Abstract
The mitochondria are the major source of chronic oxidative stress, which has been implicated in the aging process. Along with other cellular changes, aged cells accumulate mutations in both their nuclear and mitochondrial genomes, and they contain increased amounts of oxidatively damaged mutagenic bases such as 7,8-dihydro-8-oxoguanine, suggesting age-dependent inhibition of its repair. Surprisingly, the level and activity of 8-oxoguanine-DNA glycosylase (OGG1), responsible for repair of 7,8-dihydro-8-oxoguanine, was found to be higher in the liver mitochondrial extract from old rodents than in that from young ones. We addressed this paradox by analyzing OGG1 in the mitochondria of young vs. old mouse livers, as well as in replicating vs. presenescent human fibroblasts. We show here that although the total OGG1 activity is higher in old mitochondria, a large fraction of the enzyme is stuck to the membrane in the precursor form, which could not be translocated to and processed in the mitochondrial matrix. A nearly identical phenomenon was observed with the mitochondrial uracil-DNA glycosylase responsible for repair of mutagenic uracil. These results indicate an age-dependent decline in the mitochondrial import of proteins needed for DNA repair and possibly for other functions.
Keywords: aging, mitochondria, oxidative stress, 8-oxoguanine, DNA repair
Aging is a highly complex, progressive, and universal process involving multiple organ-specific macromolecular interactions (1–3). A correlation was observed between the lifespan of various mammalian species and DNA repair capacity in cells derived from them (4). Aged cells also accumulate oxidative DNA damage, which is likely to be responsible for the observed increase in mutations, particularly in the mitochondrial genome (5, 6). These results formed the basis of the mitochondrial theory of aging, which postulates that the accumulation of DNA damage and mutations in the mitochondrial genomes causes mitochondrial dysfunction, leading to chronic production of reactive oxygen species (ROS) and contributing to the development of the age-associated decline in tissue functions (7–12). The major source of ROS in the cell is the mitochondria, whose genomes are more susceptible to oxidative damage than the nuclear genome, presumably because of the physical proximity of the source of ROS and lack of histones (13). There is strong evidence for progressive mitochondrial dysfunction and accumulation of mutations in the mitochondrial genome during aging. The mutations include both deletions at a preferred region near the origin of replication and point mutations, which likely arise from oxidatively induced mutagenic base lesions such as 7,8-dihydro-8-oxoguanine (8-oxoG) and uracil (14). The 8-oxoG mispairs with A to induce G·C → T·A transversion, whereas U, formed by oxidative deamination of C, leads to G·C → A·T transition mutation (15). The damaged bases are repaired primarily by means of the DNA base excision repair pathway, which is active in both the nucleus and the mitochondria (16, 17). The first step in repair is excision of the damaged base by a specific DNA glycosylase, namely, 8-oxoguanine-DNA glycosylase (OGG1) and uracil-DNA glycosylase (UDG) for repair of 8-oxoG and U, respectively (18).
Mitochondrial DNA accumulates high levels of 8-oxoG, arguably the most important base damage induced by ROS, which plays a major role in the aging process (1, 19–22). The nuclear OGG1, OGG1-α, and the mitochondrial isoform, OGG1-β, encoded by the same gene have identical 315 amino acid residues from the N terminus. However, OGG1-α has a nuclear localization signal near the C terminus encoded by exon 7. Exon 8 replaces exon 7 in the OGG1-β mRNA. The molecular masses of mature OGG1-α and -β are 38 and 45 kDa, respectively (23). The role of OGG1 as the major repair enzyme for 8-oxoG was further underscored by the mutational studies in Ogg1-null mice and mouse cells. A 20-fold increase in 8-oxoG level was observed in liver nuclear DNA in Ogg1-null (Ogg1–/–) mice, relative to the control (24). Under chronic oxidative stress, however, this may be enhanced to 70-fold (25). Liver mitochondrial DNA of Ogg1–/– mice contained 30-fold more 8-oxoG than WT control DNA (22).
Most mitochondrial proteins are encoded by the nuclear genome as precursors with an N-terminal extension, containing a presequence or mitochondrial targeting sequence (26). After recognition of the presequence by receptors on the surface of mitochondria, the proteins are transported across the mitochondrial membrane by the translocase complexes, named TOM and TIM, present in the outer and inner membrane, respectively. The presequence is then cleaved off by the mitochondrial processing peptidase localized within the matrix (27). Alterations of these transport processes should therefore affect mitochondrial functions. Whether aging affects activities of TOM and TIM is not known. The mitochondrial targeting sequence in the 47-kDa OGG1-β precursor was identified to include residues 9–26 in both OGG1-α and OGG1-β; OGG1-β precursor is cleaved by mitochondrial processing peptidase to generate the 45-kDa mature OGG1-β. The presence of a strong nuclear localization signal in OGG1-α prevents its targeting to the mitochondrial matrix (23).
Several studies have shown that OGG1-β activity in mitochondrial lysates from rodent liver was higher when prepared from old vs. young animals (20, 22, 28–30). Surprisingly, 8-oxoG levels were also shown to increase with age in mitochondrial DNA and nuclear DNA, which would be consistent with a decline in its repair. We addressed this paradox in two model systems: hepatocytes from young vs. old mice (31, 32) and primary human diploid fibroblasts (HDF). The primary human cells undergo replicative senescence after a finite number of population doublings and have been extensively used as an in vitro model for the aging (33, 34). In the present study, we have confirmed that the level and activity of mitochondria-associated OGG1-β were higher in the livers of old mice compared to young mice and in presenescent HDF cells compared to replicating cells. However, a significant fraction of the OGG1, as well as UDG, remains localized in the outer membrane and intermembrane space in the precursor form, presumably because their import into the mitochondrial matrix is impaired because of aging.
