Abstract
A paradigm is described for recording the activity of single cortical neurons from awake, behaving macaque monkeys. Its unique features include high-density microwire arrays and multichannel instrumentation. Three adult rhesus monkeys received microwire array implants, totaling 96–704 microwires per subject, in up to five cortical areas, sometimes bilaterally. Recordings 3–4 weeks after implantation yielded 421 single neurons with a mean peak-to-peak voltage of 115 ± 3 μV and a signal-to-noise ratio of better than 5:1. As many as 247 cortical neurons were recorded in one session, and at least 58 neurons were isolated from one subject 18 months after implantation. This method should benefit neurophysiological investigation of learning, perception, and sensorimotor integration in primates and the development of neuroprosthetic devices.
The use of single, movable electrodes to record neuronal activity in macaque monkeys has proven to be an extremely productive method in neurophysiology. Although the method was originally developed by Evarts (1) to study sleep, he later noticed that when monkeys moved, neurons in motor cortex discharged “with... frequencies... reaching 60–80 per sec at certain phases of movement.” Thus, neurophysiologists could study activity in awake, behaving monkeys with methods that have changed little in the past 40 years.
Although Evarts' method was a breakthrough for neurophysiology, the use of single, movable electrodes typically limits data collection to one or two neurons at a time for brief periods. To study a neuronal population, cells must be sampled serially, over weeks. Newer systems use several movable electrodes, typically 6 to 16, but recordings remain limited to one cortical area with relatively brief recording durations. These factors impede the study of neural correlates of learning, neuronal interactions, and population activity.
Long-term, multielectrode recordings have become routine in rodent neurophysiology (2–6) and have recently been adapted to New World monkeys (7, 8). However, robust and reliable application of chronic, multiple-electrode recording methods in macaque monkeys remains a challenge. Meeting that challenge is important because macaques have been the primate model of choice for studying the neural mechanisms underlying perception, movement, control, and cognition.
Recent attempts to obtain long-lasting, single-neuron recordings from macaques have used the 100-electrode “Utah array” or arrays of individual microwires (9–14). However, these studies have provided modest neuronal yields of uncertain longevity, and, in most cases, have been limited to just one or two cortical areas per animal. This article describes a paradigm for recording simultaneously from a larger population of neurons in a larger number of cortical areas for up to 18 months. It also describes methods for evaluating the quality of single-unit isolation in multielectrode recordings.
Methods
All recordings described in this study were obtained through chronically implanted arrays of microwires. In one animal, we used commercially available (NBlabs, Dennison, TX) low-density arrays formed by 16–32 50-μm Teflon-coated stainless microwires (15). Two other animals were implanted with high-density microwire arrays developed at Duke University. The main design used in this study consists of: (i) an array of S-isonel (or Teflon)-coated tungsten (or stainless steel) microwire electrodes, (ii) a printed circuit board (PCB) connected to the microwire electrodes, and (iii) a high-density, miniature connector attached to the opposite side of the PCB (see Fig. 1 A–C). The flexibility of our PCB design allows virtually any type of microwire to be used as an electrode.
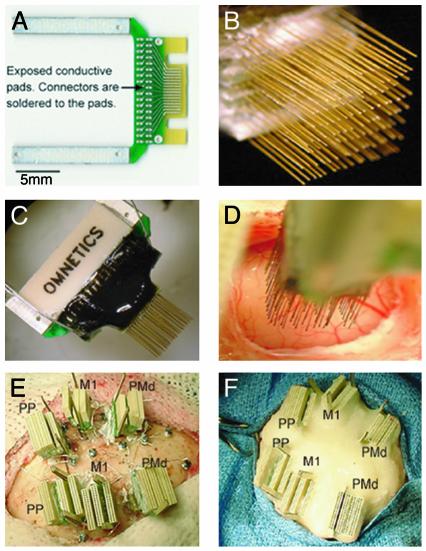
Fig. 1.
(A) Printed circuit board (PCB) used to build a high-density microwire array (spacing between traces is 300 μm). (B) Multiple boards are stacked up to form arrays with up to 128 microwires. (C) High-density connectors are attached to the other side of the board. (D) Implantation of a 128-microwire array in the M1. (E) Last stages of a 672-microwire implant. Bilateral implants included the PMd, M1, and the PP. (F) Final aspect of a multisite implant after the application of multiple layers of dental cement.
Subjects. Three rhesus monkeys (Macaca mulatta) were used in this study: two females (monkey 1, 9.7 kg and monkey 2, 9.9 kg) and one male (monkey 3, 10.2 kg). All procedures conformed to the National Academy Press Guide for the Care and Use of Laboratory Animals and were approved by the Duke University Animal Care and Use Committee.
