987 Apr;84(8):2165–2169. doi: 10.1073/pnas.84.8.2165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruss A., Ehrlich S. D. The family of highly interrelated single-stranded deoxyribonucleic acid plasmids. Microbiol Rev. 1989 Jun;53(2):231–241. doi: 10.1128/mr.53.2.231-241.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodgson A. L., Bird P., Nisbet I. T. Cloning, nucleotide sequence, and expression in Escherichia coli of the phospholipase D gene from Corynebacterium pseudotuberculosis. J Bacteriol. 1990 Mar;172(3):1256–1261. doi: 10.1128/jb.172.3.1256-1261.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodgson A. L., Krywult J., Corner L. A., Rothel J. S., Radford A. J. Rational attenuation of Corynebacterium pseudotuberculosis: potential cheesy gland vaccine and live delivery vehicle. Infect Immun. 1992 Jul;60(7):2900–2905. doi: 10.1128/iai.60.7.2900-2905.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu N., Messing J. The making of strand-specific M13 probes. Gene. 1982 Mar;17(3):271–277. doi: 10.1016/0378-1119(82)90143-3. [DOI] [PubMed] [Google Scholar]
- Hudson G. S., Davidson B. E. Nucleotide sequence and transcription of the phenylalanine and tyrosine operons of Escherichia coli K12. J Mol Biol. 1984 Dec 25;180(4):1023–1051. doi: 10.1016/0022-2836(84)90269-9. [DOI] [PubMed] [Google Scholar]
- Ilyina T. V., Koonin E. V. Conserved sequence motifs in the initiator proteins for rolling circle DNA replication encoded by diverse replicons from eubacteria, eucaryotes and archaebacteria. Nucleic Acids Res. 1992 Jul 11;20(13):3279–3285. doi: 10.1093/nar/20.13.3279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan S. A., Novick R. P. Structural analysis of plasmid pSN2 in Staphylococcus aureus: no involvement in enterotoxin B production. J Bacteriol. 1982 Feb;149(2):642–649. doi: 10.1128/jb.149.2.642-649.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kieny M. P., Lathe R., Lecocq J. P. New versatile cloning and sequencing vectors based on bacteriophage M13. Gene. 1983 Dec;26(1):91–99. doi: 10.1016/0378-1119(83)90039-2. [DOI] [PubMed] [Google Scholar]
- Krishnan B. R., Fobert P. R., Seitzer U., Iyer V. N. Mutations within the replicon of the IncN plasmid pCU1 that affect its Escherichia coli polA-independence but not its autonomous replication ability. Gene. 1990 Jul 2;91(1):1–7. doi: 10.1016/0378-1119(90)90155-k. [DOI] [PubMed] [Google Scholar]
- MONOD J., COHEN-BAZIRE G., COHN M. Sur la biosynthèse de la beta-galactosidase (lactase) chez Escherichia coli; la spécificité de l'induction. Biochim Biophys Acta. 1951 Nov;7(4):585–599. doi: 10.1016/0006-3002(51)90072-8. [DOI] [PubMed] [Google Scholar]
- Messerotti L. J., Radford A. J., Hodgson A. L. Nucleotide sequence of the replication region from the Mycobacterium-Escherichia coli shuttle vector pEP2. Gene. 1990 Nov 30;96(1):147–148. doi: 10.1016/0378-1119(90)90356-v. [DOI] [PubMed] [Google Scholar]
- Messing J. New M13 vectors for cloning. Methods Enzymol. 1983;101:20–78. doi: 10.1016/0076-6879(83)01005-8. [DOI] [PubMed] [Google Scholar]
- Monk M., Kinross J. Conditional lethality of recA and recB derivatives of a strain of Escherichia coli K-12 with a temperature-sensitive deoxyribonucleic acid polymerase I. J Bacteriol. 1972 Mar;109(3):971–978. doi: 10.1128/jb.109.3.971-978.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novick R. P., Adler G. K., Projan S. J., Carleton S., Highlander S. K., Gruss A., Khan S. A., Iordanescu S. Control of pT181 replication I. The pT181 copy control function acts by inhibiting the synthesis of a replication protein. EMBO J. 1984 Oct;3(10):2399–2405. doi: 10.1002/j.1460-2075.1984.tb02146.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novick R. P. Staphylococcal plasmids and their replication. Annu Rev Microbiol. 1989;43:537–565. doi: 10.1146/annurev.mi.43.100189.002541. [DOI] [PubMed] [Google Scholar]