Materials and Methods
Cells and Animals. Human embryonic diploid lung fibroblasts (MRC5) were grown in MEM (GIBCO) supplemented with 10% FBS (Sigma), glutamine (0.3 mg/ml), penicillin (100 units/ml), and streptomycin (100 μg/ml). To calculate population doubling level (PDL), the cells were removed by trypsinization at confluence and counted, and the number of doublings was calculated [PDL = log(number of cells obtained at subculture/105)/log 2], where the initial number of cells is 105 at each subculture (33, 35).
Four- and 20-mo-old BALB/c mice purchased from the National Institute on Aging were used in the studies. Animal experiments were performed according to the National Institutes of Health Guide for Care and Use of Laboratory Animals and were approved by the University of Texas Medical Branch Animal Care and Use Committee (no. 00-01-007).
Isolation and Fractionation of Mitochondria. Mitochondria from human fibroblasts (≈108 cells) or livers (1.5- to 2-g individual livers) from 4- and 20-mo-old mice were isolated as described (36). Final pellets were resuspended in an appropriate amount of buffer containing 10 mM Hepes–KOH (pH 7.4), 250 mM sucrose, 10 mM DTT, 0.5 mM EGTA, and 2 mM EDTA for further fractionation or for suspension in buffer containing 20 mM Hepes–KOH (pH 7.4), 1 mM EDTA, 1 mM DTT, 300 mM KCl, 5% glycerol, and 0.5% Triton X-100 (2 mg/ml). The proteins from outer membrane (OM), intermembrane space (IMS), inner membrane, and matrix fractions were isolated from intact mitochondrial pellets after suspension in buffer containing 0.7 M sucrose, 0.21 M mannitol, and 2 mM Hepes (pH 7.4), and then treated with digitonin (1.1 mg/10 mg protein) according to Landin et al. (37).
Oligonucleotide Incision Assay. OGG1 activity in mitochondrial and nuclear lysates was determined by excising 8-oxoG from a 32P-labeled oligonucleotide substrate as described (38, 39). The excision reactions (in 20 μl) were carried out in 40 mM Hepes–KOH (pH 7.6)/5 mM EDTA/1 mM DTT/75 mM KCl/10% glycerol buffer containing 0.2 pmol of 32P-labeled duplex oligonucleotide (51-mer containing 8-oxoG: 5′-GCT TAG CTT GGA ATC GTA TCA TGT A8-oxoGA CTC GTG TGC CGT GTA GAC CGT GCC-3′) and synthesized by Midland Certified Reagents (Midland, TX) and 50 or 100 μg of mitochondrial extracts. After incubation at 37°C for 3 h followed by the addition of proteinase K (250 μg/ml) and SDS (0.5%), and heating at 50°C for 15 min, DNA was precipitated with ethanol and dissolved in loading buffer (70% formamide/30 mM NaOH). To determine the activity of OGG1 in the nuclear fraction, nuclear extracts (15, 30, or 45 μg) were incubated at 37°C for 1 h, followed by addition of loading buffer. OGG1 has an intrinsic AP lyase activity that cleaves the DNA strand after excision of 8-oxoG (38). Although this AP lyase activity is weak, and OGG1 mostly remains bound to the abasic (AP, for apurinic/apyrimidinic) site product (40), treatment of assay mixtures with alkali after completion of the reaction ensures strand cleavage at such AP sites. Thus the cleaved product separated from the intact substrate is a measure of base excision activity. UDG activity was also measured by using a 51-mer oligonucleotide duplex containing a single U at position 26. 32P-labeled duplex oligonucleotide (0.2 pmol) and 15 or 30 μg of mitochondrial lysates were incubated for 1 h at 37°C. The substrate and cleaved oligonucleotide product were separated in a 20% polyacrylamide gel containing 7 M urea.
Radioactivity in the separated DNA bands was quantitated with a PhosphorImager and imagequant software (Molecular Dynamics). Preliminary enzyme activity assays were carried out to ensure linearity of product formation with respect to both time of incubation and the amount of extract. The glycosylase assay was carried out in the presence of EDTA, which inhibits Mg2+ (or Mn2+)-dependent nucleases. A small amount of nonspecific degradation was observed with nuclear extracts, which did not affect quantitation of the glycosylase product. Finally, we ensured that the OGG1 and UDG showed linear kinetics and dependence on the amount of extract from different samples.
Cytochrome c Oxidase Activity. Cytochrome c oxidase activity was measured by spectrophotometric analysis of oxidation of ferrocytochrome c at 550 nm with an assay kit (Sigma) according to the manufacturer's protocol.
Trypsin Treatment of Mitochondria. Intact mitochondria (1 mg/ml), resuspended in a buffer containing 10 mM Hepes–KOH (pH 7.4), 250 mM sucrose, 0.5 mM EGTA, 2 mM EDTA, and 1 mM DTT were treated with trypsin (10 μg/ml) for 20 min at room temperature, followed by addition of an equivalent amount of bovine trypsin inhibitor (Invitrogen) to stop proteolysis (41, 42). Protease- and mock-treated mitochondria suspensions were washed twice in the same buffer before lysis.
