Abstract
The Arabidopsis genome project assembled 15 megabases of heterochromatic sequence, facilitating investigations of heterochromatin assembly, maintenance, and structure. In many species, large quantities of methylcytosine decorate heterochromatin; these modifications are typically maintained by methyltransferases that recognize newly replicated hemimethylated DNA. We assessed the extent and patterns of Arabidopsis heterochromatin methylation by amplifying and sequencing genomic DNA treated with bisulfite, which converts cytosine, but not methylcytosine, to uracil. This survey revealed unexpected asymmetries in methylation patterns, with one helix strand often exhibiting higher levels of methylation. We confirmed these observations both by immunoprecipitating methylated DNA strands and by restriction enzyme digestion of amplified, bisulfite-treated DNA. We also developed a primer-extension assay that can monitor the methylation status of an entire chromosome, demonstrating that strand-specific methylation occurs predominantly in the centromeric regions. Conventional models for methylation maintenance do not explain these unusual patterns; instead, new models that allow for strand specificity are required. The abundance of Arabidopsis strand-specific modifications points to their importance, perhaps as epigenetic signals that mark the heterochromatic regions that confer centromere activity.
Keywords: heterochromatin, centromere, methylcytosine, DNA methylation
Animal and plant genomes often contain large quantities of methylcytosine, with genome-wide levels measuring up to 60–90% (1, 2). In euchromatin, DNA methylation is often concentrated in small regions, such as CpG islands, providing epigenetic modifications that help regulate genome imprinting, gene expression, and DNA repair (3–5). In contrast, heterochromatin is more heavily methylated, suggesting a methylation role in reducing recombination rates or forming highly condensed chromosome domains (6–8). Centromeric regions may require methylation to maintain their specialized functions: patients with ICF (immunodeficiency, centromere instability, and facial abnormalities) carry a mutation in the de novo methyltransferase Dnmt3b and undergo chromosome breaks in the heterochromatin adjacent to certain centromeres (9, 10). The methylation status of the centromere region may also be critical for the assembly of binding proteins (CENPs); for example, human cells treated with 5-aza-2′-deoxycytidine alter CENP-B localization (11).
Two distinct processes can modulate DNA methylation: (i) maintenance methylation after DNA replication, and (ii) de novo application of methyl groups in previously unmethylated regions. Cytosines in symmetrical sites (CG or CWG) are modified by methyltransferases that recognize hemimethylated daughter strands soon after replication (12). In contrast, de novo methyltransferases create new methylation patterns; the mechanisms governing the regulation of these enzymes are not clear. In plants, cytosine methylation occurs at symmetrical and nonsymmetrical (CH) sites (13). Arabidopsis genes that play a role in DNA methylation include DDM1, which encodes a member of the SNF2/SWI2 family of chromatin remodeling proteins (7, 14), histone H3 methyltransferase (15, 16), chromatin proteins such as ARGONAUTE4 (17), and enzymes that catalyze cytosine methylation, including MET1 for CG (18), CMT3 for CWG (19), and DRM1 and DRM2 that resemble Dnmt3 (20, 21). ddm1 mutations cause demethylation of repetitive centromere DNA, rDNA, and a heterochromatic “knob” within the chromosome arm (7, 22), and result in hypomethylation of regions that can alter gene activity (23). Antisense expression of MET1 also causes a ddm1-like global demethylation (18), as well as abnormal development, potentially by activating transposons or retroelements (5, 24). As in other organisms, the maintenance of Arabidopsis DNA methylation is important for endogenous gene silencing; mutations in DDM1, MET1, and CMT3 partially activate silenced genes (19, 25–27).
We measured heterochromatin methylation in Arabidopsis, which contains a limited number of heterochromatic regions: two rDNA islands (NORs), 10 telomeres, two repetitive “knobs,” and five centromeres (28). Arabidopsis centromeres have been defined cytologically as constricted regions (22) and genetically as regions that segregate in meiosis without undergoing recombination (29); presumably, only a subset of centromere DNA is essential for centromere activity. The genetically defined centromeres contain a core of 180-bp satellites surrounded by retroelements, transposons, microsatellites, various repeats, and several expressed genes (29). We monitored methylation of short sequences and an entire chromosome, focusing on the abundance and patterns of heterochromatin methylation. Unexpectedly, we discovered strand-specific biases in methylation throughout the centromere region; such biases were not observed in other heterochromatic regions. Maintenance of these patterns requires mechanisms distinct from known methyltransferase activities. The abundance of strand-specific methylation in centromeres raises the possibility that they provide epigenetic signals that contribute to kinetochore formation, sister chromatid cohesion, or spindle attachment.
Materials and Methods
Plant Material. Seeds were sterilized in 70% ethanol (30 s) and bleach (5 min), rinsed in water five times, and grown for 10 days in a 20°C growth chamber on 1/2× Murashige and Skoog basal salt medium with 1% sucrose. Several batches of DNA were prepared, as described below, from seedlings grown under identical conditions, but derived from different lots of parental seed, all of the Columbia ecotype. DNA batches 1, 3, 4, and 5 were prepared from seedlings whose parents were grown in the greenhouse (16 h of light) by using Metromix soil (batches 1 and 3) or a 1:1 mixture of Metromix and vermiculite (batches 4 and 5); DNA batches 2, 6, and 7 were from seedlings whose parents were grown in a growth chamber (20°C, constant light).