- Praszkier J., Bird P., Nikoletti S., Pittard J. Role of countertranscript RNA in the copy number control system of an IncB miniplasmid. J Bacteriol. 1989 Sep;171(9):5056–5064. doi: 10.1128/jb.171.9.5056-5064.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Projan S. J., Carleton S., Novick R. P. Determination of plasmid copy number by fluorescence densitometry. Plasmid. 1983 Mar;9(2):182–190. doi: 10.1016/0147-619x(83)90019-7. [DOI] [PubMed] [Google Scholar]
- Projan S. J., Monod M., Narayanan C. S., Dubnau D. Replication properties of pIM13, a naturally occurring plasmid found in Bacillus subtilis, and of its close relative pE5, a plasmid native to Staphylococcus aureus. J Bacteriol. 1987 Nov;169(11):5131–5139. doi: 10.1128/jb.169.11.5131-5139.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Projan S. J., Novick R. Comparative analysis of five related Staphylococcal plasmids. Plasmid. 1988 May;19(3):203–221. doi: 10.1016/0147-619x(88)90039-x. [DOI] [PubMed] [Google Scholar]
- Radford A. J., Hodgson A. L. Construction and characterization of a Mycobacterium-Escherichia coli shuttle vector. Plasmid. 1991 Mar;25(2):149–153. doi: 10.1016/0147-619x(91)90029-v. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schekman R., Weiner A., Kornberg A. Multienzyme systems of DNA replication. Science. 1974 Dec 13;186(4168):987–993. doi: 10.1126/science.186.4168.987. [DOI] [PubMed] [Google Scholar]
- Schiller J., Groman N., Coyle M. Plasmids in Corynebacterium diphtheriae and diphtheroids mediating erythromycin resistance. Antimicrob Agents Chemother. 1980 Nov;18(5):814–821. doi: 10.1128/aac.18.5.814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwinde J. W., Thum-Schmitz N., Eikmanns B. J., Sahm H. Transcriptional analysis of the gap-pgk-tpi-ppc gene cluster of Corynebacterium glutamicum. J Bacteriol. 1993 Jun;175(12):3905–3908. doi: 10.1128/jb.175.12.3905-3908.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seery L. T., Nolan N. C., Sharp P. M., Devine K. M. Comparative analysis of the pC194 group of rolling circle plasmids. Plasmid. 1993 Nov;30(3):185–196. doi: 10.1006/plas.1993.1051. [DOI] [PubMed] [Google Scholar]
- Seery L., Devine K. M. Analysis of features contributing to activity of the single-stranded origin of Bacillus plasmid pBAA1. J Bacteriol. 1993 Apr;175(7):1988–1994. doi: 10.1128/jb.175.7.1988-1994.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serwold-Davis T. M., Groman N. B., Kao C. C. Localization of an origin of replication in Corynebacterium diphtheriae broad host range plasmid pNG2 that also functions in Escherichia coli. FEMS Microbiol Lett. 1990 Jan 1;54(1-3):119–123. doi: 10.1016/0378-1097(90)90268-u. [DOI] [PubMed] [Google Scholar]
- Serwold-Davis T. M., Groman N., Rabin M. Transformation of Corynebacterium diphtheriae, Corynebacterium ulcerans, Corynebacterium glutamicum, and Escherichia coli with the C. diphtheriae plasmid pNG2. Proc Natl Acad Sci U S A. 1987 Jul;84(14):4964–4968. doi: 10.1073/pnas.84.14.4964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shine J., Dalgarno L. The 3'-terminal sequence of Escherichia coli 16S ribosomal RNA: complementarity to nonsense triplets and ribosome binding sites. Proc Natl Acad Sci U S A. 1974 Apr;71(4):1342–1346. doi: 10.1073/pnas.71.4.1342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sozhamannan S., Dabert P., Moretto V., Ehrlich S. D., Gruss A. Plus-origin mapping of single-stranded DNA plasmid pE194 and nick site homologies with other plasmids. J Bacteriol. 1990 Aug;172(8):4543–4548. doi: 10.1128/jb.172.8.4543-4548.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staudenbauer W. L. Replication of small plasmids in extracts of Escherichia coli. Mol Gen Genet. 1976 Jun 15;145(3):273–280. doi: 10.1007/BF00325823. [DOI] [PubMed] [Google Scholar]
