Abstract
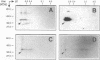
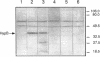
The products of the Rhizobium leguminosarum hyp gene cluster are necessary for synthesis of a functional uptake [NiFe] hydrogenase system in symbiosis with pea plants, and at least for HypB and HypF, a role in hydrogenase-specific nickel metabolism has been postulated (L. Rey, J. Murillo, Y. Hernando, E. Hidalgo, E. Cabrera, J. Imperial, and T. Ruiz-Argüeso, Mol. Microbiol. 8:471-481, 1993). The R. leguminosarum hypB gene product has been overexpressed in Escherichia coli and purified by immobilized nickel chelate affinity chromatography in a single step. The purified recombinant HypB protein was able to bind 3.9 +/- 0.1 Ni2+ ions per HypB monomer in solution. Co2+, Cu2+, and Zn2+ ions competed with Ni2+ with increasing efficiency. Monospecific HypB antibodies were raised and used to show that HypB is synthesized in R. leguminosarum microaerobic vegetative cells and pea bacteroids but not in R. leguminosarum aerobic cells. HypB protein synthesized by R. leguminosarum microaerobic vegetative cells could also be isolated by immobilized nickel chelate affinity chromatography. A histidine-rich region at the amino terminus of the protein (23-HGHHHH DGHHDHDHDHDHHRGDHEHDDHHH-54) is proposed to play a role in nickel binding, both in solution and in chelated form.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bourne H. R., Sanders D. A., McCormick F. The GTPase superfamily: conserved structure and molecular mechanism. Nature. 1991 Jan 10;349(6305):117–127. doi: 10.1038/349117a0. [DOI] [PubMed] [Google Scholar]
- Chen J. C., Mortenson L. E. Identification of six open reading frames from a region of the Azotobacter vinelandii genome likely involved in dihydrogen metabolism. Biochim Biophys Acta. 1992 Jun 15;1131(2):199–202. doi: 10.1016/0167-4781(92)90077-d. [DOI] [PubMed] [Google Scholar]
- Colbeau A., Richaud P., Toussaint B., Caballero F. J., Elster C., Delphin C., Smith R. L., Chabert J., Vignais P. M. Organization of the genes necessary for hydrogenase expression in Rhodobacter capsulatus. Sequence analysis and identification of two hyp regulatory mutants. Mol Microbiol. 1993 Apr;8(1):15–29. doi: 10.1111/j.1365-2958.1993.tb01199.x. [DOI] [PubMed] [Google Scholar]
- Dernedde J., Eitinger M., Friedrich B. Analysis of a pleiotropic gene region involved in formation of catalytically active hydrogenases in Alcaligenes eutrophus H16. Arch Microbiol. 1993;159(6):545–553. doi: 10.1007/BF00249034. [DOI] [PubMed] [Google Scholar]
- Friedrich B., Schwartz E. Molecular biology of hydrogen utilization in aerobic chemolithotrophs. Annu Rev Microbiol. 1993;47:351–383. doi: 10.1146/annurev.mi.47.100193.002031. [DOI] [PubMed] [Google Scholar]
- Hemdan E. S., Zhao Y. J., Sulkowski E., Porath J. Surface topography of histidine residues: a facile probe by immobilized metal ion affinity chromatography. Proc Natl Acad Sci U S A. 1989 Mar;86(6):1811–1815. doi: 10.1073/pnas.86.6.1811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hidalgo E., Leyva A., Ruiz-Argüeso T. Nucleotide sequence of the hydrogenase structural genes from Rhizobium leguminosarum. Plant Mol Biol. 1990 Aug;15(2):367–370. doi: 10.1007/BF00036924. [DOI] [PubMed] [Google Scholar]
- Hidalgo E., Palacios J. M., Murillo J., Ruiz-Argüeso T. Nucleotide sequence and characterization of four additional genes of the hydrogenase structural operon from Rhizobium leguminosarum bv. viciae. J Bacteriol. 1992 Jun;174(12):4130–4139. doi: 10.1128/jb.174.12.4130-4139.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hochuli E., Döbeli H., Schacher A. New metal chelate adsorbent selective for proteins and peptides containing neighbouring histidine residues. J Chromatogr. 1987 Dec 18;411:177–184. doi: 10.1016/s0021-9673(00)93969-4. [DOI] [PubMed] [Google Scholar]