Bisulfite Deamination and PCR. Ten micrograms of genomic DNA (batch 1), prepared from seedlings by using hexadecyltrimethylammonium bromide (CTAB) (30), was sheared or digested with restriction enzymes into 1- to 2-Kb fragments, denatured in 0.1 M NaOH (15 min, 20°C), neutralized with ammonium acetate, and ethanol precipitated. Nonmethylated cytosines were deaminated in 1.2 ml of 4M NaHSO3, 500 μM hydroquinone (pH 5.0) at 50°C for 24 h. DNA was purified by gel filtration, incubated in 0.3 M NaOH (10 min, 20°C), and ethanol precipitated. Purified DNA was resuspended in 100 μlofddH2O, and 1 μl was used for strand-specific PCR (ref. 31; Table 3, which is published as supporting information on the PNAS web site, www.pnas.org). One PCR fragment was cloned from each amplification (TOPO kit, Invitrogen), and automated DNA sequencing was performed. To ensure that only bisulfite-reacted DNA was amplified and to avoid biased amplification of methylated strands, primers corresponded to regions without cytosine, were degenerate, or contained thymidine corresponding to a bisulfite-converted cytosine at the 3′ end (Table 3).
Immunoprecipitation. Ten micrograms of MboI-digested DNA (batches 3–7) was melted at 100°C for 10 min, diluted 10-fold with ice-cold 0.01% SDS, 1.1% Triton X-100, 1.2 mM EDTA, 16.7 mM Tris·HCl, 167 mM NaCl (pH 8.1), and incubated 16 h at 4°C with 100 μl of 5-methyl cytosine antibodies (Fitzgerald, Concord, MA). Complexes were collected on protein A agarose, and 10 cycles of strand-specific PCR were performed with a locus-specific primer containing a 5′ non-Arabidopsis tag sequence. Double-stranded DNA was subsequently amplified with a complementary locus-specific primer and a primer corresponding to the tag.
Nick-Translation Detection of Hemimethylation. One microgram of genomic DNA (batches 1 and 2) was incubated with ddNTPs and Klenow (New England Biolabs) for 1 h at 37°C, followed by phenol extraction and ethanol precipitation. Digestion with Sau3A or MboI was performed for 3 h, followed by phenol extraction, ethanol precipitation, and nick translation with [32P]dCTP (32). Hybridization signals were measured with ImageQuant (Molecular Dynamics).
Results
To discern nucleotide methylation patterns in Arabidopsis heterochromatin, we sequenced genomic DNA treated with bisulfite, a method that converts nonmethylated cytosines to uracil (33). For each locus, the upper and lower DNA strands were amplified with unidirectional PCR; primers were designed to ensure efficient amplification of bisulfite-reacted strands. Each locus was amplified independently 10 times, and the products were sequenced (31, 34). The analyzed loci included sequences in genetically defined centromere 2 (CEN2) (genes, pseudogenes, noncoding unique sequences, and satellites) (29), other Arabidopsis centromeres (CEN1–5), noncentromeric heterochromatin (rDNA from NOR2 and the chromosome 4 repetitive knob) (22, 35), and two euchromatic genes (SUPERMAN, K14B15.1, chromosome 3; T28P16.15, chromosome 2) (23, 36). Data from the same batch of bisulfite-treated DNA (batch 1), extracted from thousands of 10-day-old seedlings of the Columbia ecotype, are compiled in Tables 1 and 2.
Table 1. Arabidopsis loci exhibiting significant strand-biased DNA methylation.
| % Methylation (n)
|
|||||||
|---|---|---|---|---|---|---|---|
| Locus | Position (BAC) | Type | CG | CWG | CH | Total | Fold difference |
| 1. CEN2 | 22783-23113 | Gene | 98 ± 5.3 (60) | 94 ± 9.7 (50)** | 75 ± 13 (520)*** | 78 ± 11 (630)*** | 1.9 |
| (T13E11) | 87 ± 23 (60) | 72 ± 19 (50) | 29 ± 32 (520) | 41 ± 28 (630) | |||
| 2. CEN2 | 89483-89824 | Noncoding | 90 ± 17 (40) | 23 ± 25 (40)*** | 14 ± 26 (560)*** | 19 ± 24 (640)*** | 4.1 |
| (T5M2) | 95 ± 11 (40) | 80 ± 16 (40) | 78 ± 8.0 (460) | 79 ± 7.1 (540) | |||
| 3. CEN2 | 13295-13596 | nMito | 83 ± 21 (60) | 58 ± 31 (60)* | 34 ± 35 (390)* | 43 ± 31 (510)* | 1.7 |
| (T5E7) | 97 ± 7.0 (60) | 83 ± 18 (60) | 67 ± 33 (550) | 71 ± 29 (670) | |||
| 4. CEN2 | 73804-74109 | Noncoding | 40 ± 38 (30)*** | 33 ± 28 (60)*** | 13 ± 22 (340)*** | 17 ± 22 (430)*** | 4.7 |
| (T12J2) | 93 ± 14 (30) | 95 ± 8.1 (60) | 80 ± 12 (460) | 83 ± 11 (550) | |||