Surgery for Implantation of Microwire Arrays. Aseptic surgical technique was used throughout. Deep surgical anesthesia was maintained throughout the procedure by using a combination of isoflurane and i.v. fentanyl. After several craniotomies were drilled, microwire array touched down the surface of the brain under visualization by using the operating microscope (Fig. 1D). The electrode array was advanced slowly at ≈100 μm per 1-min intervals by using a hydraulic micropositioner (David Kopf Instruments, Tujunga, CA). Penetration of the brain was assessed electrophysiologically by isolation, amplification, and visual and audio monitoring of neuronal activity. Up to 5 to 10 cortical implants were made per animal (Fig. 1 E and F).
Multisite, Many-Neuron Ensemble Recordings. A multichannel acquisition processor (MAP, Plexon, Dallas) cluster, formed by three 128-channel MAPs synchronized by a common (2 MHz) clock signal and an external start pulse, was specially built for the experiments reported here. This 384-channel recording system has a theoretical capacity of recording up to 1,536 single neurons simultaneously (e.g., four neurons per channel), at 25-μs precision.
Single-Unit Spike Sorting. The first step in all recording sessions involved the setting of a voltage threshold for each of the MAP channels processing signals from the implanted microwires. This threshold was set by the experimenter through visual inspection of both the original analog signal (displayed in an oscilloscope) and the digitized record displayed on the screen of the computer controlling the MAP. All events that crossed the set voltage threshold (defined as unsorted waveforms) for each channel were digitized and recorded continuously for up to 2–3 h for subsequent off-line spike sorting analysis. An Off-Line Spike Sorter (OFSS, Plexon) was used to isolate single units from all our recording sessions. Initially, principal component (PC) analysis was carried out by using all unsorted waveforms recorded. Results involving the first eight PCs were made available to the user, because they usually accounted for close to 90% of the variance of the waveforms. Next, the experimenter selected three of the available components to define 2D/3D PC spaces. These PC spaces were used to plot the values of the corresponding PC scores calculated for all unsorted waveforms. Inspection of clusters of points observed in PC space took advantage of several tools provided by OFSS. These tools allow the experimenter to visualize the waveform associated with each point in the 2D/3D PC space, measure the waveform's peak-to-peak voltage amplitude, and compare it with events immediately before or after its occurrence. This latter option involved the use of a “digital oscilloscope” that provided a continuous record of all of the stored events from a given channel. By selecting a group of points from the PC space, interspike interval (ISI) histograms were calculated. All of these tools were used to identify whether the channel contained any spurious signals (e.g., electrical noise, movement artifact). Spurious signals were classified as invalid spikes and removed from the PC space. The remainder of the waveforms, denominated valid spikes, were then scrutinized for the isolation of single units.
Based on the quantification of multiple parameters (waveform voltage, waveform shape, ISIs, etc.), the experimenter then defined the cluster partition for each channel. This was done by manually defining the clusters shown to contain similar waveforms (see an example of the off-line spike sorting procedure in Fig. 6, which is published as supporting information on the PNAS web site, www.pnas.org).
A group of similar waveforms was considered as being generated from a single neuron only if it defined a discrete cluster in 2D/3D PC space that was distinct from clusters for other units and/or multiunit activity. In addition, single units had to exhibit a clearly recognizable refractory period (>1 ms) in their ISI histograms and have a characteristic and distinct waveform shape and peak-to-peak amplitude when compared with other neuronal waveforms and multiunit activity (as seen in the digital oscilloscope) in the same channel (see Fig. 2).
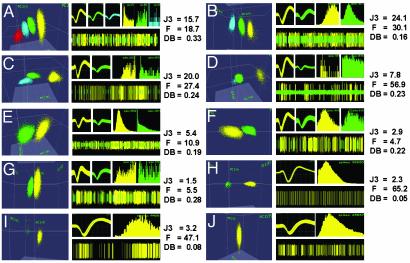
Fig. 2.
Examples of cluster separation, 30 days postimplantation. Each example contains: (Left) identified clusters in 3D PC space; (Upper Middle) waveforms; (Upper Right) ISI histograms; (Lower Right) continuous record of potentials. Different colors depict distinct clusters in all parts of each example. J3 (2D), F (2D), and DB (2D) statistics are at the far right. (A) Three single units (yellow, green, and blue) discriminated from the multiunit record (red cluster) yielded high J3 and F values, average DB, clear differences in waveforms, and ISI histograms. (B) Two single units (green and yellow) were differentiated from the multiunit activity (blue), producing high J3 and F and low DB values. (C) High separation between the multiunit cluster (blue) and two units (yellow and green). (D) One large (yellow) and one small (green) unit were isolated from the multiunit activity (blue). (E) Two clearly distinct units were isolated. J3 was above the mean, DB was good, and F value was average. (F) Two units with distinct amplitudes and ISI histograms from a channel with below average J3 and F values. The DB value indicates good cluster separation. (G) Channel with low J3 and F values, but an average DB. Two units with distinct waveforms and ISI histograms were isolated. (H) Example of a large single unit (yellow) isolated from the multiunit activity (green). J3 was below average, F was very high, and DB was very low. (I) Single unit (yellow) clearly distinct from the multiunit activity (green). The J3 value was close to its mean, the F value was high, and DB was low, indicating excellent cluster separation. (J) A single unit (yellow cluster) isolated way above the multiunit voltage level. ISI histogram time scales were 5 ms (C), 10 ms (A, D, and G), and 100 ms (B, E, F, and H–J).
