Abstract
Glycosylphosphatidylinositol (GPI) anchor is a membrane attachment mechanism for cell surface proteins widely used in eukaryotes. GPIs are added to proteins posttranslationally by a complex enzyme, GPI transamidase. Previous studies have shown that human and Saccharomyces cerevisiae GPI transamidases are similar and consist of five homologous components: GAA1, GPI8, PIG-S, PIG-T, and PIG-U in humans and Gaa1p, Gpi8p, Gpi17p, Gpi16p, and Cdc91p in S. cerevisiae. We report that GPI transamidase of Trypanosoma brucei (Tb), a causative agent of African sleeping sickness, shares only three components (TbGAA1, TbGPI8, and TbGPI16) with humans and S. cerevisiae but has two other specific components, trypanosomatid transamidase 1 (TTA1) and TTA2. GPI transamidases of both bloodstream form (growing in mammalian blood) and procyclic form (growing in tsetse fly vector) of the parasite have the same five components. Homologues of TTA1 and TTA2 are present in Leishmania and Trypanosoma cruzi but not in mammals, yeasts, flies, nematodes, plants, or malaria parasites, suggesting that these components may play unique roles in attachment of GPI anchors in trypanosomatid parasites and provide good targets for antitrypanosome drugs.
Trypanosoma brucei, a protozoan parasite, causes sleeping sickness in humans and Nagana disease in domestic animals. The parasite is transmitted by bloodsucking tsetse flies and is widely distributed in the “tsetse belt” in central Africa where trypanosomiasis is a serious medical and agricultural problem. T. brucei has two distinct proliferative forms: a bloodstream form living in mammalian bloodstream and a procyclic form living in the midgut of the tsetse fly. The cell surfaces of these forms are covered by a large amount of glycosylphosphatidylinositol (GPI)-anchored proteins, variant surface glycoproteins in the bloodstream form and procyclins in the procyclic form, corresponding to 10% and 1–3%, respectively, of total cellular proteins (1). The GPI biosynthesis pathway is a candidate target for development of chemotherapeutic agents because GPI anchoring is essential for the life of the bloodstream form (2, 3).
GPI anchoring is a ubiquitous mode of posttranslational modification in eukaryotic organisms (4). Attachment of GPI anchors to proteins is mediated by a complex enzyme GPI transamidase (4–7). Proteins destined to be GPI-anchored have a signal sequence at their C termini that directs GPI anchoring: GPI transamidase cleaves the signal sequence and replaces it with preassembled GPI anchor (5). Human and Saccharomyces cerevisiae GPI transamidases are well conserved, containing five homologous components (8). Five human components, GAA1, GPI8, PIG-S, PIG-T, and PIG-U are homologous to yeast Gaa1p, Gpi8p, Gpi17p, Gpi16p, and Cdc91p, respectively (8–15). Several lines of evidence indicate that GPI8 and Gpi8p are catalytic components responsible for cleavage of GPI-attachment signal sequences (10, 13, 16–18). PIG-T and Gpi16p stabilize the enzyme complexes (14, 15). Specific functions of the other three components are yet to be clarified.
Little is known about the composition of T. brucei GPI transamidase. TbGPI8, T. brucei homologue of the GPI8 gene, has been cloned (19) and its essential role in GPI anchoring has been elucidated by gene disruption (3). We speculated that trypanosome GPI transamidase might be significantly different from its human counterpart for three reasons. First, the GPI-attachment signals are not interchangeable between T. brucei and human cells (20). Second, the structures of the direct precursors of GPI anchors are different (21–23): Both glycan and inositolphospholipid parts are different (24). Third, human GPI transamidase processes quantitatively minor but >100 different GPI-anchored protein precursors, whereas the trypanosome enzyme processes a large amount, but a restricted number, of proteins. In the present study, we isolated trypanosome GPI transamidase to determine its subunit composition and identified two unique components.
Materials and Methods
Cell Culture and Gene Disruption. Culture of the trypanosome, gene disruption, complementation with episomal plasmids, Southern blotting, flow-cytometric analysis, and GPI biosynthesis analysis were carried out as described (2). Drug-resistant clones were selected with 50 μg/ml geneticin (Invitrogen) or 10 μg/ml blasticidin S (Funakoshi, Tokyo).
Purification of GPI transamidase. TbGPI8 was tagged with FLAG and GST at the C terminus (TbGPI8-FLAG-GST) (14, 25) or with FLAG and histidine affinity tag (HAT, CLONTECH) at the C terminus (TbGPI8-FLAG-HAT) (26). A sequence of TbGPI8-FLAG-GST or TbGPI8-FLAG-HAT, each flanked with a splice acceptor site and a poly(A) acceptor site of aldolase, was inserted into the SmaI site of pT11bs-ble (2). The resulting plasmid was transfected into the procyclic and bloodstream forms, the transfectants were selected with 10 μg/ml for procyclic form or 1 μg/ml for bloodstream form of phleomycin (Sigma), and the GPI transamidase was purified (Supporting Materials and Methods, which is published as supporting information on the PNAS web site, www.pnas.org).