- Tabor S., Richardson C. C. A bacteriophage T7 RNA polymerase/promoter system for controlled exclusive expression of specific genes. Proc Natl Acad Sci U S A. 1985 Feb;82(4):1074–1078. doi: 10.1073/pnas.82.4.1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas C. D., Balson D. F., Shaw W. V. In vitro studies of the initiation of staphylococcal plasmid replication. Specificity of RepD for its origin (oriD) and characterization of the Rep-ori tyrosyl ester intermediate. J Biol Chem. 1990 Apr 5;265(10):5519–5530. [PubMed] [Google Scholar]
- Tomizawa J. Two distinct mechanisms of synthesis of DNA fragments on colicin E1 plasmid DNA. Nature. 1975 Sep 18;257(5523):253–254. doi: 10.1038/257253a0. [DOI] [PubMed] [Google Scholar]
- Trautwetter A., Blanco C. Structural organization of the Corynebacterium glutamicum plasmid pCG100. J Gen Microbiol. 1991 Sep;137(9):2093–2101. doi: 10.1099/00221287-137-9-2093. [DOI] [PubMed] [Google Scholar]
- Vandeyar M. A., Weiner M. P., Hutton C. J., Batt C. A. A simple and rapid method for the selection of oligodeoxynucleotide-directed mutants. Gene. 1988 May 15;65(1):129–133. doi: 10.1016/0378-1119(88)90425-8. [DOI] [PubMed] [Google Scholar]
- Vieira J., Messing J. The pUC plasmids, an M13mp7-derived system for insertion mutagenesis and sequencing with synthetic universal primers. Gene. 1982 Oct;19(3):259–268. doi: 10.1016/0378-1119(82)90015-4. [DOI] [PubMed] [Google Scholar]
- Viret J. F., Alonso J. C. A DNA sequence outside the pUB110 minimal replicon is required for normal replication in Bacillus subtilis. Nucleic Acids Res. 1988 May 25;16(10):4389–4406. doi: 10.1093/nar/16.10.4389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickner R. B., Wright M., Wickner S., Hurwitz J. Conversion of phiX174 and fd single-stranded DNA to replicative forms in extracts of Escherichia coli. Proc Natl Acad Sci U S A. 1972 Nov;69(11):3233–3237. doi: 10.1073/pnas.69.11.3233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasukawa H., Hase T., Sakai A., Masamune Y. Rolling-circle replication of the plasmid pKYM isolated from a gram-negative bacterium. Proc Natl Acad Sci U S A. 1991 Nov 15;88(22):10282–10286. doi: 10.1073/pnas.88.22.10282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- del Solar G. H., Puyet A., Espinosa M. Initiation signals for the conversion of single stranded to double stranded DNA forms in the streptococcal plasmid pLS1. Nucleic Acids Res. 1987 Jul 24;15(14):5561–5580. doi: 10.1093/nar/15.14.5561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- del Solar G., Moscoso M., Espinosa M. Rolling circle-replicating plasmids from gram-positive and gram-negative bacteria: a wall falls. Mol Microbiol. 1993 May;8(5):789–796. doi: 10.1111/j.1365-2958.1993.tb01625.x. [DOI] [PubMed] [Google Scholar]
- te Riele H., Michel B., Ehrlich S. D. Single-stranded plasmid DNA in Bacillus subtilis and Staphylococcus aureus. Proc Natl Acad Sci U S A. 1986 Apr;83(8):2541–2545. doi: 10.1073/pnas.83.8.2541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Mansfeld A. D., van Teeffelen H. A., Baas P. D., Jansz H. S. Two juxtaposed tyrosyl-OH groups participate in phi X174 gene A protein catalysed cleavage and ligation of DNA. Nucleic Acids Res. 1986 May 27;14(10):4229–4238. doi: 10.1093/nar/14.10.4229. [DOI] [PMC free article] [PubMed] [Google Scholar]