Four different statistics were used to objectively quantify the overall separation between identified clusters in a given recording channel. These measurements included the classic parametric F statistic of multivariate analysis of variance (MANOVA), the J3 and PseudoF (PsF) statistics (16, 17), and the Davies-Bouldin (DB) validity index (ref. 18 and see Supporting Text, which is published as supporting information on the PNAS web site, for details on how these statistics were calculated). Control values for the four statistics were obtained from channels in which no single-unit activity was detected. To calculate control values of the four statistics, the single spheroid cloud of points centered at the origin of the PC space was divided in half. Then, two separate clusters with similar shape and number of waveforms were defined by the experimenter so that the same statistics (F, J3, PsF, and DB) could be calculated.
Mean signal-to-noise ratios were also calculated for all single units isolated by dividing the mean peak-to-peak voltage (Vpp) of the isolated single unit by the mean “noise” level observed in our microwire recordings. Mean noise was calculated in a large sample of channels by lowering the voltage threshold to zero and then measuring the Vpp of the lowest amplitude waveforms that could not be differentiated into single units (the so-called multiunit activity). Short-term single-unit isolation stability (i.e., stability over hours) was evaluated with Wavetracker (Plexon). Data collected during 1- to 2-h recording sessions were first divided in consecutive 30-min segments. For each of these data segments, single units were discriminated, and 2D/3D PC clusters, waveforms, ISIs, and cluster separation statistics were calculated for each recorded channel.
Results
Because all animals continue to be studied, the microwire-array locations reported here are based on stereotaxis, cortical landmarks observed during surgery, and high-resolution digital RX imagery (in two animals), rather than histological methods.
The mean impedance of the microwires implanted in our monkeys was 1.5 MOhms (measured in saline at 1 KHz). In monkey 1, 96 microwires were implanted in five locations: 16 in area 1 of the right primary somatosensory cortex (S1), 16 in each primary motor cortex (M1), 32 in the right dorsal premotor cortex (PMd), and 16 in the right supplementary motor area (SMA). In monkey 2, 672 microwires were implanted in three different cortical areas in each hemisphere: M1 (96 in each hemisphere), PMd (96 in the left hemisphere, 128 in the right), and posterior parietal cortex (PP, 128 in each hemisphere). In monkey 3, a total of 704 microwires were implanted in 10 cortical locations, distributed in both hemispheres: PMd (96 in the left hemisphere, 64 in the right), M1 (96 in the left, 64 in the right), S1 (64 in the left and 96 in the right hemisphere), PP (32 in the left hemisphere), and extrastriate visual cortex, near the boundary between the parietal and occipital cortex (96 in each hemisphere).
As in New World monkeys (7, 8), the number of units isolated seemed to stabilize 4–6 weeks postsurgery. High-quality single units could be isolated and recorded 30 days after the implantation surgery. Fig. 2 illustrates the entire range, in terms of quality of single-unit isolation, of single units obtained in our recordings. Each example in Fig. 2 contains: color-coded cluster in PC space (left half of each lettered part of the figure), samples of the corresponding waveforms and ISI histograms from clusters that yielded single units (top half of each plate, on the right), a brief sample of continuous recordings from each channel (digital oscilloscope, bottom half on the right), and statistics for cluster separation. Distributions for these statistics are shown in Fig. 3 B′–E′. J3 depicts the ratio of between-cluster to within-cluster scatter; high values signify good isolation. DB reflects the ratio of the sum of within-cluster scatter to between-cluster separation, and low values show good isolation. Labels in Fig. 3 B′, C′, and E′ identify each of the channels shown in Fig. 2 A–H.