| 5. CEN2 | 1845-2316 | Satellite | 100 ± 0.0 (70)* | 100 ± 0.0 (30)*** | 89 ± 4.3 (760)*** | 91 ± 3.8 (860)*** | 3.1 |
| (T14C8) | 82 ± 21 (70) | 60 ± 26 (30) | 21 ± 25 (590) | 29 ± 23 (690) | |||
| 6. CEN2 | 11257-11519 | Pseudogene | 0.0 ± 0.0 (130)*** | 1.3 ± 4.0 (80)*** | 0.5 ± 1.1 (380)*** | 0.5 ± 1.1 (590)*** | 136 |
| (T14C8) | 66 ± 28 (130) | 75 ± 26 (80) | 69 ± 26 (310) | 69 ± 25 (520) | |||
| 6a. CEN2 | 11257-11519 | Pseudogene | 9.2 ± 26 (120)* | 3.8 ± 12 (80)*** | 3.7 ± 11 (380)*** | 4.8 ± 14 (580)*** | 11 |
| (T14C8) | 40 ± 33 (120) | 56 ± 33 (80) | 56 ± 32 (310) | 53 ± 31 (510) | |||
| 7. PeriCen2 | 15754-16068 | Noncoding | 11 ± 26 (150)*** | 10 ± 32 (20)*** | 9.3 ± 25 (410)*** | 9.8 ± 25 (580)*** | 8.3 |
| (F12P23) | 74 ± 22 (150) | 90 ± 21 (20) | 82 ± 13 (600) | 81 ± 14 (770) | |||
| 8. CEN3 | 8731-8995 | Gene† | 100 ± 0.0 (20) | 100 ± 0.0 (40)** | 89 ± 7.6 (670)** | 90 ± 7.0 (730)** | 2.1 |
| (F23H6) | 90 ± 21 (20) | 58 ± 39 (40) | 37 ± 42 (290) | 43 ± 39 (350) | |||
| 9. CEN3 | 32058-32329 | Noncoding | 98 ± 5.3 (60)* | 91 ± 18 (70)*** | 80 ± 23 (530)*** | 83 ± 20 (660)*** | 2.8 |
| (T14A11) | 83 ± 19 (60) | 36 ± 39 (70) | 17 ± 32 (290) | 30 ± 28 (420) | |||
| 10. CEN5 | 3689-3857 | Noncoding | 90 ± 32 (30) | 0 (0) | 76 ± 40 (210)* | 78 ± 38 (240)* | 1.9 |
| (T3P1) | 90 ± 23 (30) | 0 (0) | 34 ± 43 (240) | 40 ± 39 (270) | |||
The analyzed DNA sequences (numbered 1-10) are indicated by locus (centromere, CEN; pericentromere, PeriCen), nucleotide position on a BAC clone, and type (nMito, an integrated portion of the mitochondrial genome; satellite, 180 bp repeat) (18, 26). Percent cytosine methylation is reported for each strand (upper and lower rows, respectively) as an average (±SD) from 10 independent clones and is divided into subcategories CG, CWG (W = A or T), or CH (H = A or T or C) and summed (total); 6a shows amplification of locus as 6 with a different batch of DNA (roots) and primers that selectively amplify bisulfite-converted strands; n is the number of occurrences of each cytosine in a given context in the sequenced region; fold difference is the ratio of average methylation on the upper and lower strands. Bold indicates statistically significant differences between strands (T test, P < 0.05); T test values of greater significance are indicated by *, P = 0.05-0.01; **, P = 0.009-0.001; ***, P < 0.001. †, Retroelement polyprotein. In the strands that showed significant methylation biases, 37% of the cytosines (n = 476) either were converted to uracil or had a neighboring cytosine that was converted, indicating overall accessibility; only 2.5% of the strands failed to react.
Table 2. Arabidopsis loci exhibiting no significant strand-biased DNA methylation.
| % Methylation (n)
|
|||||||
|---|---|---|---|---|---|---|---|
| Locus | Position (BAC) | Type | CG | CWG | CH | Total | Fold difference |
| 11. PeriCen2 | 126822-127152 | Gene | 49 ± 36 (70) | 51 ± 46 (80) | 44 ± 38 (440) | 45 ± 39 (590) | 1.4 |
| (T4E14) | 44 ± 42 (70) | 40 ± 39 (80) | 30 ± 28 (440) | 33 ± 30 (590) | |||
| 12. CEN2 | 69066-69495 | nMito | 98 ± 3.7 (130) | 92 ± 6.7 (110) | 67 ± 24 (610) | 75 ± 17 (850) | 1.1 |
| (T5M2) | 95 ± 5.4 (130) | 92 ± 12 (110) | 80 ± 16 (600) | 84 ± 12 (840) | |||
| 13. CEN2 | 50321-50723 | Gene | 43 ± 40 (170)* | 48 ± 47 (110) | 46 ± 43 (440) | 46 ± 42 (720) | 1.7 |
| (F7B19) | 76 ± 20 (170) | 76 ± 21 (110) | 75 ± 16 (670) | 76 ± 16 (950) | |||
| 14. CEN1 | 1936-2230 | Noncoding | 98 ± 5.7 (170) | 86 ± 21 (100) | 60 ± 42 (410) | 73 ± 29 (680) | 1.4 |
| (F9D18) | 84 ± 25 (170) | 68 ± 19 (100) | 36 ± 32 (370) | 54 ± 23 (320) | |||
| 15. CEN4 | 50400-50959 | Noncoding | 98 ± 3.0 (240) | 100 ± 0.0 (80)* | 87 ± 9.7 (810) | 90 ± 7.0 (1,130) | 1.0 |
| (F2112) | 99 ± 1.8 (240) | 89 ± 15 (80) | 88 ± 7.2 (1,090) | 90 ± 6.0 (1,410) | |||
| 16. CEN4 | 1836-2289 | Noncoding | 90 ± 23 (30) | 0 (0) | 41 ± 35 (370) | 44 ± 33 (400) | 1.3 |
| (F14G16) | 97 ± 11 (30) | 0 (0) | 55 ± 47 (300) | 59 ± 43 (330) | |||
| 17. PeriCen4 | 64864-65362 | Knob | 92 ± 14 (120) | 48 ± 20 (80)** | 36 ± 34 (560) | 46 ± 27 (760) | 1.4 |
| (T5H22) | 96 ± 5.9 (120) | 74 ± 20 (80) | 62 ± 30 (790) | 67 ± 25 (990) | |||
| 18. NOR2 | 2421-2999 | rDNA | 93 ± 10 (400) | 94 ± 5.6 (120) | 72 ± 17 (810) | 80 ± 12 (1,330) | 1.0 |
