Abstract
The vascular system is locally specialized to accommodate widely varying blood flow and pressure and the distinct needs of individual tissues. The endothelial cells (ECs) that line the lumens of blood and lymphatic vessels play an integral role in the regional specialization of vascular structure and physiology. However, our understanding of EC diversity is limited. To explore EC specialization on a global scale, we used DNA microarrays to determine the expression profile of 53 cultured ECs. We found that ECs from different blood vessels and microvascular ECs from different tissues have distinct and characteristic gene expression profiles. Pervasive differences in gene expression patterns distinguish the ECs of large vessels from microvascular ECs. We identified groups of genes characteristic of arterial and venous endothelium. Hey2, the human homologue of the zebrafish gene gridlock, was selectively expressed in arterial ECs and induced the expression of several arterial-specific genes. Several genes critical in the establishment of left/right asymmetry were expressed preferentially in venous ECs, suggesting coordination between vascular differentiation and body plan development. Tissue-specific expression patterns in different tissue microvascular ECs suggest they are distinct differentiated cell types that play roles in the local physiology of their respective organs and tissues.
The vascular system is a complex network of vessels connecting the heart with diverse organs and tissues to maintain their homeostasis in response to physiological and pathological changes. Endothelial cells (ECs) line the inner surface of blood and lymphatic vessels and play important roles in the development and remodeling of vasculature, maintenance of vascular tone, blood fluidity, coagulation, nutrient exchange, and organ development. Given the diversity of the vascular channels and the associated differences in hemodynamics, structure, and embryonic origins, it is not surprising that the ECs lining different vessels exhibit regional specializations in morphology and functions. Systematic identification of the specific molecular features of specialized components of the vascular network will not only enhance our understanding of vascular development, lymphocyte homing, and various disease processes but may also provide the potential for site-specific delivery of therapeutic agents. A variety of approaches have been applied to identify molecular markers for specific vessels or vascular beds, including the Stamper–Woodruff assay, mAbs, phage display, differential display, sage analysis (1), and microarrays (2).
In this article, we used DNA microarrays to explore the diversity of ECs in different types of blood vessels and different anatomic locations, as reflected in the intrinsic global gene expression programs observed in ECs from different body sites cultured under identical conditions. Our analysis shows that ECs from different blood vessels or anatomical sites are indeed distinct differentiated cell types with correspondingly characteristic gene expression programs. The analyses of these differences provide insights into the diversity of vascular beds, their differentiation programs, and distinct adaptations to physiological and pathological changes.
Materials and Methods
Reagents and Cells. Intestinal and nasal polyp ECs were obtained from the University of Oslo (3). Two of the human umbilical vein EC (HUVEC) samples were provided by the J. Swain laboratory (Stanford University). All other ECs were from Cambrex, East Rutherford, NJ. The cells were thawed and propagated on plastic flasks in EGM-2MV media (Clonetics, San Diego) containing 5% fetal sera and vascular endothelial growth factor, basic fibroblast growth factor, epidermal growth factor, hydrocortisone, ascorbic acid, and antibiotics. After the cells grew to 60–70% confluency, we changed the media 13 h before harvesting the mRNA with FastTrack (Invitrogen). The cells were harvested between the third and fifth passages (10–16 generations in culture). Antibodies against cytokeratins (C-11, Sigma), desmin (Ab-1, NeoMarkers, Fremont, CA), glial fibrillary acidic protein (ab-7, NeoMarkers), vimentin (V9, Sigma), and CD31 (Ab-2, Neomarkers or PharMingen) were obtained from the indicated sources.
Microarray Procedure and Data Analysis. Human DNA microarray production of the Stanford Functional Genomic Facility and hybridization, scanning, and analysis with the Stanford Microarray Database (4) were performed as described (5). For the Hey2 expression study, total RNAs were purified with TRIzol reagents and amplified by using a linear amplification method. Human common RNA reference (Strategene) was used in all experiments as the standard reference. Hierarchical clustering with weighted average linkage clustering (6) was performed as described. To identify genes that showed significant variations in expression between large vessels vs. microvascular ECs and artery vs. vein ECs, a Wilcoxon rank-sum test was performed by using P < 0.005 as a threshold (7). To identify genes with tissue-specific expression, we used multiclass Significance Analysis of Microarrays (SAM) (8) to analyze variations in expression in ECs from different tissues. The tissue-specific gene list was selected to have a false discovery rate of 0.2%, using 100 iterations. For detailed procedures and complete data, see http://microarray-pubs.stanford.edu/endothelial.
Retroviral Vector Production and Infection of HUVECs. The Hey2 cDNA (American Type Culture Collection) was cloned into pMIGR (gift of W. Pear, University of Pennsylvania, Philadelphia) (9) and used to transfect amphotropic Phoenix cells (gift of Gary Nolan, Stanford University) to generate retrovirus containing either GFP or Hey_GFP to infect HUVECs by spin infection (protocol detailed at www.stanford.edu/group/nolan). The HUVECs were analyzed and collected with a cell sorter 48 h after retroviral infection.