Gene Cloning. Using partial amino acid sequences, we found the gene sequences coding for the components of GPI transamidase in databases generated by the T. brucei genome project at the Sanger Institute (Hinxton, U.K.), The Institute for Genomic Research (Rockville, MD) T. brucei genome project, or the parasite genome database at the European Bioinformatics Institute. If these fragments did not cover the entire ORF, we further searched these databases with the newly found sequences. TbGAA1 was cloned in a similar way by using amino acid sequences of human and S. cerevisiae orthologues. After in silico cloning, these genes were amplified by PCR and cloned, and the sequences were confirmed.
Analysis of TbGAA1. We transfected DNA of TbGAA1 bearing FLAG-GST tags at the N terminus or an empty vector into TbGAA1-knockout (KO) procyclics and selected transfectants in 10 μg/ml phleomycin. We lysed 106 cells in TEN-NP buffer [50 mM Tris·HCl (pH 7.5)/5 mM EDTA/150 mM NaCl/Complete protease inhibitor mixture (Roche Diagnostics)/1% Nonidet P-40] immunoprecipitated tagged TbGAA1 with anti-FLAG antibodies and eluted with SDS/PAGE sample buffer with 2-mercaptoethanol. We analyzed samples equivalent to 2.5 × 107 cells by Western blotting. Proteins were visualized with biotin-conjugated anti-FLAG antibody M2 (Sigma) plus horseradish peroxidase-conjugated streptavidin (Amersham Pharmacia Biotech) or with anti-TbGPI8 or anti-CD59 5H8 (26) monoclonal antibody, plus horseradish peroxidase sheep-anti-mouse IgG F(ab′)2 (Amersham Pharmacia Biotech). Hybridoma secreting anti-TbGPI8 monoclonal antibody 7A2 (IgG1) was generated by immunizing a BALB/c mouse with affinity-purified TbGPI8-FLAG-GST and fusion with myeloma cells.
Mass Spectrometric Analysis. GPI transamidase was separated by SDS/PAGE and stained by using a SilverQuest kit (Invitrogen). Protein bands were excised, digested in-gel with trypsin, and analyzed in a QSTAR Pulser i mass spectrometer (Applied Biosystems). Data were analyzed with the mascot program (Matrix Science, London).
Results
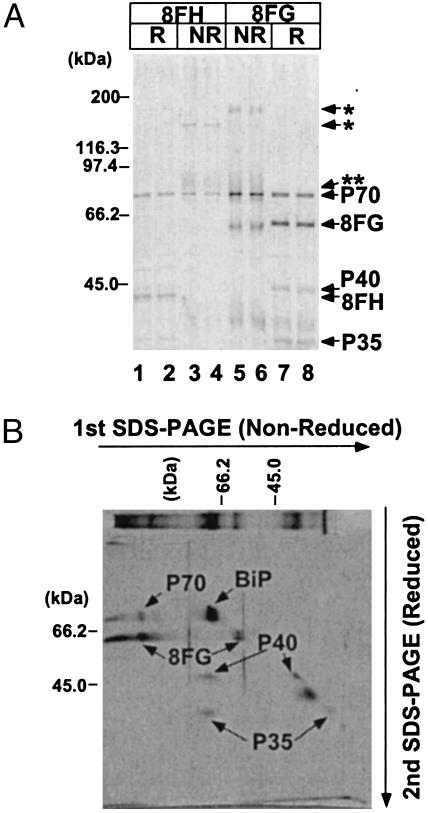
Characterization of GPI Transamidase of T. brucei. To analyze the composition of GPI transamidase of T. brucei, we prepared two cDNA constructs of TbGPI8, each bearing FLAG and GST (TbGPI8-FLAG-GST) or FLAG and HAT (TbGPI8-FLAG-HAT) tags, and transfected them into the procyclic form of T. brucei. From two each of stably transfected clones we extracted proteins with Nonidet P-40 and affinity-purified complexes containing tagged TbGPI8s. Three proteins, P70, P40 and P35, were copurified with either of the tagged TbGPI8s as shown by SDS/PAGE under reducing conditions (Fig. 1A, lanes 1, 2, 7, and 8). We determined the N-terminal sequences of three proteins and obtained new sequences for P40 and P35 but double sequences for P70. To separate the two proteins present in the P70 band, we analyzed samples of complexes under nonreducing conditions. A band of higher molecular mass appeared: 130 kDa with FLAG-HAT tags (lanes 3 and 4) and 150 kDa with FLAG-GST tags (lanes 5 and 6, *). The size difference corresponded to the different sizes of the tags. These higher molecular mass bands reacted with anti-FLAG antibodies on Western blotting (data not shown), indicating that they represent the disulfide-linked complex of TbGPI8 and some other proteins, possibly one of the P70 proteins as judged from their sizes. A diffuse band of ≈75 kDa also appeared under nonreducing conditions (**), representing another disulfide-linked complex (lanes 3–6).
Fig. 1.
SDS/PAGE analysis of the GPI transamidase complex of T. brucei procyclics. (A) GPI transamidase was affinity-purified from Nonidet P-40 extract of procyclics expressing TbGPI8-FLAG-HAT (8FH, lanes 1–4) or TbGPI8-FLAG-GST (8FG, lanes 5–8). Samples were analyzed under reducing (R, lanes 1, 2, 7, and 8) or nonreducing (NR, lanes 3–6) conditions. The identities of silver-stained bands are shown on the right, and size markers are indicated on the left. (B) Two-dimensional SDS/PAGE analysis of GPI transamidase containing TbGPI8-FLAG-GST. First- (left to right) and second- (top to bottom) dimension runs were under nonreducing and reducing conditions, respectively. The identities of silver-stained spots are shown. A similar sample stained after the first SDS/PAGE is shown at the top. Size markers are shown on the top for the first dimension and on the left for the second dimension.