Western Blot Analysis. Proteins were separated by SDS/PAGE, transferred to a nitrocellulose membrane (Protran, Schleicher & Schuell), and blocked in TBST (20 mM Tris·HCl/137 mM NaCl, pH 7.6/0.5% Tween 20) containing 5% dry milk (43). The membranes were sequentially probed with anti-mouse and antihuman OGG1 Abs (38), and Abs against cytochrome c, lamin b (Santa Cruz Biotechnology), and cytochrome oxidase subunit IV (COX IV) (Molecular Probes) at a 1:200 dilution in TBST containing 5% dry milk. In analyzing mouse Ogg1, we have used rabbit antisera raised against human OGG1-α, recognizing both nuclear and mitochondrial OGG1 of both human and mouse origin. We have also generated rabbit antisera against a human OGG1 peptide (38), which does not crossreact with mouse Ogg1, and used this Ab for Western analysis of recombinant human OGG1 used as a reference. The binding of primary Abs was detected with horseradish peroxidase-conjugated secondary Ab (1:2,000; anti-mouse or anti-rabbit IgG-horseradish peroxidase; Amersham Pharmacia). Subsequently, membranes were washed and incubated in ECL reagent (Amersham Pharmacia). Chemiluminograms were analyzed by using imagequant software (Molecular Dynamics).
Data Presentation and Analysis. Six animals were used for experiments carried out in duplicate. The experimental results are always presented as ratios of values obtained with old relative to young animals. Each experiment was repeated multiple times with a pair of young and old liver samples and the ratio of values with extracts of old vs. young was calculated for each set. Thus the relative value of the young animal was always set to 1.0. The SD in the values of young animals was 0.21. The results are presented as mean of relative values for the old ± SD. The differences among ages and population doubling levels were analyzed by the Student t test, and P = 0.05 was considered statistically significant.
Results
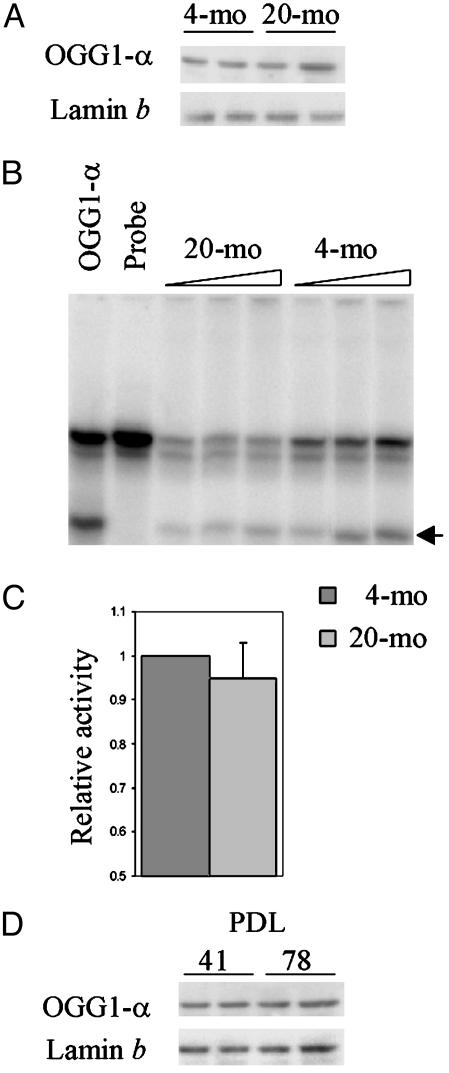
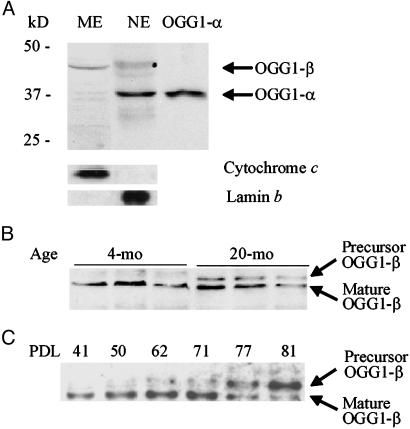
OGG1-β Levels Are Higher in the Mitochondria of Aged Liver and Senescent HDF cells. Nuclear and mitochondrial extracts from 4- and 20-mo-old mouse liver, as well as from replicating (low PDL) and presenescent (high PDL) MRC5 fibroblasts, were analyzed to determine the levels of OGG1-α and OGG1-β, respectively. A 38-kDa OGG1-α was observed in the liver nuclear extract from both 4- and 20-mo-old mice, as well as in nuclear extracts of replicating and presenescent MRC5 cells (Fig. 1 A and C). Correction of band intensities with lamin b, a nuclear protein (44), indicated no significant difference in OGG1-α levels between young vs. aged mice or high vs. low PDL cells. The specificity of our rabbit polyclonal Ab raised against a peptide common to OGG1-α and OGG1-β ensures that both OGG1-α and OGG1-β are identified with this Ab (38). These results were confirmed with commercially available mAbs against OGG1 (data not shown). We then quantitated OGG1 activity in the nuclear extracts of young vs. old cells (Fig. 1B) (39, 45). Similarly, no significant effect of PDL dependence on OGG1 activity was observed in nuclear extracts of MRC5 cells (data not shown). The absence of lamin b and cytochrome c in mitochondrial and nuclear extracts, respectively, confirmed the purity of these preparations (Fig. 2A).