Results and Discussion
Overview of the Gene Expression Patterns. Fifty-two purified EC samples, representing 14 distinct locations, were propagated in identical culture conditions. This sample set included ECs from five different arteries (aorta, coronary artery, pulmonary artery, iliac artery, and umbilical artery), two different veins (umbilical vein and saphenous vein), and seven different tissues (skin, lung, intestine, uterus myometrium, nasal polyps, bladder, and myocardium). All ECs displayed a cobblestone appearance and were free of contamination by spindle-shaped cells. The cell purity was further assayed by flow cytometry or staining with CD31 antibody, desmin, vimentin, cytokeratin, or glial fibrillary acid protein. Samples that exhibited significant non-EC cells contamination were excluded from further analysis.
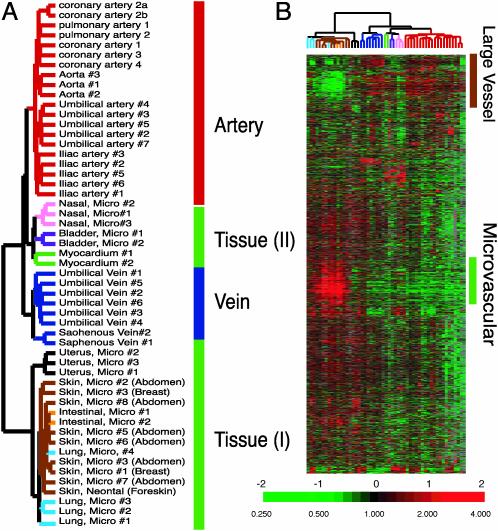
EC mRNA samples were labeled and hybridized to DNA microarrays, containing 43,000 elements representing 32,275 unique Unigene clusters (Build no. 158, released on January 18, 2003). We analyzed mRNA from 53 different cultured EC samples, including two samples from one coronary artery EC culture that was sampled twice at successive passages (coronary arteries 2a and 2b), totaling 2.4 million gene expression measurements. The first significant result was a striking order and consistency in the expression patterns, reflecting the sites of origins of the cultured ECs. Unsupervised hierarchical clustering (6) of the gene expression patterns from all 53 samples produced a consistent grouping of the cells according to their sites of origin (Fig. 1A). This finding suggests that ECs from different locations have distinct and characteristic expression patterns that persist with in vitro culture. The majority of ECs had expression patterns that clustered into discrete groups of ECs from the same location. Overall, the samples were divided into two major branches. Branch I comprises three subgroups: (i) an artery group consisting of all ECs cultured from arteries, including aorta, coronary artery, pulmonary artery, iliac artery, and umbilical artery; (ii) a vein group, consisting of all ECs cultured from umbilical vein and saphenous vein; and (iii) a group we called tissue type II consisting of ECs cultured from nasal polyps, bladder, and myocardium (Fig. 1 A). Branch II, which we designated as tissue type I (Fig. 1 A), contains all of the microvascular ECs from skin, lung, intestine, and myometrium. The most prominent differences between the two branches are defined by two large groups of genes that we label as the large vessel cluster and microvascular cluster, respectively (Fig. 1B).
Fig. 1.
Diversity of EC gene expression patterns. (A) Gene expression patterns of cultured ECs organized by unsupervised hierarchical clustering. The global gene expression patterns of 53 cultured ECs were sorted based on similarity by hierarchical clustering. Approximately 6,900 genes were selected from the total data set, based on variations in expression relative to the mean expression level across all samples >3-fold in at least two cell samples. The sites of origin of each EC culture are indicated and color-coded. The anatomic origins of skin EC are indicated. The apparent order in the grouping of EC gene expression patterns is indicated to the right of the dendrogram. (B) Overview of gene expression patterns of all EC samples. The variations in gene expression described in A are shown in matrix format (5). The scale extends from 0.25- to 4-fold over mean (–2 to +2 in log2 space) as indicated on the left. Gray represents missing data. The gene clusters characteristic of large vessel and microvascular ECs are indicated on the right. Complete data can be found at http://microarray-pubs.stanford.edu/endothelial and in the Stanford Microarray Database (http://genome-www5.stanford.edu/MicroArray/SMD).
Intrinsic Differences in Gene Expression Patterns Between Large Vessel and Microvascular ECs. We used a Wilconxon rank-sum test to identify genes with the most consistent different levels of expression between ECs from large vessels and microvascular ECs (28 large vessel ECs and 25 microvascular ECs) (7). We selected 521 large vessel EC-specific genes and 2,521 microvascular EC-specific genes with P value < 0.005. Although this P value is not corrected for multiple testing, it provides an ordering of genes in terms of their ability to differentiate between large vessel and microvascular ECs. The multiple testing correction was not performed because of heterogeneity in the samples, but the top 235 large vessel and 1,014 microvascular genes display P values < 0.5 even with the conservative Bonferroni correction.
We then performed a hierarchical cluster analysis of all of the EC samples based on expression of these genes in this combined gene list (Fig. 2A). With the exception of two saphenous vein ECs, all of the large vessels ECs were clustered in one branch, separated from all of the microvascular ECs (Fig. 2 A). The distinct gene expression patterns are likely to be related to the characteristic differences in physiological functions of these vascular channels. The differentially expressed genes play diverse roles in endothelial biology, including the biosynthesis of and interaction with extracellular matrix (ECM), neuronal signaling and migration, angiogenesis, and lipid metabolism.