To understand identities of disulfide-linked complexes, we analyzed complexes containing TbGPI8-FLAG-GST with two-dimensional SDS/PAGE, in which the first-dimension PAGE was under nonreducing conditions and the second-dimension PAGE ran at right angles to the first dimension under reducing conditions (Fig. 1B). The band of 150 kDa dissociated into tagged TbGPI8 and P70 after reduction. The diffuse band of 75 kDa dissociated into P40 and P35, indicating that P40 and P35 exist as a disulfide-linked heterodimer and might also exist individually.
We cut spots from a two-dimensional gel and determined their internal amino acid sequences. The P70 that remained at the 70-kDa position under nonreducing conditions was BiP, a molecular chaperone in the endoplasmic reticulum (spot indicated BiP in Fig. 1B). We thought that the copurification of BiP was an artifact caused by the overexpression of tagged TbGPI8 and did not analyze it further, although it was possible that BiP is involved in some step of the GPI-anchor attachment to proteins. We obtained one internal sequence of the other P70 that was disulfide-linked to TbGPI8. We also obtained four internal sequences of P40. We did not analyze a spot at the ≈37-kDa position on the diagonal (Fig. 1B) because its presence was not reproducible in repeated experiments. These results indicated that TbGPI8 makes a protein complex with at least three proteins, P70, P40, and P35, and that TbGPI8 and P70 and P40 and P35, respectively, are associated via a disulfide bond with each other.
The P70 Termed TbGPI16 Is an Orthologue of Mammalian PIG-T and Yeast Gpi16p. Using partial amino acid sequences, we found a complete ORF of P70 in the database of the European Bioinformatics Institute. P70 had significant amino acid sequence homology to human PIG-T (13% identity and 25% similarity) and S. cerevisiae Gpi16p. P70 and PIG-T/Gpi16p share similar structural characteristics, namely, the N-terminal signal sequence and one transmembrane domain near the C terminus (Fig. 7, which is published as supporting information on the PNAS web site). We recently reported that in human GPI transamidase, GPI8 and PIG-T are linked by a disulfide bond formed between C92 of GPI8 and C182 of PIG-T (25). We also reported that TbGPI8 is disulfide-linked to another protein via its C76, which corresponds to C92 of human GPI8. C182 of PIG-T corresponded to conserved C239 of P70, suggesting that C239 is involved in the disulfide linkage with TbGPI8. Therefore, in both T. brucei and human GPI transamidase the catalytic subunit (TbGPI8/GPI8) is disulfide-linked to another subunit (P70/PIG-T), indicating that P70 is functionally homologous to PIG-T. Based on these results, we termed P70 TbGPI16 (GenBank accession no. AB111856).
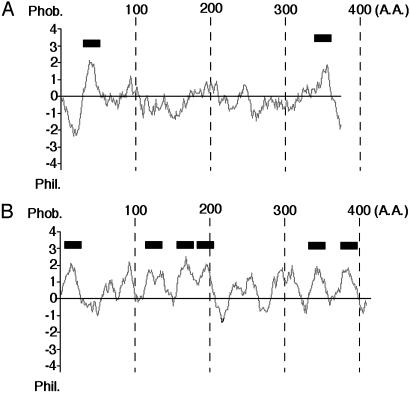
P40 [Trypanosomatid Transamidase 1 (TTA1)] and P35 (TTA2) Are Essential Components of Trypanosome GPI Transamidase. We also obtained complete ORFs of P40 and P35. Fig. 2 shows the hydrophobicity profiles of P40 and P35. P40 had transmembrane regions near the N and C termini (Fig. 2 A). It also had two N-glycosylation sites. That the size of P40 decreased after treatment with peptide N-glycanase F indicated that one or both sites were N-glycosylated (data not shown). Therefore, the central hydrophilic part should face the luminal side of the endoplasmic reticulum. The N-terminal methionine was removed, as shown by sequencing (data not shown), which is consistent with its cytoplasmic orientation. P35 had multiple (most likely six) transmembrane regions (Fig. 2B). P40 and P35 had no sequence similarity to any of the human components of GPI transamidase. They had no significant homology to other proteins with known function in the databases. Because P40 and P35 are likely components of GPI transamidase specific to trypanosomes, we termed P40 and P35 as TTA1 and TTA2, respectively (GenBank accession nos. AB111858 and AB111859, respectively).
Fig. 2.
Hydrophobicity profile of TTA1 (A) and TTA2 (B). Putative transmembrane domains are shown by thick lines above each hydrophobicity plot.