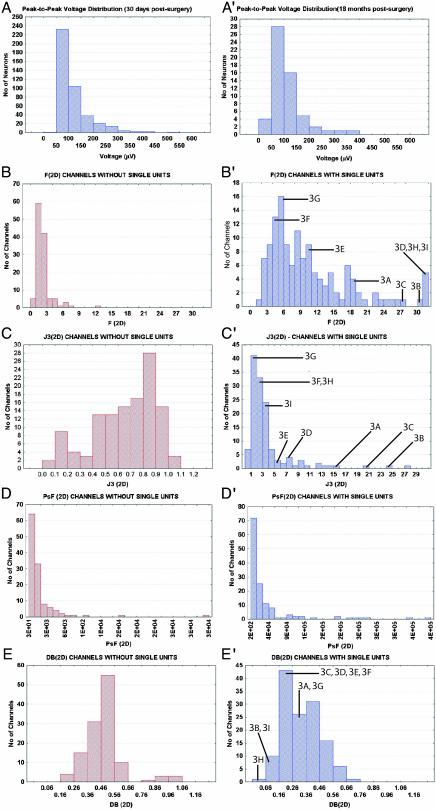
Fig. 3.
(A) Vpp distribution from 330 single neurons recorded in two monkeys, 30 days after the surgical implantation. (A′) Vpp distribution obtained for monkey 1, 18 months postsurgical implantation. Distributions of four cluster separation statistics for channels without isolated single units (B–E) and with single units (B′–E′). (B) Distribution of F (2D) values for recording channels without single units. (B′) Distribution of F (2D) values for channels with single units. The locations in the distribution of the channels are indicated by labels and lines. (C) Distribution of J3 (2D) values for channels without single units. (C′) Distribution of J3 (2D) values for channels with single units. (D) Distribution of PsF (2D) values for channels without single units. (D′) Distribution of PsF (2D) values for channels with single units. (E) Distribution of DB (2D) values for channels without single units. (E′) Distribution of DB (2D) values for channels with single units.
Fig. 2 demonstrates that clear, single-unit isolation from the macaque monkey cortex can be obtained with chronically implanted microwire arrays. Fig. 2 A–G depicts a sample of channels in which at least two well-isolated single neurons were identified, 30 days after the implantation surgery. These examples represent 48% of all channels that yielded single units at this postsurgical time. Fig. 2 A–E depicts channels with high J3 and F statistics and low DB values, indicating the presence of well-separated and defined clusters. High J3 and F combined with low DB values were also observed in channels in which only one single neuron (Fig. 2 H and I) was identified in addition to multiunit activity. Fig. 2F depicts a channel whose cluster separation was near the mean of the J3 distribution. The lower quartile of the J3 and F distributions are illustrated by the example in Fig. 2G. Interestingly, DB values for these two channels were slightly below average (see Fig. 3E′), suggesting a reasonable level of cluster separation. These examples underscore the relevance of considering multiple parameters for assessing cluster separation. Another example of this requirement is illustrated in Fig. 2I. In this channel, a clear single unit (yellow cluster) was separated from multiunit activity (green cluster). Statistical characterization of cluster separation in this channel yielded an average J3 value (3.2). A high F value and a low DB suggested better cluster separation than implied by the J3 statistics.
Fig. 2 A–D displays examples of channels in which clusters depicting single units were identified in the presence of multiunit activity. Inspection of the 3D PC plots and the continuous records in the digital oscilloscope show how single units were clearly separated from multiunit activity (red clusters in Fig. 2A, blue clusters in Fig. 2 B–D, and green clusters in Fig. 2 H and I). Channels like the ones presented in Fig. 2 A–I accounted for ≈80% of our sample. Fig. 2 J displays a representative example of the remaining 20%, channels in which a single unit was identified despite the fact that only one well-defined cluster was present in 3D PC space. Usually this occurred because the voltage threshold was set at a level higher than the background multiunit activity. These records were accepted as single units based on the existence of a well-defined cluster, regular and high-amplitude waveforms, and clear refractory periods in the ISI histograms.
Fig. 3A depicts the Vpp distribution of all single neurons isolated in monkeys 1 and 3, 30 days postsurgery. The mean ± SE value of this distribution (115 ± 3.1 μV) corresponded to a signal-to-noise ratio of ≈5.5:1. Notice that units as large as 200 μV were obtained. Fig. 3 B–E depicts the distributions of F, J3, PsF, and DB statistics calculated for channels in which no single units were isolated (control computations; see Methods) in 2D PC space. The mean (±SE) values for these channels were: F (2D) = 2.3 ± 0.1, J3 (2D) = 0.7 ± 0.02, PsF (2D) = 1,906.5 ± 314.1, and DB (2D) = 0.49 ± 0.01. For channels with single units (Fig. 3 B′–E′) the means were: F (2D) = 11.5 ± 1.0, J3 (2D) = 3.8 ± 0.4, PsF (2D) = 37,364.8 ± 6,026.9, and DB (2D) = 0.32 ± 0.01. For each of these four parameters, the distributions obtained for channels with single units differed significantly from channels without single units (t test for independent samples, P < 1 × 10–7; Mann–Whitney U test, P < 1.0 × 10–7; Kolmogorov-Smirnov two-sample test, P < 0.001).