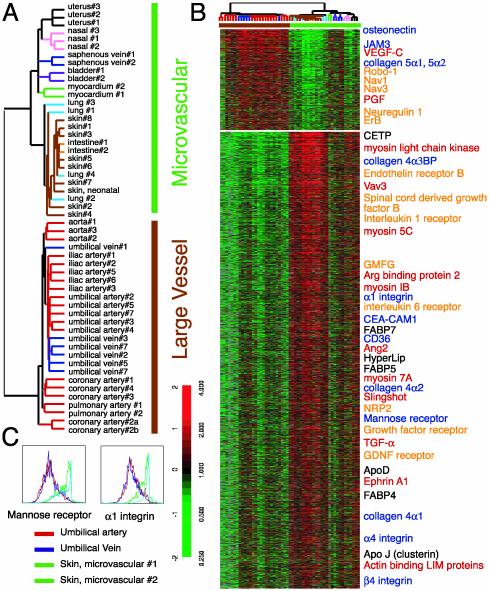
Fig. 2.
Large vessel and microvascular EC gene expression programs. (A) Identification of large vessel vs. microvascular ECs gene expression programs. Dendrogram representing the result of hierarchical clustering of EC samples, based on the similarities in their pattern of expression of the genes selected by Wilcoxon rank-sum test. (B) Features of large vessels and microvascular EC gene expression programs. A total of 521 large vessel-specific and 2,521 microvascular EC-specific genes are shown in ascending order of P values. Genes involved in ECM biosynthesis and interaction (blue), neuroglial signaling and migration (orange), angiogenesis (red), and lipid metabolism (black) are labeled by the indicated colors. (C) Validation of gene expression data by flow cytometry. Fluorescence-activated cell sorting analysis of surface expression of mannose receptor and α1 integrin on two ECs from dermal microcirculation (green, n = 2), one umbilical artery (red, n = 1), and one umbilical vein (blue, n = 1). Complete data can be found at http://microarray-pubs.stanford.edu/endothelial and in the Stanford Microarray Database (http://genome-www5.stanford.edu/MicroArray/SMD).
Large vessel ECs differentially express several genes involved in the biosynthesis and remodeling of ECM, such as fibronectin, collagen 5α1, collagen 5α2, and osteonectin (Fig. 2B, gene names shown in blue). These differences are likely to be related, in part, to the relatively thick vascular wall surrounding the endothelium of the large vessels. On the other hand, microvascular ECs express genes encoding basement membrane proteins, such as laminin, collagen 4α1, collagen 4α2, and collagen 4α-binding protein and ECM-interacting proteins, such as CD36, α1 integrin, α4 integrin, α9 integrin, and β4 integrin (Fig. 2B, gene names shown in blue), perhaps related to the intimate association of microvascular ECs with the basement membrane and ECM.
ECs present a physical barrier to both blood-borne pathogens and immune cells, which must transverse the barrier for trafficking between tissues and the bloodstream. It is interesting to note that microvascular ECs express higher levels of transcripts encoding proteins involved in the traffic of circulating blood cells and pathogens (Fig. 2B, gene names shown in blue). For example, CD36 is a cellular receptor for Plasmodium falciparum; CEACAM-1 (CD66a) interacts with the Opa proteins of Neisseria bacteria, and macrophage mannose receptors (CD206) are important for pathogen trapping and lymphocyte recruitment. “Docking” with these microvascular ECs-specific gene products proteins may help both pathogens and immune cells migrate to target tissues. We have confirmed the specific surface expression of mannose receptor and α1 integrin on skin microvascular ECs by flow cytometry (Fig. 2C).
The similarity of the migration paths of blood vessels and nerves has inspired interest in their interactions during development. Our data highlight the divergent ways in which large vessels and microvascular circulation appear designed to communicate with peripheral nerves during development and maturation. Large vessel ECs express many genes associated with neuronal cells, such as robo-1, neuron navigator 1, and neuron navigator 3 (NAV1, NAV3), neuroligin, neurogranin, and neuroregulin and its receptor ErbB (Fig. 2B, gene names shown in orange). Some of these proteins play important roles in neuronal migration during development. For example, robo-1 is a surface receptor for the slit proteins that act as migration signals. This finding suggests the possibility that large vessel ECs may respond to some of the same guidance signals that specify the paths of neural processes through the mesenchymes of different target tissues and organs (10). Recent studies have shown that when peripheral nerves are severed, large vessels still migrate to their target locations while microvascular circulation is greatly affected (11). Our results show that microvascular ECs express receptors for a wide variety of paracrine signals from neuroglial cells, such as growth hormone receptor, endothelin receptor B, glycine receptor, purinergic receptor, glial cell line derived-neurotrophic factor receptor, platelet-derived growth factor receptor, IL-1 receptor, and IL-6 receptor (Fig. 2B, gene names shown in orange.) Microvascular ECs also express secreted factors that promote the survival and differentiation of neuroglial cells, such as transforming growth factor α, glial maturation factor γ, stromal cell-derived factor 1, and spinal cord-derived growth factor. These proteins, uniquely expressed in small vessel ECs, suggest an intimate functional interaction between microvasculature and peripheral nerves. In view of the importance of Schwann cells in microvasculature development (11), the microvascular EC-specific expression of genes involved in the EC–glia cell interaction (glial cell line derived-neurotrophic factor receptor and glial maturation factor γ) is particular noteworthy.