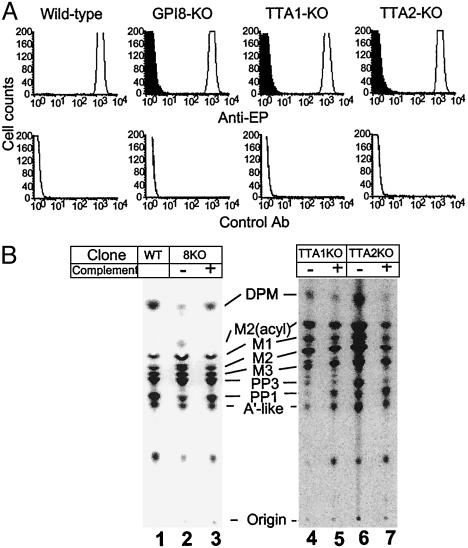
To determine whether TTA1 and TTA2 are functional components of GPI transamidase, we knocked out these genes from the procyclic form of T. brucei (Fig. 8, which is published as supporting information on the PNAS web site). Disruption of TTA1 or TTA2 resulted in slower growth (about twice the doubling time) that was normalized by transfection of corresponding cDNA. Then we studied the expression of glutamate-proline (EP) procyclins on the surface of TbGPI8-, TTA1-, and TTA2-KO clones (GPI8-KO, TTA1-KO, and TTA2-KO, respectively) (Fig. 3A). EP procyclins are GPI-anchored and are the most abundant proteins on the surface of the procyclic form of T. brucei. All of these clones lost the surface expression of EP procyclins. The plasmids bearing corresponding cDNA restored the expression of EP procyclins to a level similar to that of wild-type procyclics.
Fig. 3.
Characterization of TTA1-KO and TTA2-KO procyclics. (A) Flow-cytometric analysis of the surface expression of EP procyclin. Wild-type, TbGPI8-KO, TTA1-KO, and TTA2-KO clones transfected with an empty vector (filled) or plasmid bearing corresponding cDNA (line) were analyzed in a FACScan by using anti-EP procyclin (Anti-EP) or control antibodies. (B) TLC analysis of GPI biosynthesis of TTA1-KO and TTA2-KO. Wild-type (lane 1), GPI8KO (lanes 2 and 3), TTA1-KO (lanes 4 and 5), and TTA2-KO (lanes 6 and 7) procyclics transfected with an empty vector (lanes 2, 4, and 6) or corresponding cDNA (lanes 3, 5, and 7) were hypotonically lysed. The lysates were incubated with GDP-[3H]mannose, and mannolipids were analyzed by TLC. The identities of mannolipids are shown. DPM, dolichol-phosphate-mannose; M2(acyl), GPI bearing two mannoses with acylated inositol; M1, M2, and M3, GPI bearing one, two, and three mannoses, respectively; A′-like, GPI bearing three mannoses with ethanolamine phosphate on the third mannose; PP3, A′-like with acylated inositol; PP1, mature GPI in the procyclic form.
We also examined biosynthesis of GPI in the KO procyclics (Fig. 3B). The profiles of intermediates were similar between wild-type (lane 1) and the KO (lanes 2, 4, and 6) parasites. All generated PP1, the complete form of GPI in procyclics, indicating that TTA1 and TTA2 mutants generated but did not transfer the complete GPI to proteins. Therefore, TTA1 and TTA2 are the essential components of GPI transamidase.
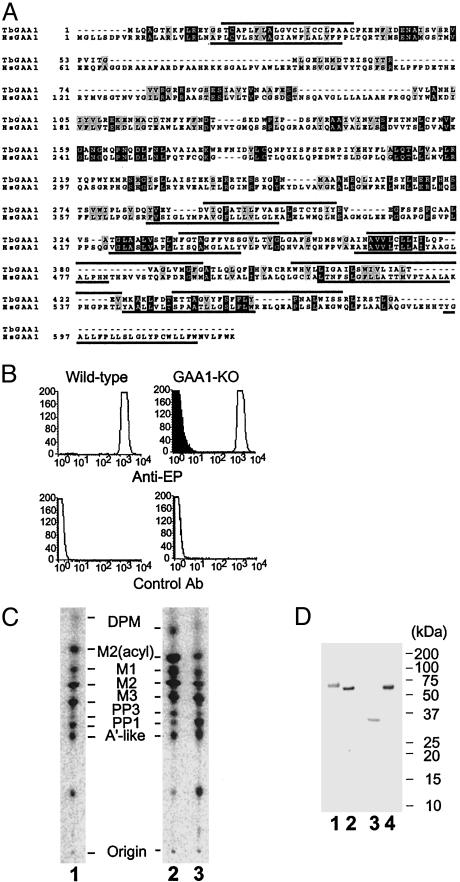
TbGAA1 Is the Fifth Component of Trypanosome GPI Transamidase. Although we did not find the T. brucei homologue of GAA1 in the analysis of the affinity-purified complexes (Fig. 1), we found it in the database and cloned it (GenBank accession no. AB111857). This homologue (TbGAA1) was smaller than human GAA1 (461 vs. 621 aa), but they had 21% amino acid identity and 38% similarity and had similar hydrophobicity profiles (Fig. 4A).
Fig. 4.