| (F23H14) | 91 ± 13 (400) | 93 ± 6.6 (120) | 80 ± 11 (980) | 84 ± 11 (1,500) | |||
| 19. EuChr3 | 1294-1505 | Gene | 0.0 ± 0.0 (20) | 0.0 ± 0.0 (90) | 0.0 ± 0.0 (360) | 0.0 ± 0.0 (470) | NA |
| (K14B15) | 0.0 ± 0.0 (20) | 0.0 ± 0.0 (90) | 0.0 ± 0.0 (360) | 0.0 ± 0.0 (470) | |||
| 20. EuChr2 | 43012-43357 | Gene | 0.0 ± 0.0 (60) | 0.0 ± 0.0 (40) | 0.5 ± 1.0 (430) | 0.4 ± 0.8 (530) | 4.8 |
| (T28P16) | 0.0 ± 0.0 (60) | 0.0 ± 0.0 (40) | 2.1 ± 5.5 (520) | 1.8 ± 4.6 (620) | |||
The analyzed DNA sequences (numbered 11-20) are indicated by locus (centromere, CEN; pericentromere, PeriCen; euchromatin, EuChr; nucleolar organizing region, NOR), nucleotide position on a BAC or P1 clone, and type (nMito, an integrated portion of the mitochondrial genome; satellite, 180 bp repeat; knob, a heterochromatic repeat) (18, 26). Percent cytosine methylation is reported for each strand (upper and lower rows, respectively) as an average (±SD) from 10 independent clones and is divided into sub-categories CG, CWG (W = A or T), or CH (H = A or T or C) and summed (total); n is the number of occurrences of each cytosine in a given context in the sequenced region; fold difference is the ratio of average methylation on the upper and lower strands. Bold indicates statistically significant differences between strands (T test, P < 0.05); T test values of greater significance are indicated by *, P = 0.05-0.01; **, P = 0.009-0.001. NA, not applicable. In the strands that showed significant methylation biases, 37% of the cytosines (n = 476) were either converted to uracil or had a neighboring cytosine that was converted, indicating overall accessibility; only 2.5% of the strands failed to react.
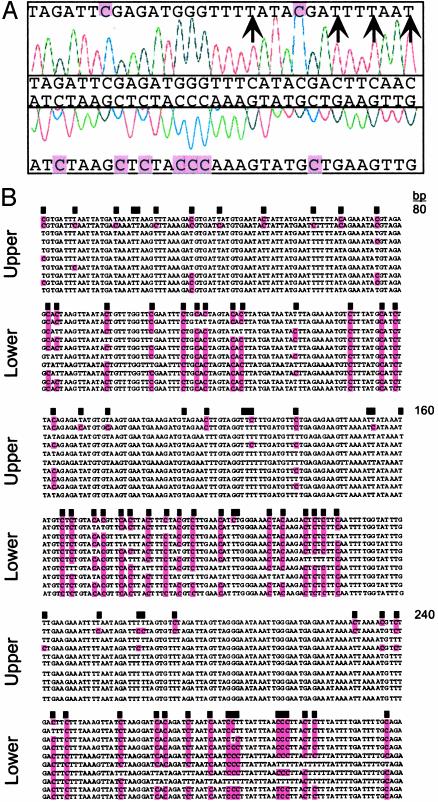
Strand-Specific Biases in DNA Methylation. Amplification of a bisulfite-treated CEN5 region showed methylation of cytosines only in a symmetrical context on the upper strand, whereas every cytosine in the lower strand was methylated (Fig. 1A). A similar disparity was observed in independent clones from CEN2: an average of 81% cytosine methylation was found on the lower strand and 17% on the upper strand (Fig. 1B). Because these products were amplified from the same DNA sample, the methylation differences reflect the average status of the cell population.
Fig. 1.
Strand-specific methylation of centromeric DNA sequences. (A) Sequence chromatogram from the CEN5 BAC T3P1, nucleotides 3690–3721 (25). The unmodified sequence (middle) is compared with sequences generated by bisulfite treatment (top and bottom); unmethylated cytosines (arrows) and cytosines protected by methylation (purple shading) are indicated. (B) Independent sequences (upper and lower strands) of a 240-bp CEN2 fragment amplified from bisulfite-treated DNA (26). Black boxes indicate the position of cytosines in the original sequence.
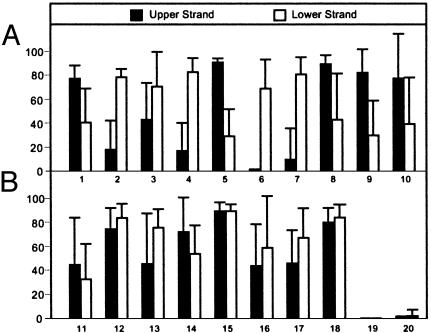
We expanded this survey to several loci (Tables 1 and 2). A t test analysis revealed highly significant differences between upper and lower strands (Table 1, Fig. 2A). Within the centromere regions, highly significant strand biases in CWG and CH methylation were detected, whereas CpG methylation was sometimes distributed on both strands (Table 1). These biases occurred in many CEN loci, whether coding or noncoding, including a recently inserted mitochondrial DNA sequence within CEN2 (29, 36). Similar strand-specific biases were found in the Ws and Columbia ecotypes at a range of developmental stages, although the extent of methylation at individual loci varied (not shown). Biases in DNA strand methylation were observed previously in the Dc8 gene of carrot (37) and in a human retrotransposon promoter (38). Because biased methylation occurs throughout Arabidopsis centromeres, it is likely that the location or context of the sequences, and not the sequences themselves, trigger strand-specific patterns.
Fig. 2.
Variation in methylation content. Average percent methylcytosine for the sequences reported in Tables 1 and 2. Sequences showing either significant (A) or insignificant (B) differences between upper and lower strands are indicated; error bars are SD; sequence numbers correspond to those of Tables 1 and 2.
