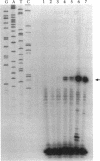
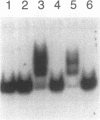
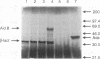
Abstract
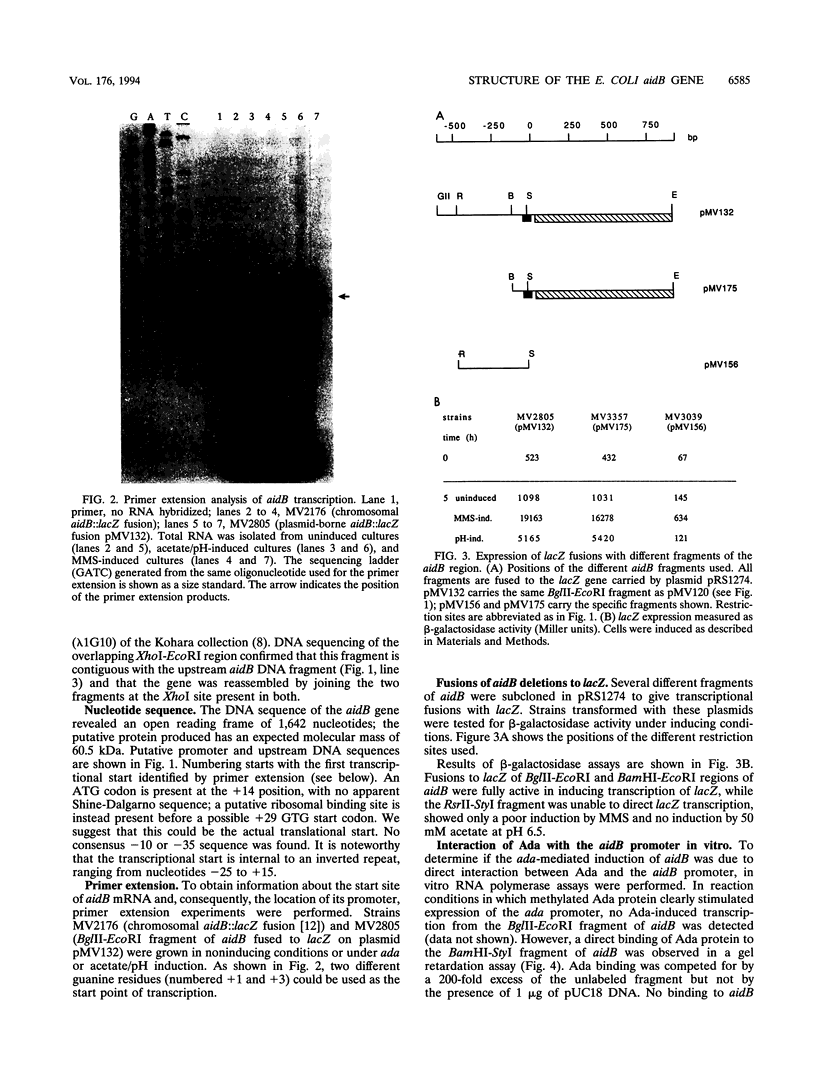
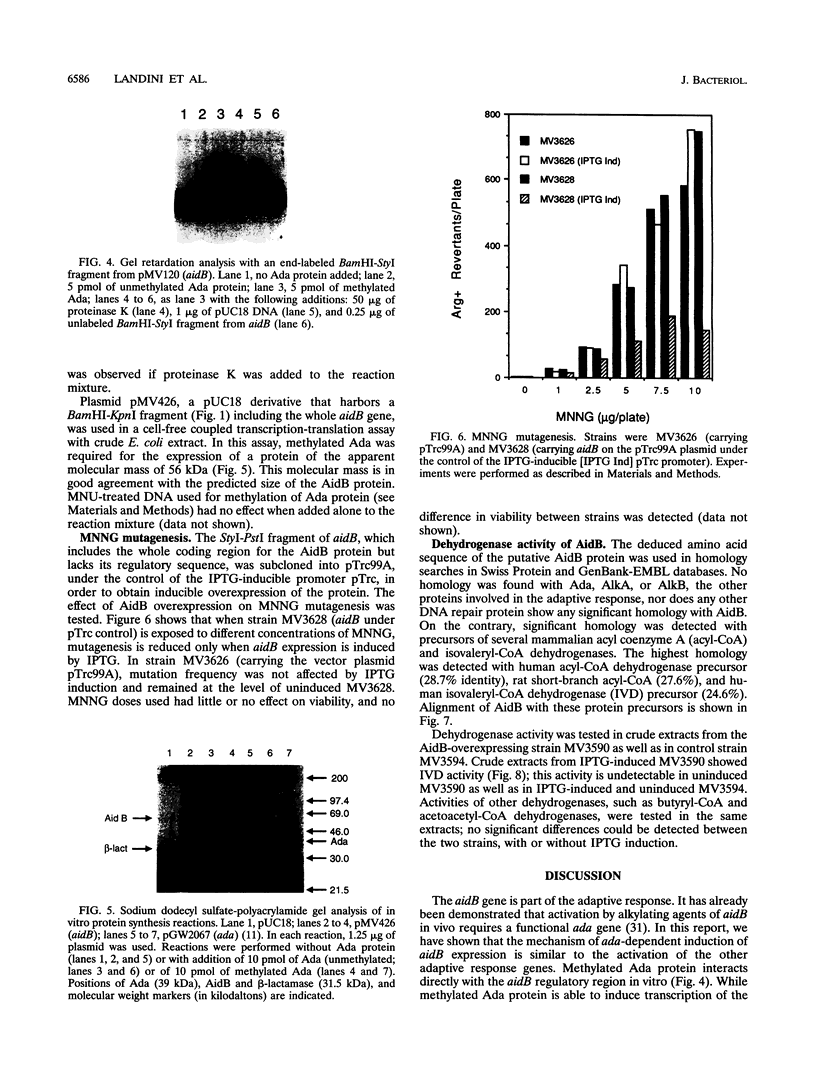
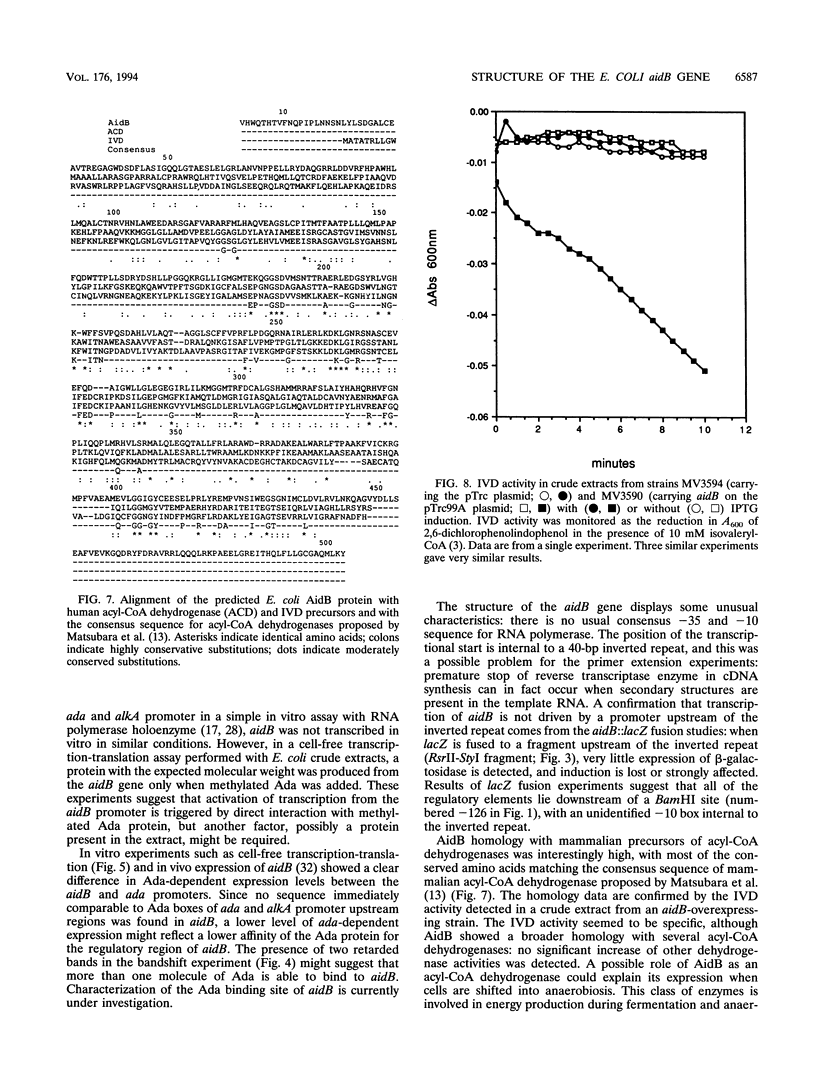
Expression of the Escherichia coli aidB gene is induced in vivo by alkylation damage in an ada-dependent pathway and by anaerobiosis or by acetate at pH 6.5 in an ada-independent fashion. In this report, we present data on aidB gene structure, function, and regulation. The aidB gene encodes a protein of ca. 60 kDa that is homologous to several mammalian acyl coenzyme A dehydrogenases. Accordingly, crude extracts from an aidB-overexpressing strain showed isovaleryl coenzyme A dehydrogenase activity. aidB overexpression also reduced N-methyl-N'-nitro-N-nitrosoguanidine-induced mutagenesis. Both ada- and acetate/pH-dependent induction of aidB are regulated at the transcriptional level, and the same transcriptional start point is used for both kinds of induction. Ada protein plays a direct role in aidB regulation: methylated Ada is able to bind to the aidB promoter region and to activate transcription from aidB in an in vitro transcription-translation system using crude E. coli extracts.