- Jacobi A., Rossmann R., Böck A. The hyp operon gene products are required for the maturation of catalytically active hydrogenase isoenzymes in Escherichia coli. Arch Microbiol. 1992;158(6):444–451. doi: 10.1007/BF00276307. [DOI] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Le Grice S. F., Grüninger-Leitch F. Rapid purification of homodimer and heterodimer HIV-1 reverse transcriptase by metal chelate affinity chromatography. Eur J Biochem. 1990 Jan 26;187(2):307–314. doi: 10.1111/j.1432-1033.1990.tb15306.x. [DOI] [PubMed] [Google Scholar]
- Lee M. H., Pankratz H. S., Wang S., Scott R. A., Finnegan M. G., Johnson M. K., Ippolito J. A., Christianson D. W., Hausinger R. P. Purification and characterization of Klebsiella aerogenes UreE protein: a nickel-binding protein that functions in urease metallocenter assembly. Protein Sci. 1993 Jun;2(6):1042–1052. doi: 10.1002/pro.5560020617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leyva A., Palacios J. M., Mozo T., Ruiz-Argüeso T. Cloning and characterization of hydrogen uptake genes from Rhizobium leguminosarum. J Bacteriol. 1987 Nov;169(11):4929–4934. doi: 10.1128/jb.169.11.4929-4934.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leyva A., Palacios J. M., Murillo J., Ruiz-Argüeso T. Genetic organization of the hydrogen uptake (hup) cluster from Rhizobium leguminosarum. J Bacteriol. 1990 Mar;172(3):1647–1655. doi: 10.1128/jb.172.3.1647-1655.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y. P., Sharer J. D., March P. E. GTPase-dependent signaling in bacteria: characterization of a membrane-binding site for era in Escherichia coli. J Bacteriol. 1994 Jan;176(1):44–49. doi: 10.1128/jb.176.1.44-49.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loddenkötter B., Kammerer B., Fischer K., Flügge U. I. Expression of the functional mature chloroplast triose phosphate translocator in yeast internal membranes and purification of the histidine-tagged protein by a single metal-affinity chromatography step. Proc Natl Acad Sci U S A. 1993 Mar 15;90(6):2155–2159. doi: 10.1073/pnas.90.6.2155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lutz S., Jacobi A., Schlensog V., Böhm R., Sawers G., Böck A. Molecular characterization of an operon (hyp) necessary for the activity of the three hydrogenase isoenzymes in Escherichia coli. Mol Microbiol. 1991 Jan;5(1):123–135. doi: 10.1111/j.1365-2958.1991.tb01833.x. [DOI] [PubMed] [Google Scholar]
- Maier T., Jacobi A., Sauter M., Böck A. The product of the hypB gene, which is required for nickel incorporation into hydrogenases, is a novel guanine nucleotide-binding protein. J Bacteriol. 1993 Feb;175(3):630–635. doi: 10.1128/jb.175.3.630-635.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsudaira P. Sequence from picomole quantities of proteins electroblotted onto polyvinylidene difluoride membranes. J Biol Chem. 1987 Jul 25;262(21):10035–10038. [PubMed] [Google Scholar]
- McPherson G. A. Analysis of radioligand binding experiments. A collection of computer programs for the IBM PC. J Pharmacol Methods. 1985 Nov;14(3):213–228. doi: 10.1016/0160-5402(85)90034-8. [DOI] [PubMed] [Google Scholar]
- Miller R. W., Eady R. R. Molybdenum nitrogenase of Azotobacter chroococcum. Tight binding of MgADP to the MoFe protein. Biochem J. 1989 Nov 1;263(3):725–729. doi: 10.1042/bj2630725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulrooney S. B., Hausinger R. P. Sequence of the Klebsiella aerogenes urease genes and evidence for accessory proteins facilitating nickel incorporation. J Bacteriol. 1990 Oct;172(10):5837–5843. doi: 10.1128/jb.172.10.5837-5843.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munson P. J., Rodbard D. Ligand: a versatile computerized approach for characterization of ligand-binding systems. Anal Biochem. 1980 Sep 1;107(1):220–239. doi: 10.1016/0003-2697(80)90515-1. [DOI] [PubMed] [Google Scholar]