Large vessel ECs expressed placental growth factor, which is involved in the establishment of collateral circulation, a response to ischemia unique to large vessels (12). They also express vascular endothelial growth factor C, a growth factor essential for the growth and differentiation of lymphatic vessels (13) (Fig. 2B, gene names shown in red). Microvascular networks are the main sites for angiogenesis in adults. Many genes associated with angiogenesis were expressed specifically in microvascular ECs (Fig. 2B, gene names shown in red). Angiopoietin 2, a marker of tumor angiogenesis, modulates the remodeling of vessels during angiogenesis by reverting vessels to a more plastic state that may facilitate the vascular sprouting necessary for subsequent remodeling (14). Lmo2 is a LIM-only transcription factor involved in angiogenesis. Sprouty is a fibroblast growth factor antagonist that participates in regulating the branching pattern of insect tracheal trees (15). Transforming growth factor α induces ephrin-A1 expression that is involved in tumor angiogenesis (16). The expression of both transforming growth factor α and ephrin-A1 in microvascular ECs raises the possibility of autocrine regulation. The higher level of expression of actin binding LIM protein 1, actinin-associated LIM protein, Arg binding protein 2, Slingshot, vav3, myosin IB, myosin 5C, myosin 7A, and myosin light chain kinase in the microvascular ECs may be related to the ability of microvascular ECs to undergo extensive cytoskeletal remodeling and migration during angiogenesis.
The microvascular EC gene clusters included genes related to lipid transport and metabolism, such as ApoD, ApoJ (clusterin), ApoL, cholesteryl ester transfer protein, FABP4, FABP5, and Hyperlip (Fig. 2B, gene names shown in black), consistent with a major role for small vessel ECs in mediating lipid transport and metabolism.
Several genes that have previously been identified as lymphatic markers were present in the microvascular gene cluster, including prox-1, desmoplakin (13), and neuropilin 2, a gene essential for the development of lymphatic vasculature (17). These results suggest that lymphatic ECs (LECs) may have been copurified in the microvascular EC cultures. Delineation of LECs vs. blood ECs (BEC) gene expression has been recently reported (2, 18). There are some discrepancies among these two reports and some proposed differences between LECs vs. BECs appear in the microvascular vs. large vessel ECs gene lists. Importantly, most of the genes that we identified comparing large vessels vs. microvascular ECs were not different between LECs and BECs. Thus, the difference between large vessel and microvascular EC gene expression could not be solely accounted for from LEC contamination in the microvascular EC preparations.
Gene Expression Patterns Difference Between Arterial and Venous ECs. Arteries and veins in the vertebrate circulatory system are functionally defined by the direction of blood flow relative to the heart. Recent evidence indicates that the artery-vein identities of ECs are established before blood circulation begins (19). Several molecular markers specifically expressed in arteries or veins have been identified in model organisms (19). In the unsupervised clustering analysis, all of the arterial and venous ECs were separated into two different branches (arterial branch and venous branch in Fig. 1 A), reflecting extensive differences in their expression patterns. Because the microvascular ECs could contain different ECs from a complex mixture of vessels of different kinds, such as small arteries, arterioles, capillaries, veins, venules or lymphatic vessels, they were excluded from analysis. We used a rank-sum test to identify genes with the largest, consistent differences in expression between two groups of samples: 8 venous ECs (6 umbilical veins and 2 sapheonous veins) and 20 arterial ECs (3 aortas, 2 pulmonary arteries, 5 coronary arteries, 5 umbilical arteries, and 5 iliac arteries). We selected 817 vein-specific genes and 59 artery-specific genes (Fig. 3A) with P values < 0.005. The multiple testing correction was not performed because of heterogeneity in the samples, but the top 7 arterial and 65 venous genes display P values < 0.5 even with the conservative Bonferroni correction.
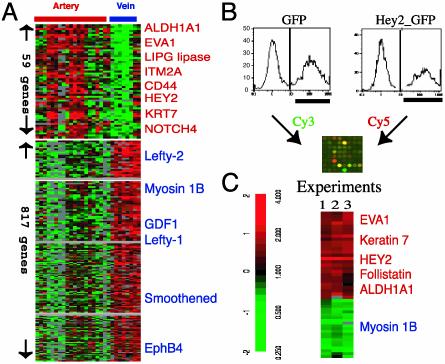
Fig. 3.