Further quantification revealed that 56 of 96 implanted microwires (58%) in monkey 1 yielded at least one single neuron 30 days after the surgery. This included 25 of 32 (78%) microwires in right PMd, 13 of 16 (81%) in right M1, 11 of 16 (69%) in right S1, and 7 of 16 (69%) in left M1. The overall mean of neurons isolated per microwire implanted was 0.9 (83 neurons from 96 microwires). Divided by cortical area, we recorded 1.3, 1.4, 0.8, and 0.4 neurons per microwire in right PMd, right M1, right S1, and left M1, respectively. When only the microwires containing at least one single unit were considered (56 microwires), these values changed to 1.5 overall, and by area: PMD = 1.6, right M1 = 1.7, right S1 = 1.2, and left M1 = 1.0.
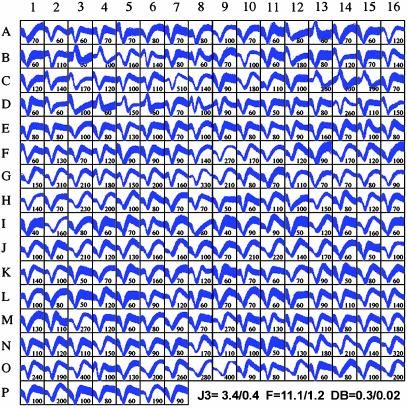
The largest sample of neurons recorded in a single session was obtained in monkey 3. Using a 384-channel MAP cluster, we were able to record the extracellular activity of 247 single cortical neurons from 384 microwires (see Fig. 5B, mean = 0.6 neurons per implanted microwire), 30 days after the implantation surgery (Fig. 4). The mean ± SE Vpp of the waveforms displayed in Fig. 4 was 126 ± 4.2 μV, which corresponded to a mean signal-tonoise ratio of ≈6:1. Single units with Vpp as high as 300–500 μV were present in monkey 3. The mean cluster separation statistics for the recording session depicted in Fig. 4 were: F (2D) = 11.1 ± 1.2, J3 (2D) = 3.4 ± 0.4, PsF (2D) = 35,471 ± 7,401, and DB (2) = 0.34 ± 0.02. Single units were recorded from the left M1 (87 neurons from 96 microwires, A1-F7), left PMd (53 neurons from 96 microwires, F8-H7 and J3-K7), left S1 (27 neurons from 64 microwires, H8-J2), the right S1 (52 from 96 microwires, K8-N11), and the right M1 (28 neurons from 32 microwires, N12-P7). In this recording session, the percentage of microwires yielding at least one single neuron was 35% (135 of 384 microwires). This percentage was higher in the M1 (51%, 65 of 128 microwires) than in the PMd (26%, 25 of 96 microwires) and the S1 (28%, 45 of 160 microwires). The mean of neurons isolated per implanted microwire varied significantly according to cortical area. Again, the M1 mean (0.9 neurons per implanted microwire) was higher than those obtained for PMd (0.6) and S1 (0.5). When only microwires containing single units were considered (135 microwires), the overall mean rose to 1.8 neurons per microwire. When this calculation was broken down according to cortical area, recordings in PMd yielded 2.1 neurons per viable microwire; M1 (1.8) and S1 recordings (1.8) produced slightly smaller values.
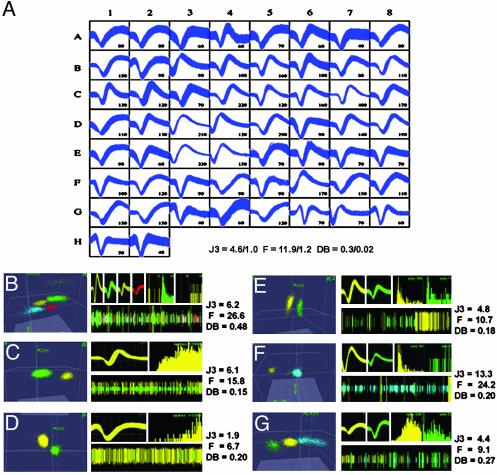
Fig. 5.