Fig. 1.
Age-dependent change in OGG1-α. (A) Western blot analysis of OGG1-α polypeptide levels in nuclei of 4- and 20-mo-old mouse livers. Two animals were used for each age group. The blots were reprobed with antilamin b Ab. (B) OGG1-α activity in liver nuclear extracts (15, 30, and 45 μg for each) of 4- and 20-mo-old mice. (C) The average activity for six animals of each group relative to the young animal. (D) Western analysis of OGG1-α levels in nuclear extracts (in duplicate) from proliferating (PDL 41) and presenescent (PDL 78) MRC5 cells. Other details are given in Materials and Methods.
Fig. 2.
Western blot analysis of precursor and mature OGG1-β in mitochondria and submitochondrial fractions of young and old mouse livers. (A) Identification of 38-kDa OGG1-α and 45-kDa OGG1-β in the mitochondrial extract (ME) and nuclear extract (NE), respectively, from 4-mo-old mouse livers. Lamin b and cytochrome c were analyzed in these extracts to confirm the lack of cross-contamination between nuclear and mitochondrial preparations. (B) Presence of 47-kDa OGG1-β precursor in mitochondrial extracts of 20-mo-old mouse livers. (C) Presence of OGG1-β precursor in high PDL (>50) MRC5 cells. Mitochondrial extracts from MRC5 cells collected at various PDL were analyzed for OGG1.
Presence of Nonprocessed OGG1-β in Mitochondria from Aged Liver and Presenescent HDF. The mitochondrial extracts from aged liver and presenescent MRC5 cells contained two distinct OGG1 bands of ≈45 and 47 kDa, respectively (Fig. 2B). As mentioned, the 47-kDa OGG1-β band should correspond to the precursor form of mitochondrial OGG1-β, whereas the 45-kDa band corresponds to mature OGG1-β (23, 46). To test whether the level of OGG1-β precursor is affected as a function of PDL, we isolated mitochondria from serially passaged MRC5 cells (Fig. 2C). We detected the 47-kDa precursor form in cells with PDL >50. Thus the majority of OGG1-β was present as the precursor in high PDL cells, which suggests that its import into the mitochondrial matrix was inhibited during replicative senescence.
To localize the precursor and mature OGG1-β forms, mitochondria were freshly isolated from mouse livers and were fractionated into OM/IMS proteins, as well as mitoplasts, as described in Materials and Methods. The 47-kDa OGG1-β precursor was abundant in the OM/IMS from 20-mo-old mice, but barely detectable in that from mitochondria of 4-mo-old mice (Fig. 3A). Mitoplasts were disrupted by repeated sonication and fractionated by ultracentrifugation for membrane and matrix proteins. As shown in Fig. 3A, the level of OGG1-β (45 kDa) in the matrix was consistently lower in the mitochondria of livers from 20- vs. 4-mo-old mice. These results indicate that a significant amount of OGG1-β polypeptide was trapped in the OM/IMS in a precursor form. Similar results were obtained with fractionated mitochondria isolated from MRC5 cells (data not shown). We conclude that the lower level of processed OGG1-β resulted from inefficient targeting of OGG1-β into the mitochondria matrix in aged mouse hepatocytes and presenescent human fibroblasts.
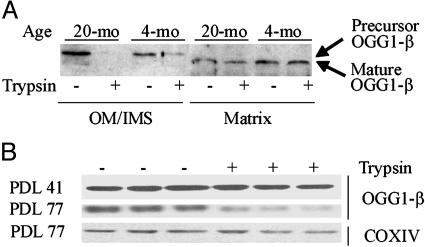
Fig. 3.
Sensitivity of mitochondrial OGG1-β to trypsin. (A) Presence of precursor OGG1-β in OM/IMS and its sensitivity to trypsin. The precursor and mature OGG1-β bands are indicated. (B) Trypsin sensitivity of OGG1-β in presenescent PDL 77 vs. PDL 41 MRC5 cells. The 47-kDa OGG1-β precursor was not separated from the mature 45-kDa band. Cytochrome c oxidase subunit IV (COX IV) analyzed in the same blot served as an internal control.
Protease Sensitivity of OGG1-β Associated with Mitochondria of Aged Cells. The susceptibility of the precursor protein to proteolytic cleavage suggests that the precursor protein is attached to the membrane as a translocation intermediate, which spans the OM (47). We tested this possibility by trypsin treatment of mitochondria isolated from hepatocytes of 4- and 20-mo-old mice as well as from replicating and presenescent MRC5 cells (41, 42). Western blot analysis showed that limited trypsin digestion reduced the level of OGG1-β associated with the mitochondria from aged liver (Fig. 3A) and presenescent MRC5 (PDL 77) cells by 30–40% (Fig. 3B) (in this analysis precursor and mature form of OGG1-β of MRC5 cells were not separated). In contrast, the level of OGG1-β in liver mitochondria of 4-mo-old mice or from replicating MRC5 (PDL 41) was only slightly reduced (≈5–10%) after proteolytic digestion. We assayed the level of subunit IV of cytochrome c oxidase for normalizing the intensity of OGG1 bands (Fig. 3B).