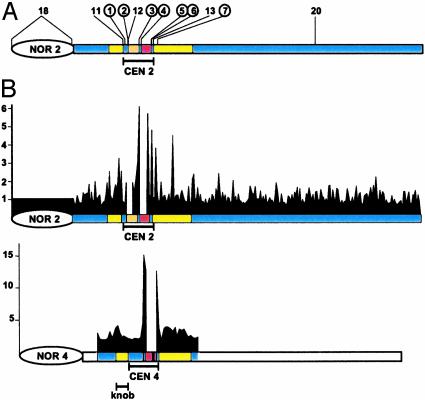
Strand-Biased Methylation Is Restricted to Centromeric Heterochromatin. To explore whether the centromere methylation patterns were a consequence of their heterochromatic states, we examined the 18S-25S rDNA intergenic spacer in NOR2 and the knob on chromosome 4 (22, 35). These sequences did not show significant differences (t test, P > 0.05) in complementary DNA strand methylation, (rDNA, 80 and 84%, respectively; knob, 46% and 67%, respectively) (Table 2 and Fig. 2B). Similarly, both DNA strands carried substantial levels of methylation at centromeric and pericentromeric loci (Table 2 and Fig. 2B). As expected, euchromatin contained little methylation. Overlaying these data on the Arabidopsis physical and genetic maps showed high levels of methylation in all heterochromatic regions, with the genetically defined centromere and the nearby pericentromeric regions (29) uniquely showing strong strand asymmetries (Fig. 3A). These observations indicate that heterochromatin is not a sufficient signal for biased strand methylation and implicate strand asymmetries as unique features of the centromere region.
Fig. 3.
Strand-biased methylation across chromosomes 2 and 4. Shown are scale drawings of chromosome 2 and a portion of chromosome 4, depicting rDNA (NOR2,4), the genetically defined centromeres (CEN2,4), pericentromeric regions including the chromosome 4 knob (yellow), a mitochondrial insertion in CEN2 (orange), and the 180-bp repeat array (red) (25). (A) Circles, regions with significantly different methylation levels between the two complementary strands. (B) Abundance of hemimethylated Sau3A sites along 255 BAC and P1 clones spanning chromosome 2 and 43 BAC clones surrounding CEN4; gaps correspond to unsequenced portions of the chromosome (25, 33). Relative signals from nick translation products after a Sau3A and MboI digests are plotted on the vertical axis.
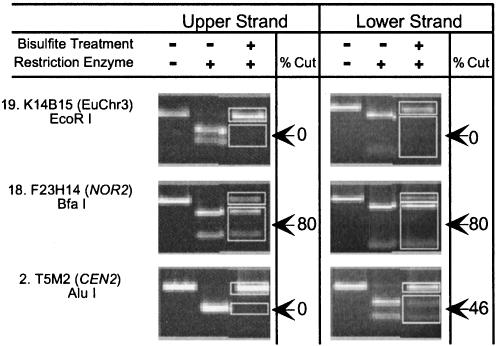
A Restriction Enzyme Assay for Strand-Specific Methylation. To alleviate the time consuming, labor intensive, and costly requirements of PCR product cloning and sequencing, we used an alternative approach: (i) genomic DNA (batch 1) was treated with bisulfite, (ii) loci were amplified with strand-specific PCR, and (iii) the amplified fragments were digested with restriction enzymes that contain cytosine in their recognition sites. In unmethylated regions, bisulfite treatment destroys the restriction site; for example, bisulfite treatment completely disrupted the EcoRI (GAATTC) site within the euchromatic SUPERMAN locus, eliminating the ability to digest the PCR products (Fig. 4). Further, because methylation protects cytosines from bisulfite, the extent of digestion can be used to assess the degree of methylation. For example, we found ≈80% of the NOR2 PCR products amplified from bisulfite-treated DNA could be digested with Bfa I (CTAG), regardless of whether the product was derived from the upper or lower strand (Fig. 4). We used this assay to assess relative methylation levels at an AluI site (AGCT) on two complementary DNA strands within CEN2, finding 46% methylation on the lower strand and no detectable methylation on the upper strand (Fig. 4). Similar results were obtained for a T14C8 (CEN2) fragment (not shown).
Fig. 4.
Restriction digestion assay for strand-specific methylation. PCR products amplified from selected regions (Tables 1 and 2) of bisulfite-treated DNA are numbered, and the extent of digestion with the indicated enzyme was measured with NIH image software. Primers survey the same restriction site on the upper and lower strands; differences in primer location sometimes resulted in different product lengths.
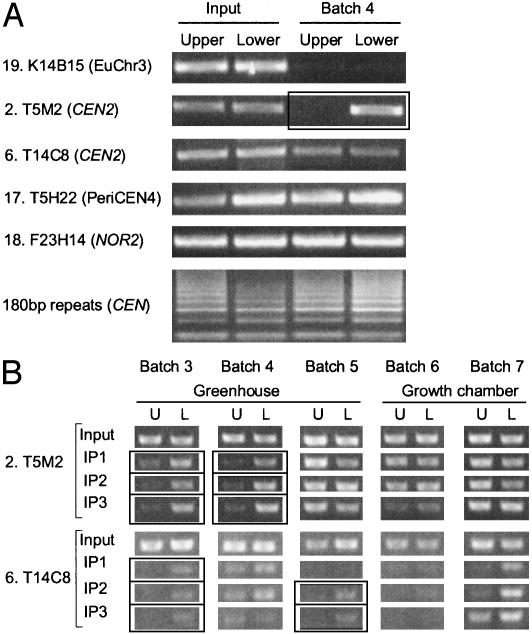
An Immunoprecipitation Assay for Strand-Specific Methylation. We developed a bisulfite-independent method for detecting strand-specific methylation, melting genomic DNA fragments and using 5-methyl cytosine antibodies to purify methylated strands (Fig. 5A). The abundance of purified fragments was assessed with strand-specific PCR amplification. Amplified products corresponding to the euchromatic SUPERMAN locus were not recovered, consistent with its low methylation content. In contrast, we efficiently purified DNA from both strands of NOR2, the chromosome 4 knob, and the 180-bp satellites; in the latter case, complex amplification patterns likely reflect purified tandem copies of satellite DNA (Fig. 5A). Consistent with the data in Tables 1 and 2, one DNA strand was preferentially precipitated from centromeric loci. Results for a given batch of seedling DNA were typically reproducible, but strand-specific methylation patterns varied when seedling batches were compared (Fig. 5B). For example, strand-specific methylation was detected at T5M2 (CEN2) in seedling batches 3 and 4, and T14C8 (CEN2) in batches 3 and 5, indicating that it is not always present at a particular locus (Fig. 5B). Nonetheless, even in samples with modest levels of strand-specific methylation, such modifications could be detected, provided sufficient numbers of centromere DNA loci were surveyed (Fig. 3B and data not shown). Interestingly, strand-specific modifications were most common in seedlings derived from seeds produced in a greenhouse, but not in a growth chamber, suggesting that its distribution is established early in development and is dictated by environmental cues.