Full text
PDF
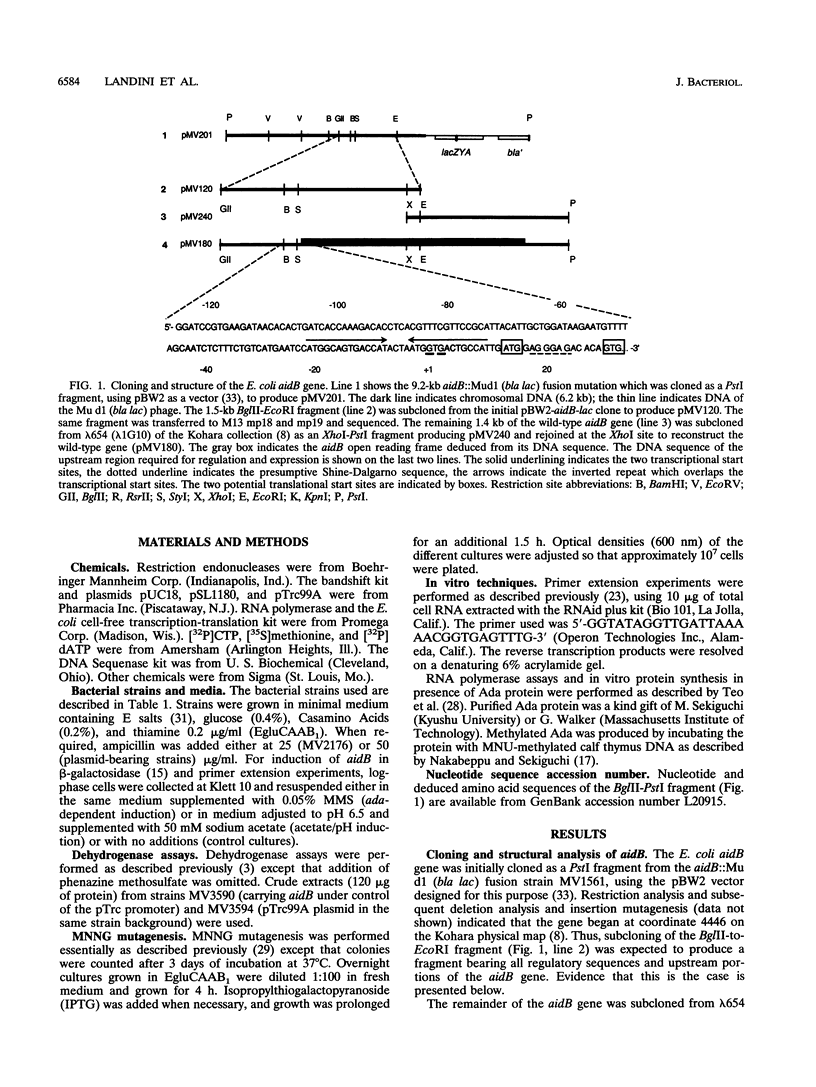


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bakker E. P., Mangerich W. E. The effects of weak acids on potassium uptake by Escherichia coli K-12 inhibition by low cytoplasmic pH. Biochim Biophys Acta. 1983 May 5;730(2):379–386. doi: 10.1016/0005-2736(83)90355-3. [DOI] [PubMed] [Google Scholar]
- Brockman H. L., Wood W. A. Electron-transferring flavoprotein of Peptostreptococcus elsdenii that functions in the reduction of acrylyl-coenzyme A. J Bacteriol. 1975 Dec;124(3):1447–1453. doi: 10.1128/jb.124.3.1447-1453.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evensen G., Seeberg E. Adaptation to alkylation resistance involves the induction of a DNA glycosylase. Nature. 1982 Apr 22;296(5859):773–775. doi: 10.1038/296773a0. [DOI] [PubMed] [Google Scholar]
- Greenberg J. T., Demple B. A global response induced in Escherichia coli by redox-cycling agents overlaps with that induced by peroxide stress. J Bacteriol. 1989 Jul;171(7):3933–3939. doi: 10.1128/jb.171.7.3933-3939.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeggo P. Isolation and characterization of Escherichia coli K-12 mutants unable to induce the adaptive response to simple alkylating agents. J Bacteriol. 1979 Sep;139(3):783–791. doi: 10.1128/jb.139.3.783-791.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kataoka H., Yamamoto Y., Sekiguchi M. A new gene (alkB) of Escherichia coli that controls sensitivity to methyl methane sulfonate. J Bacteriol. 1983 Mar;153(3):1301–1307. doi: 10.1128/jb.153.3.1301-1307.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohara Y., Akiyama K., Isono K. The physical map of the whole E. coli chromosome: application of a new strategy for rapid analysis and sorting of a large genomic library. Cell. 1987 Jul 31;50(3):495–508. doi: 10.1016/0092-8674(87)90503-4. [DOI] [PubMed] [Google Scholar]
- Lange R., Hengge-Aronis R. Identification of a central regulator of stationary-phase gene expression in Escherichia coli. Mol Microbiol. 1991 Jan;5(1):49–59. doi: 10.1111/j.1365-2958.1991.tb01825.x. [DOI] [PubMed] [Google Scholar]
- Lawley P. D. Some chemical aspects of dose-response relationships in alkylation mutagenesis. Mutat Res. 1974 Jun;23(3):283–295. doi: 10.1016/0027-5107(74)90102-x. [DOI] [PubMed] [Google Scholar]