(A) Waveform sample from 58 single neurons recorded 18 months after the implantation surgery. Numbers next to waveforms depict the mean Vpp (μV) of the action potentials. Right PMd (A1–B2), right M1 (B3–E6), right S1 (E7–F7), and right SMA (F8–H2). Mean ± SE J3 (2D), F (2D), and DB (2D) statistics for the sample are plotted at the bottom right. (B–G) Single neuron isolation quality is illustrated as described in Fig. 3. (B) Four single units discriminated from the multiunit activity (blue). (C) Clear isolation of a single neuron (yellow) from the multiunit activity (green) is confirmed by high J3 and F values and a low DB. (D) Channel with a clear single unit, but with a smaller signal-to-noise ratio than C. ISI histograms show a clear refractory period. (E) Two clearly distinct single units produced clusters with average J3, close to average F, and low DB. (F) Two units (green and yellow) discriminated from the multiunit activity (blue) yielding high J3, F, and low DB statistics. Distinct ISIs, waveform shapes, and amplitudes further supported their designation as single units. (G) Channel with average J3 and slightly lower than average F and DB statistics. Clear differences in ISI histograms and waveform shape and amplitude confirm nice separation of units (green and yellow) from multiunit activity (blue). ISI histogram time scales are 10 ms (B–F) and 100 ms (G).
Fig. 4.
Samples of action potentials from 247 single neurons recorded in monkey 3, 30 days after implantation. Sample includes left M1 (A1–F7), left PMd (F8–H7 and J3–K7), left S1 (H8–J2), right S1 (K8–N11), and right M1 (N12–P7). Numbers next to the sampled waveforms display their Vpp (μV). Mean ± SE J3 (2D), F (2D), and DB (2D) statistics for the sample are plotted at the bottom right.
By combining results from the three individual recording sessions, we verify that 3–4 weeks after the implantation surgery a total of 421 neurons were recorded from 576 implanted microwires (a mean of 0.7 neurons per implanted microwire) in the three monkeys. On average, 54% of the implanted microwires in the three monkeys yielded at least one isolated cell. Thus, if one considers only the total number of viable microwires in these recording sessions, the overall mean of neurons discriminated per viable microwire was 1.6.
Short- and Long-Term Stability of Single-Unit Isolation. In addition to providing a very reasonable yield, chronic microwire recordings were stable at both short- and long-term time scales (see Figs. 7 and 8, which are published as supporting information on the PNAS web site). Quantitative analysis revealed that PC clustering depicting single-unit isolation remained stable throughout recording sessions that lasted for 1–2 h. This finding was confirmed by the fact that neither the ISI histograms calculated for these single units nor the cluster separation statistics calculated for these channels varied significantly during the recording sessions.
Fig. 5A displays waveform samples from 58 simultaneously recorded neurons in monkey 1, 18 months after the initial implantation surgery. The Vpp distribution (112.6 ± 8.9 μV) for the action potentials of these neurons is presented in Fig. 3A′.No statistical difference was found between this Vpp distribution and that obtained from data collected 30 days postsurgery. Means ± SE of three cluster separation statistics for this recording session are displayed at the bottom of Fig. 5A. The obtained values [F (2D) = 11.9 ± 1.2, J3 (2D) = 4.6 ± 1.0, and DB (2D) = 0.3 ± 0.02] did not differ from those obtained 30 days postsurgery. These 58 neurons were discriminated from 34 viable microwires, which corresponded to 35% of the 96 microwires originally implanted. The number of neurons isolated per implanted microwire in different cortical areas was the following: PMd = 0.3, M1 = 1.8, S1 = 0.8, and SMA = 0.7, total = 0.6. When only the number of viable microwires was considered, the neuronal yield was: PMd = 1.7 neurons per microwire, M1 = 1.9, S1 = 1.3, and SMA = 1.8. The mean (1.7 neurons per viable microwire) was the same as the value obtained for monkeys 1 and 3, 30 days postsurgery. As in recordings obtained 30 days postsurgery (Fig. 3), Fig. 5 B–G demonstrates that 18 months after implantation, recordings from single microwires allowed multiple single units to be clearly separated from background multiunit activity (blue clusters in Fig. 5 B and F–G, and green clusters in Fig. 5 C and D).
Discussion
This article describes a paradigm for obtaining long-term, multielectrode recordings in macaque monkeys. In each of three rhesus monkeys, simultaneous recordings were obtained from 83–247 neurons, distributed across five cortical fields. These neuronal samples, which were each obtained in a single day, are comparable to those obtained with single-electrode methods in weeks of recording. Long-term experiments in one subject revealed that high-quality single-unit recordings could be obtained for 18 months.
Methodology. The principal issue addressed in this study was whether the methods we have previously applied to rodents and New World primates, which have few and shallow sulci, can be applied to macaques, with their larger brains, deeper sulci, greater brain pulsations, and lower cell-packing densities. Our findings suggest that the methods used here are robust and reliable in macaque monkeys. Indeed, the yield of single neurons in the present study exceeds dramatically those reported with previous methods (9–14). In addition, we have shown the capability to record simultaneously from many cortical areas in both hemispheres for many months.