Artery and vein EC-specific gene expression programs. (A) Artery- or vein-specific genes identified by a Wilcoxon rank-sum test are shown in ascending order of P value within each gene list, and names of select artery-specific genes (red) and vein-specific genes (blue) are shown. Complete data can be found at http://microarray-pubs.stanford.edu/endothelial and in the Stanford Microarray Database (http://genome-www5.stanford.edu/MicroArray/SMD). (B) Strategy for identification of Hey2 target genes. HUVECs were infected by retrovirus expressing GFP or Hey2-IRES-GFP. The GPF-positive cells (labeled black bar) were sorted by fluorescence-activated cell sorting, and RNA from the sorted cells was reverse-transcribed with either Cy3 (GFP) or Cy5 (Hey2_GFP) and used for competitive hybridization to cDNA microarrays. (C) Induction of artery-specific genes by Hey2. Genes exhibiting >2-fold variation in expression in two of three independent experiments as described in B are shown. Genes previously identified as artery-specific in A are colored red, and vein-specific genes are colored blue. As an internal positive control, Hey2 expression was always higher in cells infected with Hey2_GFP retrovirus compared with cells infected with GFP retrovirus.
The higher number of vein-specific genes compared with artery-specific genes may reflect the relatively low diversity of vein samples (2 types, 8 samples) compared with artery samples (5 types, 20 samples). EphB4, as previously reported, was among the venous EC-specific genes (Fig. 3A). Genes involved in determining left/right (L-R) asymmetry of the body plan, including smoothened, growth differentiation factor 1, lefty-1, and lefty-2 were particularly noteworthy members of the venous-specific group. During development, right-sided looping of the developing cardiac tube is the first sign of the L-R asymmetry, and the L-R determination is intimately connected to the development of the heart and vasculature. Defects in L-R asymmetry, such as situs inversus, are characteristically associated with vascular anomalies. Disruption of lefty-1 in mice leads to malpositioning of venous vessels and anomalies in the heart and its connection with major vessels (20). Lefty-1 is an antagonist of Nodal through the activin-like receptor. Mutations in activin receptor-like kinase-1 result in persistent arterial-venous shunts and early loss of anatomical, molecular, and functional distinctions between arteries and veins (21). The persistent expression of these genes in venous ECs suggests a molecular connection between L-R determination and the distinct differentiation programs of arterial and venous ECs.
The arterial EC-specific gene cluster includes cell surface proteins (Notch 4, EVA1, CD44, Ephrin-B1, and integral membrane protein 2A), metabolic enzymes (aldehyde dehydrogenase A1 and endothelial lipase), C17, keratin 7, and a transcription factor termed Hairy/Enhancer of split-related basic helix–loop–helix protein 2 (Hey2). We have confirmed the arterial-specific expression of Hey2 and C17 transcript with real-time PCR and CD44 surface expression with flow cytometry (Figs. 5 and 6, which are published as supporting information on the PNAS web site, www.pnas.org). Several of these genes have already been implicated in vascular development. The Notch family of receptors and ligand Delta-like 4 show arterial expression in zebrafish/mouse and are essential for arterial cell fate determination (22). Hey2, a member of the Hairy-related transcription factor family of transcription factors, is induced by Notch signaling (23). The zebrafish homolog of Hey2 is the gene targeted by the gridlock (grl) mutation (24), which leads to a localized defect in vascular patterning of dorsal aorta, consistent with specific expression of grl in dorsal aorta. The apparently conserved expression pattern of the Notch pathway (Notch 4 and Hey2) in human arterial ECs highlights the potential importance of this pathway in human arterial EC differentiation.
Hey2 Activates Expression of Arterial-Specific Genes. To further examine the possible role of Hey2 in EC differentiation, we tested whether Hey2 expression would be sufficient to confer features of arterial EC gene expression on vein-derived ECs. We infected HUVECs with a retroviral vector carrying the Hey2 gene, along with a GFP marker, or with a control vector expressing GFP alone, and selected the infected ECs by positive GFP expression by using flow cytometry. Typically, 40–60% of the infected HUVECs exhibited elevated levels of GFP (Fig. 3B). RNA isolated from these GFP-positive cells was labeled with either Cy3 (GFP only) or Cy5 (Hey2_GFP) and the global expression profiles were analyzed by comparative hybridization to DNA microarrays (Fig. 3B). Forty-nine genes were found to be consistently affected by Hey2 expression in three independent experiments (Fig. 3C). The Hey2 transcript was consistently elevated in the Hey2-transduced cells, serving as positive control for the retroviral transduction. Hey2 transduction leads to elevated expression of follistatin, an antagonist of activin, and several artery-specific genes identified in our previous analysis (Fig. 3A), including ADHA1, EVA1, and keratin 7 (gene names in red in Fig. 3C), and down-regulation of myosin I (colored in blue in Fig. 3C). These results suggest a pathway of arterial EC differentiation in which Hey2 turns on features of artery-specific gene expression program. Hey2 induction of follistatin may also contribute to arterial differentiation by antagonizing transforming growth factor β family members (such as GDF, lefty-1, and lefty-2) expressed by veinous ECs.