Characterization of TbGAA1. (A) Comparison of amino acid sequences of T. brucei and human GAA1. Putative transmembrane domains are overlined for T. brucei GAA1 (TbGAA1) and underlined for human GAA1 (HsGAA1). Identical and similar amino acids are in black and gray boxes, respectively. (B) Flow-cytometric analysis of the surface expression of EP procyclin on TbGAA1-KO mutant (GAA1-KO). For GAA1-KO, filled and open lines are transfectants with an empty vector and TbGAA1 cDNA, respectively. (C) GPI biosynthesis of GAA1-KO. Lane 1, wild type; lane 2, GAA1-KO transfected with an empty vector; lane 3, GAA1-KO transfected with TbGAA1 cDNA. The identities of mannolipids are the same as those in Fig. 3B. (D) Association of TbGAA1 with TbGPI8. The Nonidet P-40 extracts of GAA1-KO procyclics transfected with FLAG-GST-TbGAA1 (lanes 1 and 3) and GPI8-KO procyclics transfected with TbGPI8-FLAG-GST, as a control for Western blotting (lanes 2 and 4), were immunoprecipitated with anti-FLAG antibody and Western-blotted with anti-FLAG (lanes 1 and 2) or anti-TbGPI8 (lanes 3 and 4) antibodies. Size markers are shown on the right.
We knocked out TbGAA1 to determine whether it is involved in GPI-anchor attachment. The TbGAA1-KO clone (GAA1-KO) completely lost the surface expression of EP procyclin, which was restored by transfection of the plasmid bearing TbGAA1 (Fig. 4B). The profile of GPI biosynthesis was similar to those of wild-type (Fig. 4C) and other mutants. These results indicated that TbGAA1 is essential for GPI transamidase.
To test whether TbGAA1 is associated with the GPI transamidase complex, we constructed TbGAA1 tagged with FLAG-GST at the N terminus and transfected it into GAA1-KO procyclics. The transfectant restored the surface expression of EP procyclins, indicating that the tagged TbGAA1 was functional (data not shown). We then immunoprecipitated TbGAA1 with anti-FLAG antibodies and subjected it to Western blotting. We detected FLAG-GST-tagged TbGAA1 with anti-FLAG antibodies (Fig. 4D, lane 1). The size of the tagged TbGAA1 was ≈70 kDa, being smaller than the calculated size of 80 kDa. It is possible that a cleavage occurred near the C terminus. With an anti-TbGPI8 monoclonal antibody, we detected a band of ≈35 kDa, indicating that TbGPI8 was associated with tagged TbGAA1 (Fig. 4D, lane 3).
To confirm the presence of TbGAA1 in GPI transamidase complexes, we affinity-purified the complex again. This time we transfected TbGPI8-FLAG-GST into TbGPI8-KO procyclics and purified the complex similarly. Analysis with SDS/PAGE and silver staining revealed four similar bands (Fig. 5, lane 1). Because we predicted that the molecular sizes of nontagged TbGAA1 and TTA1 are similar based on the size of tagged TbGAA1, we analyzed the band at a 40-kDa position with in-gel digestion and mass spectrometry and found peptides derived from TbGAA1 and TTA1, showing that TbGAA1 is a component of GPI transamidase complex.
Fig. 5.
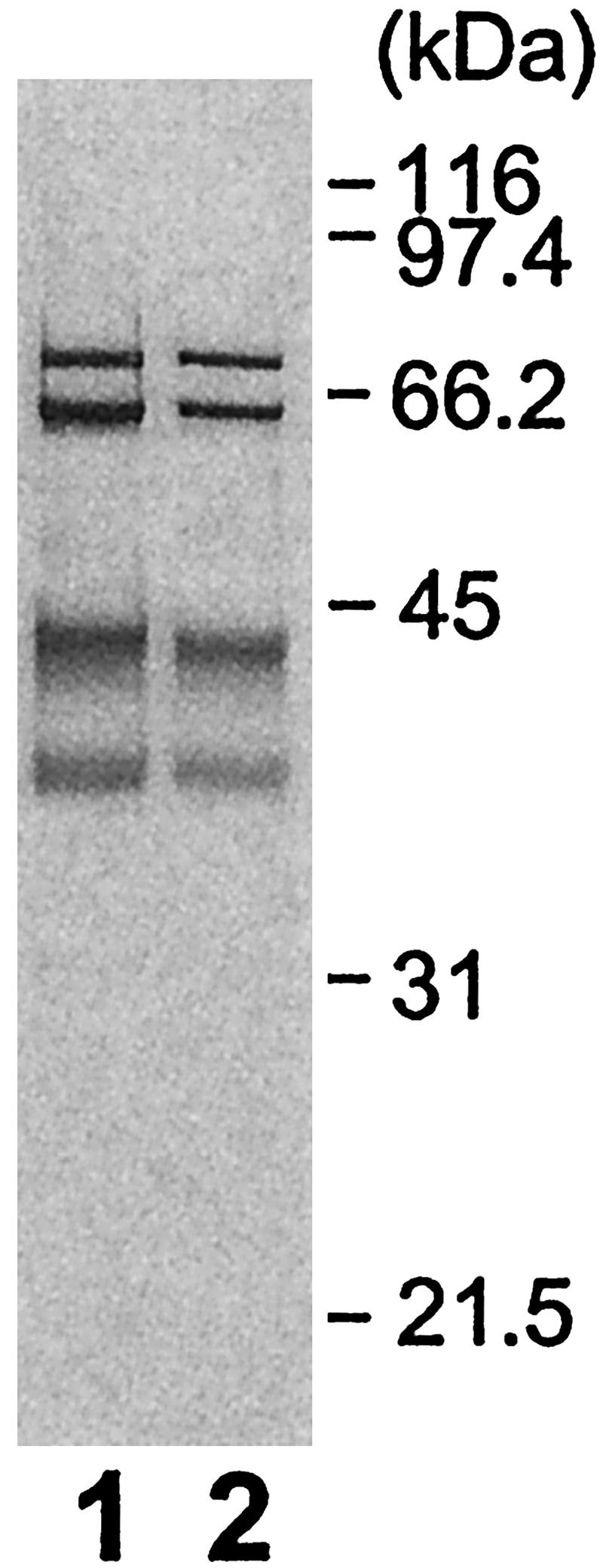
GPI transamidase of the bloodstream form of T. brucei. GPI transamidase complexes were affinity-purified from the procyclic (lane 1) and bloodstream (lane 2) forms of T. brucei expressing TbGPI8-FLAG-GST. Samples were analyzed by SDS/PAGE under reducing conditions. Size markers are shown on the right.