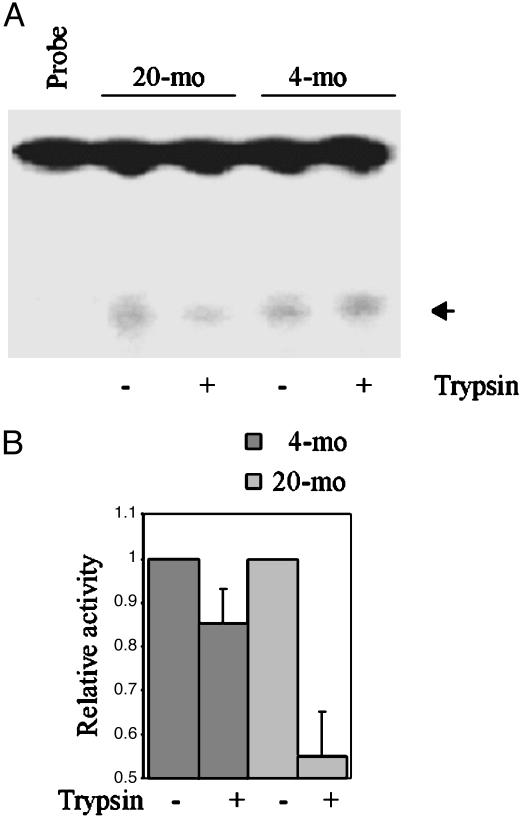
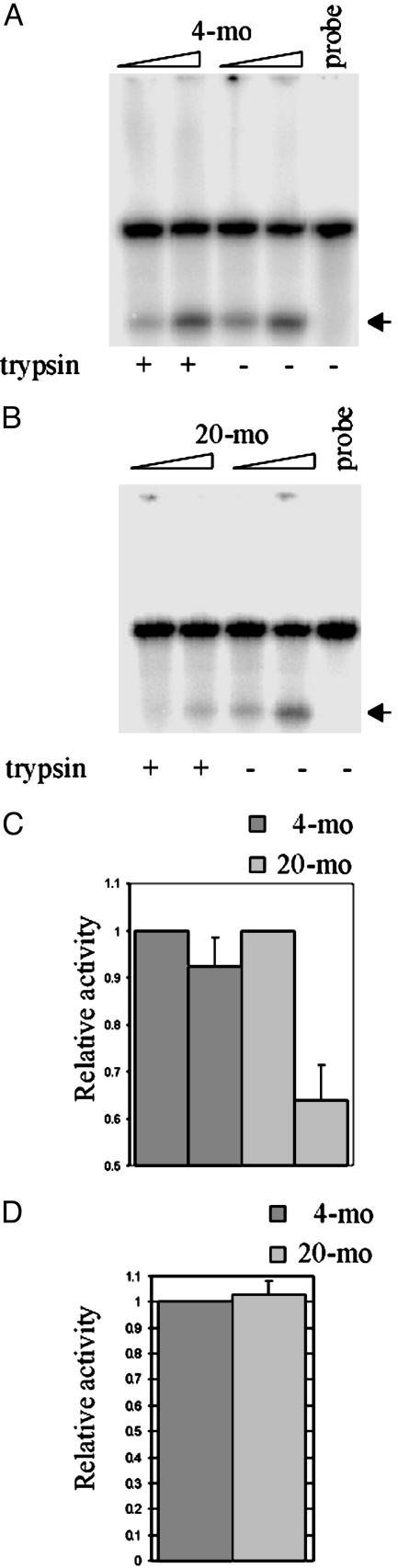
Liver mitochondria from aged rodents were previously shown to possess higher OGG1 activity relative to young (29, 30). We confirmed this age dependence. Higher OGG1 activity (by ≈30%) in mitochondrial extracts from 20-mo-old mouse liver was observed (data not shown). However, several experiments showed a significant reduction in OGG1 activity after limited trypsin digestion of intact mitochondria isolated from 20-mo-old mouse livers, but not from 4-mo-old mouse livers (Fig. 4). The decrease as a fraction of the untreated control is shown in Fig. 4B. Similar observations were obtained for replicating vs. presenescent MRC5 cells (data not shown).
Fig. 4.
Effect of mitochondrial trypsin treatment on OGG1-β activity. (A) 8-oxoG excision activity in extracts (50 μg each) of mitochondria pretreated with or without trypsin. Probe indicates oligonucleotide substrate without extract. (B) The average OGG activity from six animals expressed relative to untreated samples is shown in a bar graph.
Trypsin-Sensitive UDG in Mitochondria of Aged Cells. In both nuclear and mitochondrial DNA of aged cells, uracil can arise from spontaneous or oxidative deamination of cytosine or incorporation of dUMP during DNA replication (48). When left unrepaired, U generated in situ will give rise to G·C → A·T transition mutations. Mitochondrial UDG, like OGG1, is synthesized from alternatively spliced mRNA in the cytoplasm (36, 49) and is targeted to the mitochondrial matrix. No significant age-dependent increase was observed for mitochondrial UDG activity (30).