Genes Differentially Expressed in ECs from Different Tissues. Previous studies with phage display have revealed molecular heterogeneity in the vascular beds in different organs/tissues (25), but little is known about the detailed origins of this heterogeneity. ECs are also implicated in the development of pancreas (26) and liver (27), suggesting their ability to deliver specific paracrine inductive signals during development. Our survey of different fibroblasts from diverse anatomical sites revealed intrinsic differences in gene expression programs, related to the anatomic site of origin (10). We were therefore interested in exploring the possibility that there might be similar consistent differences among ECs derived from different organs and tissues. We used a permutation-based technique termed Significance Analysis of Microarrays (SAM) (8) to systematically identify genes whose expression in cultured ECs varied according to the tissues of origin. SAM was used for multiclass classification because rank-sum test is not able to perform mutliclass classification. The analysis identified >2,000 such genes, with an estimated false discovery rate <0.2% (Fig. 4A). Some of the identified genes varied in expression among ECs from different groups of tissues, such as the tissue type I and II branches in Fig. 1 A, and some were highly specific to ECs from a single tissue. Groups of genes specifically expressed in one or two of the tissues we examined are marked and expanded in Fig. 4B.
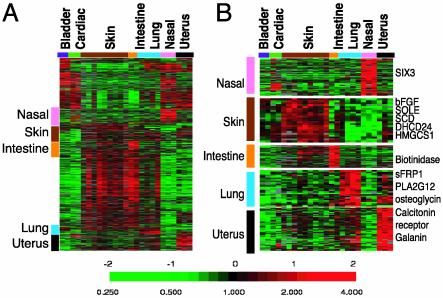
Fig. 4.
Identification of tissue-specific EC genes. (A) The expression patterns of tissue-specific genes as identified by Significance Analysis of Microarrays among all of the tissue microvascular ECs are shown. The clusters of genes with unique tissue expression in nasal polyps (pink), skin (brown), intestine (orange), lung (blue), and uterus (black) are marked by the indicated color and expanded on the right (B) with selected gene names. Complete data can be found at http://microarray-pubs.stanford.edu/endothelial and in the Stanford Microarray Database (http://genome-www5.stanford.edu/MicroArray/SMD).
Nasal polyp ECs expressed SIX3, a homeodomain protein with forebrain-specific expression (28) (Fig. 4B). The specific expression of SIX3 in ECs from the nasal cavity suggests that developmentally patterned transcriptional programs in ECs may preserve the positional information consistent with their sites of origins even after transfer to tissue culture. Diverse tissue-specific genes point to different physiological properties of ECs from each site. The skin ECs expressed basic fibroblast growth factor and a set of genes involved in cholesterol biosynthesis, including squalene epoxidase (SOLE), 24-dehydrocholesterol reductase (DHCR24), stearoyl-CoA desaturase (SCD), fatty acid desaturase (FADS2), and 3-hydroxy-3-metheylglutaryl-CoA synthase 1 (HMGCS1). The intestinal ECs express biotinidase, which is involved in biotin recycling. The lung ECs specifically expressed phospholipase A2 group XII, an enzyme involved in surfactant secretion, and the developmental regulators, secreted frizzled related protein 1 (sFRP1), and osteoglycin. Myometrium ECs specifically expressed the calcitonin receptor and gallanin. Calcitonin is important for the implantation of embryos (29), and gallanin is a peptide hormone that stimulates the contraction of the uterus myometrium (30). Their specific expression in myometrium EC may point to active roles played by ECs in the function and physiology of the uterus. Together, these results show that ECs from different tissues are distinct differentiated cell types, with specialized roles in the functions and physiology of the respective tissues/organs from which they were derived. It will be an important challenge to trace the steps by which angioblasts adopt a distinct differentiated fate in each different tissue or organ during development and tissue repair.
Conclusions
There is no doubt that our vascular system is extremely diverse in structure, architecture, and physiology. The branching architectures, histological structures, and hemodynamic characteristics are clearly specified and regulated in great detail and with exquisite anatomical specificity, during development and in physiological adaptation. Recent studies have provided evidence that the “stromal cells” that make up the infrastructure of our bodies may be more diverse and specialized than previously apparent (10). We investigated the possibility that ECs might comprise a diversity of specialized differentiated cell types and that understanding the nature and the basis of this diversity might provide new insights into vascular physiology, organogenesis, and vascular diseases. Our studies provide strong evidence that ECs actually comprise many distinct differentiated cell types, with distinct, intrinsic gene expression programs, that persist for many generations in cell culture. It is important to note that, although the gene expression patterns we saw in cultured cells clearly reveal the molecular heterogeneity of these cells, their correspondence with the gene expression programs in their in vivo counterparts remains to be defined. Our results do suggest at least three broad themes in the specialization of ECs. First, ECs from large vessels share many differences from microvascular ECs in global gene expression patterns. Some of these differences may be related to differences in the mechanical and structural characteristics that accompany differences in vessel calibers, others may be related to different physiological roles in coagulation and hemodynamics. An understanding of the basis of the differences and their implications will require much more investigation. Second, ECs from arteries and veins appear to share many distinct differences in their expression programs. The persistence of molecular distinction between arterial and venous ECs in culture is consistent with the accumulating evidences that their identities are established before patent vessel formation and independent of environmental differences. Interestingly, some of these differences suggest an evolutionarily conserved early developmental divergence, in which the Notch signaling pathway and Hey2 transcription factor appear to have a role. Third, we find extensive specialization of ECs related to anatomical locations. The differences in expression strongly suggest the possibility that there will be important differences in their physiology and developmental responses in those locations, their behavior in disease processes, and their responses to drugs.