GPI Transamidase of the Bloodstream Form of T. brucei. The above compositional analysis of GPI transamidase was done with the procyclic form. We next tested whether the components are the same in the bloodstream form. We affinity-purified the GPI transamidase complex from the bloodstream form expressing TbGPI8-FLAG-GST and compared it with the complex from the procyclic form (Fig. 5). The complexes of the bloodstream form showed four similar bands. We also analyzed the band at a 40-kDa position with in-gel digestion and mass spectrometry and found peptides of TbGAA1 and TTA1. Therefore, the bloodstream and procyclic forms use the same five components of GPI transamidase.
GPI Transamidases of Various Eukaryotes Are Grouped into Two. A search of databases of eukaryotes with complete genome projects revealed that five components of human GPI transamidases are conserved in Schizosaccharomyces pombe, Drosophila melanogaster, Caenorhabditis elegans, and Arabidopsis thaliana as well as S. cerevisiae (Table 1). We did not find homologues of TTA1 and TTA2 in these databases, suggesting that mammalian, insect, nematode, yeast, and plant GPI transamidases are similar, sharing five homologous components. We found homologues of GPI8, GAA1, PIG-T, and PIG-U but not PIG-S in the Plasmodium falciparum database. Using the sequence of the S. pombe PIG-S homologue, we found a Plasmodium PIG-S homologue (Table 1). We did not find Plasmodium homologues of TTA1 and TTA2, suggesting that P. falciparum has GPI transamidase similar to mammals and yeasts.
Table 1. Components of GPI transamidases of various organisms.
| GPI8 | GAA1 | PIG-T | PIG-S | PIG-U | TTA1 | TTA2 | |
|---|---|---|---|---|---|---|---|
| Homo sapiens | + | + | + | + | + | - | - |
| T. brucei | + | + | + | - | - | + | + |
| S. cerevisiae | 2 × 10-94* | 6 × 10-40 | 4 × 10-59 | 7 × 10-20 | 6 × 10-49 | (-)† | (-) |
| S. pombe | 3 × 10-93 | 2 × 10-42 | 4 × 10-55 | 3 × 10-39 | 1 × 10-10 | (-) | (-) |
| D. melanogaster | 1 × 10-114 | 3 × 10-72 | 1 × 10-107 | 8 × 10-46 | 5 × 10-82 | (-) | (-) |
| C. elegans | 2 × 10-90 | 3 × 10-25 | 1 × 10-39 | 9 × 10-13 | 8 × 10-21 | (-) | (-) |
| A. thaliana | 1 × 10-91 | 4 × 10-36 | 1 × 10-82 | 3 × 10-26 | 2 × 10-44 | (-) | (-) |
| P. falciparum | 4 × 10-31 | 1 × 10-4 | 0.028 | (-)‡ | 7 × 10-8 | (-) | (-) |
| L. major | 5.1 × 10-77§ | 6.9 × 10-76§ | 9.1 × 10-48§ | (-) | (-) | 4.4 × 10-20§ | 2.7 × 10-19§ |
The genome databases of S. cerevisiae, S. pombe, D. melanogaster, C. elegans, A. thaliana, and P. falciparum were searched by using the blastp program using amino acid sequences of human GPI8, GAA1, PIG-T, PIG-S, and PIG-U and Tb TTA1 and TbTTA2. E values of homologues are shown.
Homologue with an E value <0.1 was not found.
Sequence of S. pombe PIG-S hit a homologue with an E value of 0.059.
The L. major databases in the European Bioinformatics Institute (updated on 03/31/2003) were searched by the tblastn program by using sequences of TbGPI8, TbGAA1, TbGPI16, TTA1, and TTA2. E values were against partial gene sequences.
We found Leishmania major homologues of TTA1 and TTA2 and Trypanosoma cruzi TTA2 homologue in databases. L. major had homologues of TbGPI8, TbGAA1, and TbGPI16 (Table 1). It is very likely, therefore, that GPI transamidases of trypanosomatid protozoa form a different group.