To test whether mitochondrial UDG molecules are also trapped in the OM of mitochondria from aged hepatocytes, UDG activity was measured in extracts of intact mitochondria before or after limited trypsin treatment. Fig. 5A shows that UDG activity in liver mitochondrial extract from 4-mo-old mice was not significantly affected by limited trypsin digestion. However, trypsin treatment reduced this activity by 30% in the liver mitochondria of 20-mo-old mice (Fig. 5B). Fig. 5C shows the difference of these activities relative to the control. Similar results were obtained for replicating vs. presenescent MRC5 cells (data not shown).
Fig. 5.
Effects of trypsin on UDG activity associated with mitochondria from young and old mice. Uracil excision activity was measured, as described in Materials and Methods, with extracts (15 and 30 μg) of mitochondria with or without trypsin pretreatment from young (A) and old (B) mouse livers. The relative activity is shown in C. (D) Cytochrome c oxidase activity in mitochondrial extracts from young and old mouse livers.
To rule out the possibility that the observed difference in OGG1 and UDG activities between young vs. old mice groups and low vs. high PDL MRC5 cells is not related to differences in mitochondrial enrichment, we determined cytochrome c oxidase activity, which showed no significant difference (P < 0.05) between mitochondrial extracts prepared from the livers of 4- and 20-mo-old mice (Fig. 5D). Taken together, these data indicate that not only OGG1-β, but also a significant portion of the UDG, was trapped in OM and is trypsin-sensitive, suggesting their import deficiency in aged cells.
Discussion
The higher steady-state level of base damage in the aged mitochondrial genome may have multiple causes, including inability of the repair system to handle an ever-increasing load. This would suggest that the observed increase in the level of 8-oxoG in the mitochondrial (and nuclear) genome with age results from decreased repair capacity of the aged cells. Thus the contrary observation that the OGG1 level increased in the mitochondria, but not in the nucleus, of aged rodent livers, compared with the young rodent livers, was quite surprising and warranted an in-depth examination. The mechanism of mitochondrial uptake of cytoplasmic proteins is distinct from nuclear uptake and not just because of the distinct protein sequence motifs required for organelle-specific import. Nearly all of the mitochondrial proteins, including the repair proteins, are encoded by the nuclear genes and then transported to mitochondria. DNA glycosylases such as OGG1 and UDG, differentially targeted to the nucleus and mitochondria, are products of the same gene (46). The mitochondrial targeting sequence, a 20- to 80-residue oligopeptide usually present at the N terminus of the precursor protein, is cleaved off by the mitochondrial precursor peptidase presented in the mitochondrial matrix (26, 27). Thus the mature form is formed irreversibly and is present only in the mitochondrial matrix, whereas the precursor polypeptide transits through the outer membrane and intermembrane space (26).
A method, based on protease sensitivity, has been developed to distinguish between the precursor and mature forms. Thus with intact mitochondria, trypsin degrades the protein bound to the outer membrane without affecting the proteins internalized in the matrix. We used this approach to identify the 47-kDa precursor form of mitochondrial OGG1-β bound to the OM, whereas the mature 45-kDa OGG1-β was present mostly in the mitochondrial matrix. Our results showing the presence of a larger abundance of the precursor form in the mitochondria of old mouse liver, and also presenescent cells, suggested that the processing of OGG1 is impaired with aging. This conclusion was confirmed by our subsequent results showing that trypsin treatment significantly reduced the level of mitochondria-specific OGG1-β in the old mouse liver compared with young liver. Interestingly, a similar age-dependent decline in import of OGG1 was not observed for the nucleus.
Our additional studies, showing an excellent correlation in the age-dependent polypeptide vs. activity of OGG1-β within the mitochondrial matrix, provides strong evidence for our conclusion that the OGG1 activity in mitochondria indeed declines with age. We propose an explanation for the paradox of association of enhanced 8-oxoG level with increased OGG1 activity in aged mitochondria by showing that a significant fraction of the enzyme is located not in the matrix, but rather in the OM, where it is sensitive to trypsin treatment.
Because nearly identical results were obtained with both aging models, namely, young vs. old mouse livers and replicating vs. presenescent HDF, and a similar situation was observed with mitochondrial UDG, it is highly likely that the age-dependent decline in mitochondrial import is a general phenomenon. de Souza-Pinto et al. (29) observed an increase in OGG1-β but not UDG in old liver mitochondria. Our results showing that the total UDG activity of control extract from mitochondria purified from young vs. old mouse liver are in agreement with those results. However, the results after trypsin treatment indicated that the UDG level and activity also declined as a function of aging. Thus there appears to be a general deficiency in import of base excision repair proteins into the mitochondria matrix, which may contribute to the age-dependent increase in the levels of damaged bases and increased frequency of mutations in mitochondria. However, there may not be a global age-dependent decline in import of proteins into mitochondria because the cytochrome c oxidase activity was not affected by aging.
Import of proteins into mitochondrial matrix has been extensively studied. The key transporters TOM and TIM complexes are present in the outer and inner membrane, respectively. The defect in processing of OGG1-β precursor polypeptide before its import into the matrix is likely to be in the TOM complex because of the sensitivity to trypsin proteolysis. Oxidative modification of one or more subunits of TOM could be among many possible mechanisms for such a defect. Some of the TOM subunits, including TOM20 and TOM70, with domains exposed to the cytosol and able to bind mitochondrial precursor proteins, may play a key role in recognizing the mitochondrial targeting sequence (26). It is also known that the mitochondrial heat shock protein, Hsp70, is essential for translocation of nuclear-encoded mitochondrial proteins (50). Probably its oxidative modification increasing with age could cause the deficiency in protein uptake. In any event, our results raise the strong possibility that deficiency in mitochondrial import machinery could make a major contribution to the aging syndrome.