Fig. 5.
Immunoprecipitation assay for strand-specific methylation. Genomic DNA was melted and immunoprecipitated with α-5-methylcytosine antibodies; strand-specific PCR-amplified selected regions (Table 1 and 2) from independent batches of DNA as indicated; amplification of input DNA verified primer quality. U, upper strand; L, lower strand; boxes indicate strands with at least a 5-fold difference in methylation levels. In B, three independent immunoprecipitations (IP1-IP3) were performed, demonstrating reproducibility within a given sample.
Chromosome-Wide Analysis of Strand-Biased DNA Methylation. To expand the analysis of strand biases in cytosine methylation to an entire chromosome, we took advantage of the ability of Sau3A to preferentially nick the unmethylated strand of hemimethylated GATC sites (39). Before digestion with Sau3A, nicks that occurred naturally or during DNA purification were blocked by incubation with Klenow and dideoxynucleotide triphosphates. Subsequently, genomic DNA was digested with either Sau3A or its methylation-insensitive isoschizomer, MboI, and E. coli DNA polymerase I was used to produce 32P-labeled probes by nick translation. The portion of the genome corresponding to the nick translation products was determined by hybridization to an ordered BAC and P1 clone array from chromosomes 2 and 4 (Fig. 3B) (36). Because MboI produces double strand breaks at methylated or hemimethylated sites, the nick-translation products formed after MboI digestion provided a normalization standard for the Sau3A digests. Thus, the ratio of signal after Sau3A and MboI cleavage and nick translation can detect hemimethylated sites throughout the genome. By using this method, nick translation fragments from the chromosome arm, from NOR2, and from the heterochromatic chromosome 4 knob were detected in equal abundance whereas nick translation fragments were generated at a much higher frequency from Sau3A digested DNA in the vicinity of the centromere (Fig. 3B); two repetitions of this experiment gave similar patterns (not shown). These results strongly suggest that strand-specific DNA methylation is a unique feature of the centromeric region.
Discussion
We used four methods to demonstrate strand-biased DNA methylation of heterochromatic Arabidopsis centromeres. Unlike the hemimethylation that occurs when methylated DNA is replicated, the patterns reported here are characterized by nearly complete modification of one strand and limited modification of its complement. Such biases have not been widely observed, perhaps because the most common methylation assay (genomic DNA digestion with a methylation-sensitive enzyme followed by Southern blot analysis) cannot unambiguously detect hemimethylation. Nonetheless, reports of strand-specific methylation in carrot and human DNA suggest the presence of these patterns in many genomes (37, 38).
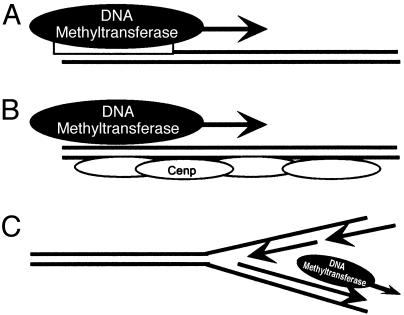
Arabidopsis enzymes that methylate cytosine have been identified (40), yet methyltransferases capable of biased modification of complementary strands are unknown. Such enzymes must transfer methyl groups to only one strand of the helix in a pattern that extends over hundreds of bases. Mechanistically, this may involve: (i) specific binding of de novo methyltransferases that processively modify one strand; (ii) assembly of centromere binding proteins that limit methyltransferase access to one strand of newly replicated DNA; or (iii) differential access of methyltransferases to the leading or lagging strand during DNA synthesis (Fig. 6). Mutants that alter strand-specific methylation would be valuable in discriminating among these models, as would the identification of proteins that bind to hemimethylated DNA. Further, whereas our initial studies surveyed regions <700 bp, determining the size of methylation tracts may reveal boundary sequences that regulate biased methylation. Because the extent of strand-specific methylation at a locus can vary, these boundaries may be influenced by developmental or environmental cues. Finally, interactions between replication complexes and methyltransferases may be clarified by identifying replication origins, an undertaking that has not been launched in Arabidopsis.
Fig. 6.
Models for generating strand-biased DNA methylation. Strand-specific biases in methylation could be generated by DNA methyltransferase binding at specific sites (A), interference with DNA methyltransferase activity by centromere binding proteins (Cenp) (B), or closely coordinating DNA methyltransferase activity with leading or lagging strand synthesis (C).