Discussion
The major finding of this study is that GPI transamidase of T. brucei contains two unique components. Human and S. cerevisiae GPI transamidases are very well conserved, consisting of five homologous components. It is highly likely that GPI transamidases of other eukaryotic organisms such as D. melanogaster, C. elegans, A. thaliana, and P. falciparum also consist of the same five components. T. brucei does share three of these components, namely GAA1, GPI8, and PIG-T homologues, but it does not have PIG-S and PIG-U homologues; instead it has TTA1 and TTA2. Homologues of TTA1 and TTA2 are found in other trypanosomatids (L. major and T. cruzi) but not in the wider range of the other eukaryotes, indicating that TTA1 and TTA2 are unique components of trypanosomatid GPI transamidases. Therefore, GPI transamidases have two groupings: one containing mammalian, insect, nematode, yeast, plant, and malaria enzymes, and the other containing T. brucei, T. cruzi, and L. major.
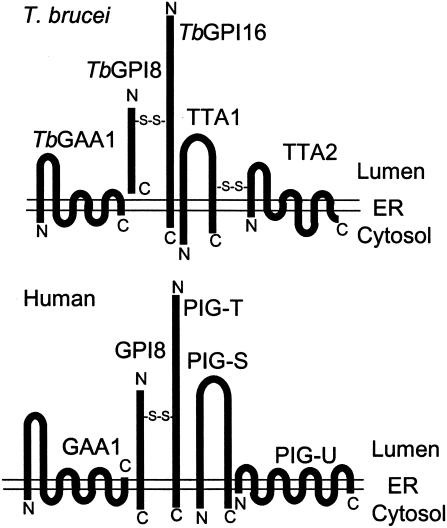
Fig. 6 shows the predicted membrane orientation of the components of T. brucei and the human GPI transamidases. Among three common components, a structural difference is seen in the catalytic component GPI8. Human GPI8 is a type I membrane protein having a transmembrane domain near the C terminus (13), whereas TbGPI8 lacks a transmembrane domain (3, 19). The transmembrane domain of human GPI8 is not essential for the enzyme activity (13). GPI8 homologues of S. cerevisiae, S. pombe, A. thaliana, and P. falciparum are type I transmembrane proteins. In contrast, D. melanogaster, C. elegans, and L. major GPI8 homologues do not have a transmembrane domain (27). Therefore, the presence or absence of a transmembrane domain does not correspond to two groups of GPI transamidases.
Fig. 6.
Comparison of five components of human and trypanosome GPI transamidases. Schematic representation of three common components (GAA1, GPI8, and PIG-T/GPI16) and two unique components (TTA1 and TTA2 of T. brucei and PIG-S and PIG-U of humans). The membrane orientation and N and C termini are shown.
In the human enzyme, GPI8 is linked to PIG-T via a disulfide bond formed between C92 of GPI8 and C182 of PIG-T (Fig. 6). This disulfide linkage is important for the activity of GPI transamidase (25). Similarly, TbGPI8 is disulfide-linked to TbGPI16 in that C76 of TbGPI8, corresponding to C92 of human GPI8, is involved in the disulfide formation (25). C239 of TbGPI16 is homologous to C182 of PIG-T, suggesting that it is the responsible cysteine. Because both of these cysteines are conserved in other eukaryotes, it is likely that the covalent linkage of GPI8 to PIG-T is common among all GPI transamidases, contributing to the stability of the complexes.
TTA1 and TTA2 have no significant sequence homology to PIG-S and PIG-U and are linked via a disulfide bond, whereas there is no covalent linkage between PIG-S and PIG-U. Nevertheless, hydrophobicity profiles and membrane orientations of TTA1 and TTA2 are similar to those of PIG-S and PIG-U, respectively (Fig. 6); they might even have similar functions. As described in the introduction, GPI-attachment signal sequences are not interchangeable between human and T. brucei (20). Also, the structures of the direct precursors of GPI anchors are different in both glycan and inositolphospholipid parts. It is likely, therefore, that components that recognize the GPI-attachment signals and GPI are different. The two different components of GPI transamidases may be involved in such functions.
Five components are shared by the procyclic and bloodstream forms. In the procyclic form, the direct precursor of the GPI anchor, PP1, has lysoPI with acylated inositol (23). In contrast, the direct precursor of the GPI anchor in the bloodstream form, glycolipid A, has strictly dimyristoylated phosphatidylinositol with nonacylated inositol (21). If the heteropentameric complex represents a complete, fully active GPI transamidase, the fact that the direct GPI-anchor precursors of the procyclic and bloodstream forms have different inositolphospholipid structures suggests that the common glycan part might be the main determinant of substrate specificity. Alternatively, components that recognize GPI are still missing, and the procyclic and bloodstream forms use different components.
As revealed by gene disruption, both TTA1 and TTA2 are essential components of GPI transamidase. Because GPI transamidase is essential for life of the bloodstream form (3), drugs that inhibit TTA1 or TTA2 should kill the bloodstream form of T. brucei. Therefore, TTA1 and TTA2 may be potential targets for antitrypanosome drug development. Furthermore, GPI transamidase is critical for the procyclic form to survive in the tsetse fly (3), suggesting that drugs that inhibit TTA1 or TTA2 may also be useful to inhibit spreading of trypanosomes from one human to another.
Supplementary Material
Acknowledgments
We thank Yosuke Fujinaga for contribution to the initial stage of this work; Jenny Stephens for contribution to the analysis of TbGAA1; and Fumiko Ishii and Keiko Kinoshita for technical assistance. This work was supported by grants from the Ministry of Education, Science, Sports, Culture, and Technology of Japan.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: GPI; glycosylphosphatidylinositol; Tb, Trypanosoma brucei; HAT, histidine affinity tag; KO, knockout; EP, glutamate-proline; TTA, trypanosomatid transamidase.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. AB111856–AB111859).