Acknowledgments
We thank Dr. D. Konkel for careful editing and Ms. Wanda Smith for expert secretarial help. Dr. T. Izumi assisted in initial animal studies. This research was supported by U.S. Public Health Service Grants P01 AG10514, CA84461, CA81063, and ES06676.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: OGG1, 8-oxoguanine-DNA glycosylase; 8-oxoG, 7,8-dihydro-8-oxoguanine; UDG, uracil-DNA glycosylase; PDL, population doubling level; HDF, human diploid fibroblasts; IMS, intermembrane space; OM, outer membrane.
References
- 1.Beckman, K. B. & Ames, B. N. (1998) Physiol. Rev. 78, 547–581. [DOI] [PubMed] [Google Scholar]
- 2.Cortopassi, G. A. & Wong, A. (1999) Biochim. Biophys. Acta 1410, 183–193. [DOI] [PubMed] [Google Scholar]
- 3.Sohal, R. S., Mockett, R. J. & Orr, W. C. (2002) Free Radical Biol. Med. 33, 575–586. [DOI] [PubMed] [Google Scholar]
- 4.Hart, R. W. & Setlow, R. B. (1974) Proc. Natl. Acad. Sci. USA 71, 2169–2173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kowald, A. & Kirkwood, T. B. (1993) Mutat. Res. 295, 93–103. [DOI] [PubMed] [Google Scholar]
- 6.Shenkar, R., Navidi, W., Tavare, S., Dang, M. H., Chomyn, A., Attardi, G., Cortopassi, G. & Arnheim, N. (1996) Am. J. Hum. Genet. 59, 772–780. [PMC free article] [PubMed] [Google Scholar]
- 7.Sohal, R. S., Sohal, B. H. & Orr, W. C. (1995) Free Radical Biol. Med. 19, 499–504. [DOI] [PubMed] [Google Scholar]
- 8.Perez-Campo, R., Lopez-Torres, M., Cadenas, S., Rojas, C. & Barja, G. (1998) J. Comp. Physiol. B 168, 149–158. [DOI] [PubMed] [Google Scholar]
- 9.Sastre, J., Pallardo, F. V., Garcia de la Asuncion, J. & Vina, J. (2000) Free Radical Res. 32, 189–198. [DOI] [PubMed] [Google Scholar]
- 10.Miquel, J., Economos, A. C., Fleming, J. & Johnson, J. E., Jr. (1980) Exp. Gerontol. 15, 575–591. [DOI] [PubMed] [Google Scholar]
- 11.Osiewacz, H. D. (2002) Gene 286, 65–71. [DOI] [PubMed] [Google Scholar]
- 12.Wei, Y. H. & Lee, H. C. (2002) Exp. Biol. Med. 227, 671–682. [DOI] [PubMed] [Google Scholar]
- 13.Yakes, F. M. & Van Houten, B. (1997) Proc. Natl. Acad. Sci. USA 94, 514–519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dizdaroglu, M., Jaruga, P., Birincioglu, M. & Rodriguez, H. (2002) Free Radical Biol. Med. 32, 1102–1115. [DOI] [PubMed] [Google Scholar]
- 15.Moriya, M. (1993) Proc. Natl. Acad. Sci. USA 90, 1122–1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rachek, L. I., Grishko, V. I., Musiyenko, S. I., Kelley, M. R., LeDoux, S. P. & Wilson, G. L. (2002) J. Biol. Chem. 277, 44932–44937. [DOI] [PubMed] [Google Scholar]
- 17.Dianov, G. L., Souza-Pinto, N., Nyaga, S. G., Thybo, T., Stevnsner, T. & Bohr, V. A. (2001) Prog. Nucleic Acid Res. Mol. Biol. 68, 285–297. [DOI] [PubMed] [Google Scholar]
- 18.Mitra, S., Hazra, T. K., Roy, R., Ikeda, S., Biswas, T., Lock, J., Boldogh, I. & Izumi, T. (1997) Mol. Cells 7, 305–312. [PubMed] [Google Scholar]
- 19.Hamilton, M. L., Van Remmen, H., Drake, J. A., Yang, H., Guo, Z. M., Kewitt, K., Walter, C. A. & Richardson, A. (2001) Proc. Natl. Acad. Sci. USA 98, 10469–10474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hudson, E. K., Hogue, B. A., Souza-Pinto, N. C., Croteau, D. L., Anson, R. M., Bohr, V. A. & Hansford, R. G. (1998) Free Radical Res. 29, 573–579. [DOI] [PubMed] [Google Scholar]
- 21.Shigenaga, M. K., Hagen, T. M. & Ames, B. N. (1994) Proc. Natl. Acad. Sci. USA 91, 10771–10778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.de Souza-Pinto, N. C., Eide, L., Hogue, B. A., Thybo, T., Stevnsner, T., Seeberg, E., Klungland, A. & Bohr, V. A. (2001) Cancer Res. 61, 5378–5381. [PubMed] [Google Scholar]