This diversity of differentiated cell types, and their retention of distinct differentiated gene expression programs in homogenous cell culture, has implications for the design of research studies using isolated ECs, and for tissue engineering using cultured ECs, and surgical treatments that rely on autologous vessel grafts. The differences in gene expression programs strongly suggest that ECs from different sites will have corresponding differences in their developmental potential, their response to experimental manipulation, and their interactions with other cells. Therefore, the precise identity of ECs should be carefully considered in future research and therapeutic applications involving manipulations of ECs or vascular structures.
Our studies have also offered insight into the EC's role in different disease processes affecting vascular systems. It has long been recognized that diseases that affect vascular structures do not do so uniformly. We are used to thinking that these differences are largely caused by differences extrinsic to the ECs (such as local inflammatory responses and hemodynamic differences in the corresponding vessels), which differentially affect what would otherwise be a relatively homogenous cell type. Our studies have clearly shown that ECs from different blood vessels have distinct expression profiles and these differences might account for the selective involvement in different disease processes. For example, the preferentially arterial involvement of atherosclerosis is usually attributed to the hemodynamic characteristics of arteries. We have found that artery ECs have higher levels of CD44 and endothelial lipase (Fig. 3A). Disruption of CD44 in mice results in a dramatic decrease in atherosclerosis (31). Elevated level of endothelial lipase is associated with reduced high-density lipoprotein and increased atherosclerosis (32). Therefore, the higher level of CD44 and endothelial lipase of artery ECs may underlie the preferential arterial involvement of atherosclerosis.
The diversity and regional specificity in gene expression programs in ECs provides insight into the well-recognized regional variations in physiological properties and pathological processes that affect the vascular system. It will be important now to extend these studies to define the full range of diversity of ECs and other cells found in the vascular system and to dissect the regulatory mechanisms that lead to the diversification and specialization of these cells during development. It is also imperative to investigate the changes in gene expression of ECs undergoing physiological (wound healing, menstrual cycles) and pathological (cancer angiogenesis, diabetic retinopathy, arteryvein fistula) adaptations to analyze their roles in these processes and explore the potentials for therapeutic interventions.
Supplementary Material
Acknowledgments
We thank members of the Brown and Botstein laboratories and staff at the Stanford Microarray Database for their support and comments on the manuscript. We are indebted to Mike Fero and the staff at the Stanford Functional Genomic Facility for their support. We are also grateful to the Hutchison Foundation and Cambrex Corporation. This work was supported by the Howard Hughes Medical Institute and a grant from the National Cancer Institute (to P.O.B.). P.O.B. is an Investigator of the Howard Hughes Medical Institute.
Abbreviations: EC, endothelial cell; HUVEC, human umbilical vein EC; LEC, lymphatic EC; ECM, extracellular matrix; L-R, left/right.
References
- 1.St Croix, B., Rago, C., Velculescu, V., Traverso, G., Romans, K. E., Montgomery, E., Lal, A., Riggins, G. J., Lengauer, C., Vogelstein, B. & Kinzler, K. W. (2000) Science 289, 1197–1202. [DOI] [PubMed] [Google Scholar]
- 2.Podgrabinska, S., Braun, P., Velasco, P., Kloos, B., Pepper, M. S., Jackson, D. G. & Skobe, M. (2002) Proc. Natl. Acad. Sci. USA 99, 16069–16074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jahnsen, F. L., Brandtzaeg, P., Haye, R. & Haraldsen, G. (1997) Am. J. Pathol. 150, 2113–2123. [PMC free article] [PubMed] [Google Scholar]
- 4.Sherlock, G., Hernandez-Boussard, T., Kasarskis, A., Binkley, G., Matese, J. C., Dwight, S. S., Kaloper, M., Weng, S., Jin, H., Ball, C. A., et al. (2001) Nucleic Acids Res. 29, 152–155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Alizadeh, A. A., Eisen, M. B., Davis, R. E., Ma, C., Lossos, I. S., Rosenwald, A., Boldrick, J. C., Sabet, H., Tran, T., Yu, X., et al. (2000) Nature 403, 503–511. [DOI] [PubMed] [Google Scholar]