References
- 1.Ferguson, M. A. J. (2000) Proc. Natl. Acad. Sci. USA 97, 10673–10675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nagamune, K., Nozaki, T., Maeda, Y., Ohishi, K., Fukuma, T., Hara, T., Schwarz, R. T., Sutterlin, C., Brun, R., Riezman, H. & Kinoshita, T. (2000) Proc. Natl. Acad. Sci. USA 97, 10336–10341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lillico, S., Field, M. C., Blundell, P., Coombs, G. H. & Mottram, J. C. (2003) Mol. Biol. Cell 14, 1182–1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.McConville, M. J. & Menon, A. K. (2000) Mol. Membr. Biol. 17, 1–16. [DOI] [PubMed] [Google Scholar]
- 5.Udenfriend, S. & Kodukula, K. (1995) Annu. Rev. Biochem. 64, 563–591. [DOI] [PubMed] [Google Scholar]
- 6.Tiede, A., Bastisch, I., Schubert, J., Orlean, P. & Schmidt, R. E. (1999) Biol. Chem. 380, 503–523. [DOI] [PubMed] [Google Scholar]
- 7.Ikezawa, H. (2002) Biol. Pharm. Bull. 25, 409–417. [DOI] [PubMed] [Google Scholar]
- 8.Hong, Y., Ohishi, K., Kang, J. Y., Tanaka, S., Inoue, N., Nishimura, J., Maeda, Y. & Kinoshita, T. (2003) Mol. Biol. Cell 14, 1780–1789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hamburger, D., Egerton, M. & Riezman, H. (1995) J. Cell Biol. 129, 629–639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Benghezal, M., Benachour, A., Rusconi, S., Aebi, M. & Conzelmann, A. (1996) EMBO J. 15, 6575–6583. [PMC free article] [PubMed] [Google Scholar]
- 11.Yu, J., Nagarajan, S., Knez, J. J., Udenfriend, S., Chen, R. & Medof, M. E. (1997) Proc. Natl. Acad. Sci. USA 94, 12580–12585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hiroi, Y., Komuro, I., Chen, R., Hosoda, T., Mizuno, T., Kudoh, S., Georgescu, S. P., Medof, M. E. & Yazaki, Y. (1998) FEBS Lett. 421, 252–258. [DOI] [PubMed] [Google Scholar]
- 13.Ohishi, K., Inoue, N., Maeda, Y., Takeda, J., Riezman, H. & Kinoshita, T. (2000) Mol. Biol. Cell 11, 1523–1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ohishi, K., Inoue, N. & Kinoshita, T. (2001) EMBO J. 20, 4088–4098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fraering, P., Imhof, I., Meyer, U., Strub, J. M., van Dorsselaer, A., Vionnet, C. & Conzelmann, A. (2001) Mol. Biol. Cell 12, 3295–3306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Meyer, U., Benghezal, M., Imhof, I. & Conzelmann, A. (2000) Biochemistry 39, 3461–3471. [DOI] [PubMed] [Google Scholar]
- 17.Vidugiriene, J., Vainauskas, S., Johnson, A. E. & Menon, A. K. (2001) Eur. J. Biochem. 268, 2290–2300. [DOI] [PubMed] [Google Scholar]
- 18.Spurway, T. D., Dalley, J. A., High, S. & Bulleid, N. J. (2001) J. Biol. Chem. 276, 15975–15982. [DOI] [PubMed] [Google Scholar]
- 19.Kang, X., Szallies, A., Rawer, M., Echner, H. & Duszenko, M. (2002) J. Cell Sci. 115, 2529–2539. [DOI] [PubMed] [Google Scholar]
- 20.Moran, P. & Caras, I. W. (1994) J. Cell Biol. 125, 333–343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Masterson, W. J., Raper, J., Doering, T. L., Hart, G. W. & Englund, P. T. (1990) Cell 62, 73–80. [DOI] [PubMed] [Google Scholar]
- 22.Englund, P. T. (1993) Annu. Rev. Biochem. 62, 121–138. [DOI] [PubMed] [Google Scholar]
- 23.Treumann, A., Zitzmann, N., Hulsmeier, A., Prescott, A. R., Almond, A., Sheehan, J. & Ferguson, M. A. (1997) J. Mol. Biol. 269, 529–547. [DOI] [PubMed] [Google Scholar]
- 24.Mehlert, A., Zitzmann, N., Richardson, J. M., Treumann, A. & Ferguson, M. A. (1998) Mol. Biochem. Parasitol. 91, 145–152. [DOI] [PubMed] [Google Scholar]
- 25.Ohishi, K., Nagamune, K., Maeda, Y. & Kinoshita, T. (2003) J. Biol. Chem. 278, 13959–13967. [DOI] [PubMed] [Google Scholar]
- 26.Watanabe, R., Murakami, Y., Marmor, M. D., Inoue, N., Maeda, Y., Hino, J., Kangawa, K., Julius, M. & Kinoshita, T. (2000) EMBO J. 19, 4402–4411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hilley, J. D., Zawadzki, J. L., McConville, M. J., Coombs, G. H. & Mottram, J. C. (2000) Mol. Biol. Cell 11, 1183–1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.