The abundance of strand-biased methylation in centromeric heterochromatin suggests a role in centromere function, perhaps to mark regions that provide centromere activity. Alternatively, strand biases in methylation could modulate the expression of the many genes residing in the heterochromatic Arabidopsis centromeres (29). Interestingly, Activator/Dissociation (Ac/Ds) transposons exhibit “chromatid selectivity,” in which transposition is preferentially launched from a nonmethylated DNA strand (41). Thus, large tracts of strand-specific methylation in the Arabidopsis centromeres could serve to regulate transposition from individual strands.
In response to developmental and environmental changes, the distribution of DNA methylation in many eukaryotic systems can vary. Genome-wide reprogramming of DNA methylation is important in the early stages of mouse embryogenesis (42). Environmental factors can also affect gene expression by altering DNA methylation in animal cells (43). In the plant Bryonia dioica, mechanical stress causes DNA methylation levels to drop from 25% to nearly 0% in <1 h (44). When maize seedlings are exposed to cold stress, methylation decreases by >10% genome-wide (45), and cultured maize cells have large variations in DNA methylation levels (46). In this study, DNA from different batches of seedlings showed considerable variation in the extent of total and strand-specific methylation. Because the seedlings themselves were grown under identical conditions, this variation was induced during the formation of the seeds, perhaps as a result of different environmental conditions. Seeds produced in a more stressful greenhouse setting produced seedlings with enhanced strand-specific methylation; such patterns are likely established early in development and heritable over several days of seedling growth. Despite the variation in the extent and location of strand-specific methylation, these modifications could always be detected when several centromeric loci were analyzed (see for example, Fig. 3B).
In contrast to Saccharomyces cerevisiae centromeres, where 125 bp are sufficient for centromere function, the role of centromere DNA sequences in higher organisms has remained controversial. The abundance of repetitive centromere sequences in many organisms suggests that satellites are important for function. However, the ability of nonrepetitive sequences from chromosome arms to form neocentromeres raises the possibility that higher order structures instead confer centromere activity (47, 48). Other studies have implicated a role for DNA replication in specifying centromere identity. For example, portions of the Drosophila centromere replicate earlier than other heterochromatin (49, 50). Hemimethylated DNA marks newly replicated DNA sequences and might inhibit replication initiation (51); our observations raise the possibility that strand-biased methylation alters the replication timing or other functions of centromeric loci. Methylcytosine is abundant in the genomes of mammals and plants; determining whether biases in strand methylation occur specifically in the centromeres of other species will clarify its role.
Supplementary Material
Acknowledgments
We thank members of the Preuss laboratory and L. Mets for helpful comments, W. Buikema for DNA sequencing, J. Bender for technical advice, and the Arabidopsis Biological Resource Center for clones. This work was supported by the National Science Foundation (Grant 9872641), the Howard Hughes Medical Institute, and the David and Lucile Packard Foundation.
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Jones, P. L. & Wolffe, A. P. (1999) Semin. Cancer Biol. 9, 339-347. [DOI] [PubMed] [Google Scholar]
- 2.Gruenbaum, Y., Stein, R., Cedar, H. & Razin, A. (1981) FEBS Lett. 124, 67-71. [DOI] [PubMed] [Google Scholar]
- 3.Ng, H.-H. & Bird, A. P. (1999) Curr. Opin. Genet. Dev. 9, 158-163. [DOI] [PubMed] [Google Scholar]
- 4.Robertson, K. D. & Jones, P. A. (2000) Carcinogenesis 21, 461-467. [DOI] [PubMed] [Google Scholar]
- 5.Singer, T., Yordan, C. & Martienssen, R. A. (2001) Genes Dev. 15, 591-602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bender, J. (1998) Trends Biochem. Sci. 23, 252-256. [DOI] [PubMed] [Google Scholar]
- 7.Vongs, A., Kakutani, T., Martienssen, R. A. & Richards, E. J. (1993) Science 260, 1926-1928. [DOI] [PubMed] [Google Scholar]
- 8.Yoder, J. A., Walsh, C. P. & Bestor, T. H. (1997) Trends Genet. 13, 335-340. [DOI] [PubMed] [Google Scholar]
- 9.Hansen, R. S., Wijmenga, C., Luo, P., Stanek, A. M., Canfield, T. K., Weemaes, C. M. & Gartler, S. M. (1999) Proc. Natl. Acad. Sci. USA 96, 14412-14417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xu, G. L., Bestor, T. H., Bourc'his, D., Hsieh, C. L., Tommerup, N., Bugge, M., Hulten, M., Qu, X., Russo, J. J. & Viegas-Pequignot, E. (1999) Nature 402, 187-191. [DOI] [PubMed] [Google Scholar]
- 11.Mitchell, A. R., Jeppesen, P., Nicol, L., Morrison, H. & Kipling, D. (1996) J. Cell Sci. 109, 2199-2206. [DOI] [PubMed] [Google Scholar]
- 12.Riggs, A. D. (1989) Cell Biophys. 15, 1-13. [DOI] [PubMed] [Google Scholar]