- O'Farrell P. H. High resolution two-dimensional electrophoresis of proteins. J Biol Chem. 1975 May 25;250(10):4007–4021. [PMC free article] [PubMed] [Google Scholar]
- Palacios J. M., Murillo J., Leyva A., Ditta G., Ruiz-Argüeso T. Differential expression of hydrogen uptake (hup) genes in vegetative and symbiotic cells of Rhizobium leguminosarum. Mol Gen Genet. 1990 May;221(3):363–370. doi: 10.1007/BF00259401. [DOI] [PubMed] [Google Scholar]
- Reinard T., Jacobsen H. J. An inexpensive small volume equilibrium dialysis system for protein-ligand binding assays. Anal Biochem. 1989 Jan;176(1):157–160. doi: 10.1016/0003-2697(89)90286-8. [DOI] [PubMed] [Google Scholar]
- Rey L., Hidalgo E., Palacios J., Ruiz-Argüeso T. Nucleotide sequence and organization of an H2-uptake gene cluster from Rhizobium leguminosarum bv. viciae containing a rubredoxin-like gene and four additional open reading frames. J Mol Biol. 1992 Dec 5;228(3):998–1002. doi: 10.1016/0022-2836(92)90886-o. [DOI] [PubMed] [Google Scholar]
- Rey L., Murillo J., Hernando Y., Hidalgo E., Cabrera E., Imperial J., Ruiz-Argüeso T. Molecular analysis of a microaerobically induced operon required for hydrogenase synthesis in Rhizobium leguminosarum biovar viciae. Mol Microbiol. 1993 May;8(3):471–481. doi: 10.1111/j.1365-2958.1993.tb01591.x. [DOI] [PubMed] [Google Scholar]
- Short J. M., Fernandez J. M., Sorge J. A., Huse W. D. Lambda ZAP: a bacteriophage lambda expression vector with in vivo excision properties. Nucleic Acids Res. 1988 Aug 11;16(15):7583–7600. doi: 10.1093/nar/16.15.7583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith P. K., Krohn R. I., Hermanson G. T., Mallia A. K., Gartner F. H., Provenzano M. D., Fujimoto E. K., Goeke N. M., Olson B. J., Klenk D. C. Measurement of protein using bicinchoninic acid. Anal Biochem. 1985 Oct;150(1):76–85. doi: 10.1016/0003-2697(85)90442-7. [DOI] [PubMed] [Google Scholar]
- Studier F. W., Moffatt B. A. Use of bacteriophage T7 RNA polymerase to direct selective high-level expression of cloned genes. J Mol Biol. 1986 May 5;189(1):113–130. doi: 10.1016/0022-2836(86)90385-2. [DOI] [PubMed] [Google Scholar]
- Studier F. W., Rosenberg A. H., Dunn J. J., Dubendorff J. W. Use of T7 RNA polymerase to direct expression of cloned genes. Methods Enzymol. 1990;185:60–89. doi: 10.1016/0076-6879(90)85008-c. [DOI] [PubMed] [Google Scholar]
- Tibelius K. H., Du L., Tito D., Stejskal F. The Azotobacter chroococcum hydrogenase gene cluster: sequences and genetic analysis of four accessory genes, hupA, hupB, hupY and hupC. Gene. 1993 May 15;127(1):53–61. doi: 10.1016/0378-1119(93)90616-b. [DOI] [PubMed] [Google Scholar]
- Vignais P. M., Toussaint B. Molecular biology of membrane-bound H2 uptake hydrogenases. Arch Microbiol. 1994;161(1):1–10. doi: 10.1007/BF00248887. [DOI] [PubMed] [Google Scholar]
- Waugh R., Boxer D. H. Pleiotropic hydrogenase mutants of Escherichia coli K12: growth in the presence of nickel can restore hydrogenase activity. Biochimie. 1986 Jan;68(1):157–166. doi: 10.1016/s0300-9084(86)81080-x. [DOI] [PubMed] [Google Scholar]
- Wülfing C., Lombardero J., Plückthun A. An Escherichia coli protein consisting of a domain homologous to FK506-binding proteins (FKBP) and a new metal binding motif. J Biol Chem. 1994 Jan 28;269(4):2895–2901. [PubMed] [Google Scholar]
- Xu H. W., Wall J. D. Clustering of genes necessary for hydrogen oxidation in Rhodobacter capsulatus. J Bacteriol. 1991 Apr;173(7):2401–2405. doi: 10.1128/jb.173.7.2401-2405.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]