- 23.Nishioka, K., Ohtsubo, T., Oda, H., Fujiwara, T., Kang, D., Sugimachi, K. & Nakabeppu, Y. (1999) Mol. Biol. Cell 10, 1637–1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Minowa, O., Arai, T., Hirano, M., Monden, Y., Nakai, S., Fukuda, M., Itoh, M., Takano, H., Hippou, Y., Aburatani, H., et al. (2000) Proc. Natl. Acad. Sci. USA 97, 4156–4161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Arai, T., Kelly, V. P., Minowa, O., Noda, T. & Nishimura, S. (2002) Carcinogenesis 23, 2005–2010. [DOI] [PubMed] [Google Scholar]
- 26.Pfanner, N. (2000) Curr. Biol. 10, R412–R415. [DOI] [PubMed] [Google Scholar]
- 27.Gakh, O., Cavadini, P. & Isaya, G. (2002) Biochim. Biophys. Acta 1592, 63–77. [DOI] [PubMed] [Google Scholar]
- 28.Bohr, V. A. & Anson, R. M. (1995) Mutat. Res. 338, 25–34. [DOI] [PubMed] [Google Scholar]
- 29.de Souza-Pinto, N. C., Hogue, B. A. & Bohr, V. A. (2001) Free Radical Biol. Med. 30, 916–923. [DOI] [PubMed] [Google Scholar]
- 30.Souza-Pinto, N. C., Croteau, D. L., Hudson, E. K., Hansford, R. G. & Bohr, V. A. (1999) Nucleic Acids Res. 27, 1935–1942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rabek, J. P., Scott, S., Hsieh, C. C., Reisner, P. D. & Papaconstantinou, J. (1998) Biochim. Biophys. Acta 1398, 137–147. [DOI] [PubMed] [Google Scholar]
- 32.Hsieh, C. C. & Papaconstantinou, J. (2002) Mech. Ageing Dev. 123, 1423–1435. [DOI] [PubMed] [Google Scholar]
- 33.Cristofalo, V. J., Allen, R. G., Pignolo, R. J., Martin, B. G. & Beck, J. C. (1998) Proc. Natl. Acad. Sci. USA 95, 10614–10619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen, Q., Fischer, A., Reagan, J. D., Yan, L. J. & Ames, B. N. (1995) Proc. Natl. Acad. Sci. USA 92, 4337–4341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Saito, H., Hammond, A. T. & Moses, R. E. (1995) Exp. Cell Res. 217, 272–279. [DOI] [PubMed] [Google Scholar]
- 36.Domena, J. D. & Mosbaugh, D. W. (1985) Biochemistry 24, 7320–7328. [DOI] [PubMed] [Google Scholar]
- 37.Landin, J. S., Cohen, S. D. & Khairallah, E. A. (1996) Toxicol. Appl. Pharmacol. 141, 299–307. [DOI] [PubMed] [Google Scholar]
- 38.Hazra, T. K., Izumi, T., Maidt, L., Floyd, R. A. & Mitra, S. (1998) Nucleic Acids Res. 26, 5116–5122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hazra, T. K., Hill, J. W., Izumi, T. & Mitra, S. (2001) Prog. Nucleic Acid Res. Mol. Biol. 68, 193–205. [DOI] [PubMed] [Google Scholar]
- 40.Hill, J. W., Hazra, T. K., Izumi, T. & Mitra, S. (2001) Nucleic Acids Res. 29, 430–438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gordon, D. M., Wang, J., Amutha, B. & Pain, D. (2001) Biochem. J. 356, 207–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schulke, N., Sepuri, N. B., Gordon, D. M., Saxena, S., Dancis, A. & Pain, D. (1999) J. Biol. Chem. 274, 22847–22854. [DOI] [PubMed] [Google Scholar]
- 43.Boldogh, I., Ramana, C. V., Chen, Z., Biswas, T., Hazra, T. K., Grosch, S., Grombacher, T., Mitra, S. & Kaina, B. (1998) Cancer Res. 58, 3950–3956. [PubMed] [Google Scholar]
- 44.Imai, S., Nishibayashi, S., Takao, K., Tomifuji, M., Fujino, T., Hasegawa, M. & Takano, T. (1997) Mol. Biol. Cell 8, 2407–2419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hazra, T. K., Izumi, T., Venkataraman, R., Kow, Y. W., Dizdaroglu, M. & Mitra, S. (2000) J. Biol. Chem. 275, 27762–27767. [DOI] [PubMed] [Google Scholar]
- 46.Takao, M., Aburatani, H., Kobayashi, K. & Yasui, A. (1998) Nucleic Acids Res. 26, 2917–2922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gordon, D. M., Dancis, A. & Pain, D. (2000) Essays Biochem. 36, 61–73. [DOI] [PubMed] [Google Scholar]
- 48.Nilsen, H., Rosewell, I., Robins, P., Skjelbred, C. F., Andersen, S., Slupphaug, G., Daly, G., Krokan, H. E., Lindahl, T. & Barnes, D. E. (2000) Mol. Cell 5, 1059–1065. [DOI] [PubMed] [Google Scholar]
- 49.Nilsen, H., Otterlei, M., Haug, T., Solum, K., Nagelhus, T. A., Skorpen, F. & Krokan, H. E. (1997) Nucleic Acids Res. 25, 750–755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Geissler, A., Rassow, J., Pfanner, N. & Voos, W. (2001) Mol. Cell. Biol. 21, 7097–7104. [DOI] [PMC free article] [PubMed] [Google Scholar]