- 13.Finnegan, E. J., Genger, R. K., Peacock, W. J. & Dennis, E. S. (1998) Annu. Rev. Plant Physiol. Plant Mol. Biol. 49, 223-247. [DOI] [PubMed] [Google Scholar]
- 14.Jeddeloh, J. A., Stokes, T. L. & Richards, E. J. (1999) Nat. Genet. 22, 94-97. [DOI] [PubMed] [Google Scholar]
- 15.Jackson, J. P., Lindroth, A. M., Cao, X. & Jacobsen, S. E. (2002) Nature 416, 556-560. [DOI] [PubMed] [Google Scholar]
- 16.Malagnac, F., Bartee, L. & Bender, J. (2002) EMBO J. 21, 6842-6852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zilberman, D., Cao, X. & Jacobsen, S. E. (2003) Science 299, 716-719. [DOI] [PubMed] [Google Scholar]
- 18.Finnegan, E. J., Peacock, W. J. & Dennis, E. S. (1996) Proc. Natl. Acad. Sci. USA 93, 8449-8454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lindroth, A. M., Cao, X., Jackson, J. P., Zilberman, D., McCallum, C. M., Henikoff, S. & Jacobsen, S. E. (2001) Science 292, 2077-2080. [DOI] [PubMed] [Google Scholar]
- 20.Cao, X., Springer, N. M., Muszynski, M. G., Phillips, R. L., Kaeppler, S. & Jacobsen, S. E. (2000) Proc. Natl. Acad. Sci. USA 97, 4979-4984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cao, X. & Jacobsen, S. E. (2002) Curr. Biol. 12, 1138-1144. [DOI] [PubMed] [Google Scholar]
- 22.Fransz, P. F., Armstrong, S., de Jong, J. H., Parnell, L. D., van Drunen, C., Dean, C., Zabel, P., Bisseling, T. & Jones, G. H. (2000) Cell 100, 367-376. [DOI] [PubMed] [Google Scholar]
- 23.Jacobsen, S. E. & Meyerowitz, E. M. (1997) Science 277, 1100-1103. [DOI] [PubMed] [Google Scholar]
- 24.Miura, A., Yonebayashi, S., Watanabe, K., Toyama, T., Shimada, H. & Kakutani, T. (2001) Nature 411, 212-214. [DOI] [PubMed] [Google Scholar]
- 25.Jeddeloh, J. A., Bender, J. & Richards, E. J. (1998) Genes Dev. 12, 1714-1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bartee, L. & Bender, J. (2001) Nucleic Acids Res. 29, 2127-2134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bartee, L., Malagnac, F. & Bender, J. (2001) Genes Dev. 15, 1753-1758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.The Arabidopsis Genome Initiative (2000) Nature 408, 796-815. [DOI] [PubMed] [Google Scholar]
- 29.Copenhaver, G. P., Nickel, K., Kuromori, T., Benito, M. I., Kaul, S., Lin, X., Bevan, M., Murphy, G., Harris, B., Parnell, L. D., McCombie, W. R., Martienssen, R. A., Marra, M. & Preuss, D. (1999) Science 286, 2468-2474. [DOI] [PubMed] [Google Scholar]
- 30.Gelvin, S. B., Schilperoort, R. A. & Verma, D. P. (1991) Plant Molecular Biology Manual (Kluwer, Dordrecht, The Netherlands), Vol. 1, A6: 1-10. [Google Scholar]
- 31.Nagane, Y., Utsugisawa, K. & Tohgi, H. (2000) Brain Res. Protoc. 5, 167-171. [DOI] [PubMed] [Google Scholar]
- 32.Sambrook, J. & Russell, D. W. (2001) Molecular Cloning (Cold Spring Harbor Lab. Press, Plainview, NY), 3rd Ed.
- 33.Frommer, M., McDonald, L. E., Millar, D. S., Collis, C. M., Watt, F., Grigg, G. W., Molloy, P. L. & Paul, C. L. (1992) Proc. Natl. Acad. Sci. USA 89, 1827-1831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Luff, B., Pawlowski, L. & Bender, J. (1999) Mol. Cell 3, 505-511. [DOI] [PubMed] [Google Scholar]
- 35.Copenhaver, G. P. & Pikaard, C. S. (1996) Plant J. 9, 259-272. [DOI] [PubMed] [Google Scholar]
- 36.Lin, X., Kaul, S., Rounsley, S., Shea, T. P., Benito, M. I., Town, C. D., Fujii, C. Y., Mason, T., Bowman, C. L., Barnstead, M., et al. (1999) Nature 402, 761-768. [DOI] [PubMed] [Google Scholar]
- 37.Zhou, Y., Magill, C. W., Magill, J. M. & Newton, R. J. (1998) Genome 41, 23-33. [DOI] [PubMed] [Google Scholar]
- 38.Woodcock, D. M., Lawler, C. B., Linsenmeyer, M. E., Doherty, J. P. & Warren, W. D. (1997) J. Biol. Chem. 272, 7810-7816. [DOI] [PubMed] [Google Scholar]
- 39.Streeck, R. E. (1980) Gene 12, 267-275. [DOI] [PubMed] [Google Scholar]
- 40.Genger, R. K., Kovac, K. A., Dennis, E. S., Peacock, W. J. & Finnegan, E. J. (1999) Plant Mol. Biol. 41, 269-278. [DOI] [PubMed] [Google Scholar]
- 41.Ros, F. & Kunze, R. (2001) Genetics 157, 1723-1733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Reik, W., Dean, W. & Walter, J. (2001) Science 293, 1089-1093. [DOI] [PubMed] [Google Scholar]
- 43.Minamoto, T., Mai, M. & Ronai, Z. (1999) Carcinogenesis 20, 519-527. [DOI] [PubMed] [Google Scholar]
- 44.Galaud, J., Gaspar, T. & Boyer, N. (1993) Physiol. Plant 87, 25-30. [Google Scholar]
- 45.Steward, N., Ito, M., Yamaguchi, Y., Koizumi, N. & Sano, H. (2002) J. Biol. Chem. 277, 37741-37746. [DOI] [PubMed] [Google Scholar]
- 46.Kaeppler, S. M. & Phillips, R. L. (1993) Proc. Natl. Acad. Sci. USA, 90, 8773-8776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Choo, K. H. (2000) Trends Cell Biol. 10, 182-188. [DOI] [PubMed] [Google Scholar]
- 48.Willard, H. F. (2001) Proc. Natl. Acad. Sci. USA 98, 5374-5376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ahmad, K. & Henikoff, S. (2001) J. Cell Biol. 153, 101-110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sullivan, B. & Karpen, G. (2001) J. Cell Biol. 154, 683-690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Taylor, J. H. (1984) in DNA Methylation and Cellular Differentiation (Springer, New York), p. 109.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.