- 6.Eisen, M. B., Spellman, P. T., Brown, P. O. & Botstein, D. (1998) Proc. Natl. Acad. Sci. USA 95, 14863–14868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Troyanskaya, O. G., Garber, M. E., Brown, P. O., Botstein, D. & Altman, R. B. (2002) Bioinformatics 18, 1454–1461. [DOI] [PubMed] [Google Scholar]
- 8.Tusher, V. G., Tibshirani, R. & Chu, G. (2001) Proc. Natl. Acad. Sci. USA 98, 5116–5121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pear, W. S., Miller, J. P., Xu, L., Pui, J. C., Soffer, B., Quackenbush, R. C., Pendergast, A. M., Bronson, R., Aster, J. C., Scott, M. L. & Baltimore, D. (1998) Blood 92, 3780–3792. [PubMed] [Google Scholar]
- 10.Chang, H. Y., Chi, J. T., Dudoit, S., Bondre, C., van de Rijn, M., Botstein, D. & Brown, P. O. (2002) Proc. Natl. Acad. Sci. USA 99, 12877–12882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mukouyama, Y. S., Shin, D., Britsch, S., Taniguchi, M. & Anderson, D. J. (2002) Cell 109, 693–705. [DOI] [PubMed] [Google Scholar]
- 12.Luttun, A., Tjwa, M., Moons, L., Wu, Y., Angelillo-Scherrer, A., Liao, F., Nagy, J. A., Hooper, A., Priller, J., De Klerck, B., et al. (2002) Nat. Med. 8, 831–840. [DOI] [PubMed] [Google Scholar]
- 13.Oliver, G. & Detmar, M. (2002) Genes Dev. 16, 773–783. [DOI] [PubMed] [Google Scholar]
- 14.Yancopoulos, G. D., Davis, S., Gale, N. W., Rudge, J. S., Wiegand, S. J. & Holash, J. (2000) Nature 407, 242–248. [DOI] [PubMed] [Google Scholar]
- 15.Hacohen, N., Kramer, S., Sutherland, D., Hiromi, Y. & Krasnow, M. A. (1998) Cell 92, 253–263. [DOI] [PubMed] [Google Scholar]
- 16.Cheng, N., Brantley, D. M. & Chen, J. (2002) Cytokine Growth Factor Rev. 13, 75–85. [DOI] [PubMed] [Google Scholar]
- 17.Yuan, L., Moyon, D., Pardanaud, L., Breant, C., Karkkainen, M. J., Alitalo, K. & Eichmann, A. (2002) Development (Cambridge, U.K.) 129, 4797–4806. [DOI] [PubMed] [Google Scholar]
- 18.Hirakawa, S., Hong, Y. K., Harvey, N., Schacht, V., Matsuda, K., Libermann, T. & Detmar, M. (2003) Am. J. Pathol. 162, 575–586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lawson, N. D. & Weinstein, B. (2002) Nat. Rev. Genet. 3, 674–682. [DOI] [PubMed] [Google Scholar]
- 20.Meno, C., Shimono, A., Saijoh, Y., Yashiro, K., Mochida, K., Ohishi, S., Noji, S., Kondoh, H. & Hamada, H. (1998) Cell 94, 287–297. [DOI] [PubMed] [Google Scholar]
- 21.Urness, L. D., Sorensen, L. K. & Li, D. Y. (2000) Nat. Genet. 26, 328–331. [DOI] [PubMed] [Google Scholar]
- 22.Lawson, N. D., Scheer, N., Pham, V. N., Kim, C. H., Chitnis, A. B., Campos-Ortega, J. A. & Weinstein, B. M. (2001) Development (Cambridge, U.K.) 128, 3675–3683. [DOI] [PubMed] [Google Scholar]
- 23.Nakagawa, O., McFadden, D. G., Nakagawa, M., Yanagisawa, H., Hu, T., Srivastava, D. & Olson, E. N. (2000) Proc. Natl. Acad. Sci. USA 97, 13655–13660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhong, T. P., Rosenberg, M., Mohideen, M. A., Weinstein, B. & Fishman, M. C. (2000) Science 287, 1820–1824. [DOI] [PubMed] [Google Scholar]
- 25.Arap, W., Kolonin, M. G., Trepel, M., Lahdenranta, J., Cardo-Vila, M., Giordano, R. J., Mintz, P. J., Ardelt, P. U., Yao, V. J., Vidal, C. I., et al. (2002) Nat. Med. 8, 121–127. [DOI] [PubMed] [Google Scholar]
- 26.Lammert, E., Cleaver, O. & Melton, D. (2001) Science 294, 564–567. [DOI] [PubMed] [Google Scholar]
- 27.Matsumoto, K., Yoshitomi, H., Rossant, J. & Zaret, K. S. (2001) Science 294, 559–563. [DOI] [PubMed] [Google Scholar]
- 28.Oliver, G., Mailhos, A., Wehr, R., Copeland, N. G., Jenkins, N. A. & Gruss, P. (1995) Development (Cambridge, U.K.) 121, 4045–4055. [DOI] [PubMed] [Google Scholar]
- 29.Zhu, L. J., Bagchi, M. K. & Bagchi, I. C. (1998) Endocrinology 139, 330–339. [DOI] [PubMed] [Google Scholar]
- 30.Niiro, N., Nishimura, J., Hirano, K., Nakano, H. & Kanaide, H. (1998) Br. J. Pharmacol. 124, 1623–1632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cuff, C. A., Kothapalli, D., Azonobi, I., Chun, S., Zhang, Y., Belkin, R., Yeh, C., Secreto, A., Assoian, R. K., Rader, D. J. & Pure, E. (2001) J. Clin. Invest. 108, 1031–1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Choi, S. Y., Hirata, K., Ishida, T., Quertermous, T. & Cooper, A. D. (2002) J. Lipid Res. 43, 1763–1769. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.