Various factors contributed to the robustness and reliability of the present method. First, high-quality chronic recordings were most reliably obtained (i) when the surgical procedure involves very slow implantation of the microwire arrays and (ii) when single-unit recordings observed during the implantation procedure were of high quality. This slow implantation procedure differs dramatically from that used for the Utah array, for example, which enters the cortex at high speed (9, 10). Another important factor seems to be the configuration of the electrode tip. We found that, like in other species, blunt-tip microwires work well in a variety of cortical areas in macaques.
Not only do the methods described here work well in macaques, in some ways they exceed previous results in other species. For example, the 247 neurons isolated from one subject represent a 2-fold increase in yield compared with our previous studies in rats (15) and New World monkeys (7, 8). This improvement results mainly from the ability to implant a larger number of electrodes in macaque brains. Furthermore, our previous multielectrode recordings in rats have usually involved arrays of 16 microwires. In the present study, we used arrays of 32–128 microwires. Thus, one can isolate neurons from up to 40 mm2 of cortex per implant, which is more than a third of the forelimb representation in the macaque M1 (19).
Single-Unit Sorting and Statistics. The larger number of electrodes and the presence of more than one single unit on many of those electrodes presents some challenges. With traditional neurophysiological methods, subjective judgment is used to decide whether the records consist of single units. The present use of PC analysis is only one among many methods for sorting spikes from multiunit noise and from each other. PC analysis appears to be very useful, when used in conjunction with other criteria. Similarly, the precise statistical methods used to validate single-unit isolation have not yet been standardized in neurophysiology. There are probably alternatives to the four statistics used here, which would work as well or better. Yet, no one statistic seems to provide a unique solution to this problem. Instead, we propose that using several different statistics in conjunction with other criteria for cell isolation (see Methods), showing the distribution of those statistics for channels with and without single units (Fig. 3), and illustrating examples that span the entire range of single neuron isolated in a given study (see Fig. 3) should be used to document the quality of multielectrode recordings.
Potential Applications. This methodology should be of value whenever a large sample size is important for testing a hypothesis. For example, simultaneously recording neuronal populations on the order of hundreds of cells should enable better tests of ideas about the role of precise timing and large-scale neuronal interactions in cortical computations (20–24). The stability of single-unit recording during a single recording session will enable investigators to study a larger set of independent variables. And by inserting cannulae in the arrays, pharmacological methods for inactivation or stimulation (25) could be combined with recordings. Moreover, the present methods should result in an improved ability to study neuronal activity during learning. A modest beginning has been made with single-electrode methods (e.g., ref. 26), but such studies required extraordinarily rapid learning and yielded relatively small neuronal samples. At a minimum, it should be possible to study anything that can be learned within a few days or even weeks.
Finally, the methods described here could have an impact on testing cortical neuroprosthetic devices (8, 13, 27). The main long-term goal behind this research is to allow neurological patients to use spared brain tissue to restore function. To this end, we have trained owl monkeys to use their own cortical activity to control either the movements of a robot arm or a cursor (8). Extension of this experimental design should also allow real-time implementations of computational models that predict the animal's perceptual, motor, or cognitive capabilities from brain activity and allow these predictions to be tested in real time, before an animal's response (28).
Supplementary Material
Acknowledgments
This article is dedicated to Dr. Cesar Timo-Iaria for his >50 years of continuous contributions to neurophysiology. We thank Drs. P. Lange, J. Harer, and R. S. Williams for their support and E. Phelps, L. Oliveira, S. Halkiotis, L. Spence, D. Schmid, and S. Wiebe for outstanding technical assistance. This work was supported by the National Institutes of Health, the National Science Foundation, the Defense Advanced Research Projects Agency, the James S. McDonnell Foundation, and a Duke University instrumentation award (to M.A.L.N.).
Abbreviations: MAP, multichannel acquisition processor; PC, principal component; ISI, interspike interval; PsF, PseudoF; DB, Davies-Bouldin; Vpp, peak-to-peak voltage; S1, primary somatosensory cortex; M1, primary motor cortex; PMd, dorsal premotor cortex; SMA, supplementary motor area.
References
- 1.Evarts, E. V. (1960) Fed. Proc. 19, Suppl. 4, 828–837. [PubMed] [Google Scholar]
- 2.Wilson, M. A. & McNaughton, B. L. (1993) Science 261, 1055–1058. [DOI] [PubMed] [Google Scholar]
- 3.Nicolelis, M. A. L., Lin, R. C. S., Woodward, D. J. & Chapin, J. K. (1993) Nature 361, 533–536. [DOI] [PubMed] [Google Scholar]
- 4.Nicolelis, M. A. L., Lin, R. C., Woodward, D. J. & Chapin, J. K. (1993) Proc. Natl. Acad. Sci. USA 90, 2212–2216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nicolelis, M. A. L., Baccala, L. A., Lin, R. C. S. & Chapin, J. K. (1995) Science 268, 1353–1358. [DOI] [PubMed] [Google Scholar]
- 6.Welsh, J. P., Lang, E. J., Suglhara, I. & Llinas, R. (1995) Nature 374, 453–457. [DOI] [PubMed] [Google Scholar]
- 7.Nicolelis, M. A. L., Ghazanfar, A. A., Stambaugh, C. R., Oliveira, L. M. O., Laubach, M., Chapin, J. K., Nelson, R. J. & Kaas, J. H. (1998) Nat. Neurosci. 1, 621–630. [DOI] [PubMed] [Google Scholar]
- 8.Wessberg, J., Stambaugh, C. R., Kralik, J. D., Beck, P. D., Laubach, M., Chapin, J. K., Kim, J., Biggs, S. J., Srinivasan, M. A. & Nicolelis, M. A. (2000) Nature 408, 361–365. [DOI] [PubMed] [Google Scholar]
- 9.Hatsopoulos, N. G., Ojakangas, C. L., Paninski, L. & Donoghue, J. P. (1998) Proc. Natl. Acad. Sci. USA 95, 15706–15711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Maynard, E. M., Hatsopoulos, N. G., Ojakangas, C. L., Acuna, B. D., Sanes, J. N., Normann, R. A. & Donaghue, J. P. (1999) J. Neurosci. 19, 8083–8093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Isaacs, R. E., Weber, D. J. & Schwartz, A. B. (2000) IEE Trans. Rehab. Engin. 8, 196–198. [DOI] [PubMed] [Google Scholar]
- 12.Serruya, M. D., Hatsopoulos, N. G., Paninski, L., Fellows, M. R. & Donoghue, J. P. (2002) Nature 416, 141–142. [DOI] [PubMed] [Google Scholar]
- 13.Taylor, D. M., Helms-Tillery, S. I. & Schwartz, A. B. (2002) Science 296, 1829–1832. [DOI] [PubMed] [Google Scholar]
- 14.Hoffman, K. L. & McNaughton, B. L. (2002) Science 297, 2070–2073. [DOI] [PubMed] [Google Scholar]
- 15.Nicolelis, M. A. L., Ghazanfar, A. A., Faggin, B. M., Votaw, S. & Oliveira, L. M. O. (1997) Neuron 18, 529–537. [DOI] [PubMed] [Google Scholar]
- 16.Wheeler, B. C. (1999) in Methods for Neural Ensemble Recordings, ed. Nicolelis, M. A. L. (CRC, Boca Raton, FL), pp. 61–77.
- 17.Spath, H. (1980) Cluster Analysis Algorithms (Ellis Horwood, New York).
- 18.Davies, D. L. & Bouldin, D. W. (1979) IEEE Trans. Pattern Anal. Machine Intell. 1, 224–227. [PubMed] [Google Scholar]
- 19.Woolsey, C. N., Settlage, P., Meyer, D. R., Sencer, W., Hamuy, T. P. & Travis, A. M. (1952) Res. Publ. Assoc. Res. Nerv. Ment. Dis. 30, 238–264. [PubMed] [Google Scholar]
- 20.Singer, W. & Gray C. M. (1995) Annu. Rev. Neurosci. 18, 555–586. [DOI] [PubMed] [Google Scholar]
- 21.Vaadia, E., Haalman, I., Abeles, M., Bergman, H., Prut, Y. Slovin, H. & Aertsen, A. (1995) Nature 373, 515–518. [DOI] [PubMed] [Google Scholar]
- 22.Newsome, W. T., Shadlen, M. N., Zohary, E., Britten, K. H. & Movshon, J. A. (1995) in The Cognitive Neurosciences, ed. Gazzaniga, M. S. (MIT Press, Cambridge, MA), pp. 401–414.
- 23.Ahissar, E. & Arieli, A. (2001) Neuron 32, 185–201. [DOI] [PubMed] [Google Scholar]
- 24.Nicolelis, M. A. L. & Fanselow, E. E. (2002) Nat. Neurosci. 5, 517–523. [DOI] [PubMed] [Google Scholar]
- 25.Krupa, D. J., Ghazanfar, A. A. & Nicolelis, M. A. L. (1999) Proc. Natl. Acad. Sci. USA 96, 8200–8205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mitz, A. R., Godschalk, M. & Wise, S. P. (1991) J. Neurosci. 1, 1855–1872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nicolelis, M. A. L. (2001) Nature 409, 403–407. [DOI] [PubMed] [Google Scholar]
- 28.Nicolelis, M. A. L. & Ribeiro, S. (2002) Curr. Opin. Neurobiol. 12, 602–606. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.