- Lemotte P. K., Walker G. C. Induction and autoregulation of ada, a positively acting element regulating the response of Escherichia coli K-12 to methylating agents. J Bacteriol. 1985 Mar;161(3):888–895. doi: 10.1128/jb.161.3.888-895.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matijasević Z., Hajec L. I., Volkert M. R. Anaerobic induction of the alkylation-inducible Escherichia coli aidB gene involves genes of the cysteine biosynthetic pathway. J Bacteriol. 1992 Mar;174(6):2043–2046. doi: 10.1128/jb.174.6.2043-2046.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsubara Y., Indo Y., Naito E., Ozasa H., Glassberg R., Vockley J., Ikeda Y., Kraus J., Tanaka K. Molecular cloning and nucleotide sequence of cDNAs encoding the precursors of rat long chain acyl-coenzyme A, short chain acyl-coenzyme A, and isovaleryl-coenzyme A dehydrogenases. Sequence homology of four enzymes of the acyl-CoA dehydrogenase family. J Biol Chem. 1989 Sep 25;264(27):16321–16331. [PubMed] [Google Scholar]
- Nakabeppu Y., Miyata T., Kondo H., Iwanaga S., Sekiguchi M. Structure and expression of the alkA gene of Escherichia coli involved in adaptive response to alkylating agents. J Biol Chem. 1984 Nov 25;259(22):13730–13736. [PubMed] [Google Scholar]
- Nakabeppu Y., Sekiguchi M. Regulatory mechanisms for induction of synthesis of repair enzymes in response to alkylating agents: ada protein acts as a transcriptional regulator. Proc Natl Acad Sci U S A. 1986 Sep;83(17):6297–6301. doi: 10.1073/pnas.83.17.6297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura T., Tokumoto Y., Sakumi K., Koike G., Nakabeppu Y., Sekiguchi M. Expression of the ada gene of Escherichia coli in response to alkylating agents. Identification of transcriptional regulatory elements. J Mol Biol. 1988 Aug 5;202(3):483–494. doi: 10.1016/0022-2836(88)90280-x. [DOI] [PubMed] [Google Scholar]
- Navon G., Ogawa S., Shulman R. G., Yamane T. High-resolution 31P nuclear magnetic resonance studies of metabolism in aerobic Escherichia coli cells. Proc Natl Acad Sci U S A. 1977 Mar;74(3):888–891. doi: 10.1073/pnas.74.3.888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oktyabrsky O. N., Golyasnaya N. V., Smirnova G. V., Demakov V. A., Posokhina N. Kh, Kholstova T. A. Acidification of Escherichia coli and Salmonella typhimurium cytoplasm reduces the mutagenic effect of N-methyl-N'-nitro-N-nitrosoguanidine. Mutat Res. 1993 Mar;293(3):197–204. doi: 10.1016/0921-8777(93)90070-w. [DOI] [PubMed] [Google Scholar]
- Olsson M., Lindahl T. Repair of alkylated DNA in Escherichia coli. Methyl group transfer from O6-methylguanine to a protein cysteine residue. J Biol Chem. 1980 Nov 25;255(22):10569–10571. [PubMed] [Google Scholar]
- Sakumi K., Sekiguchi M. Regulation of expression of the ada gene controlling the adaptive response. Interactions with the ada promoter of the Ada protein and RNA polymerase. J Mol Biol. 1989 Jan 20;205(2):373–385. doi: 10.1016/0022-2836(89)90348-3. [DOI] [PubMed] [Google Scholar]
- Samson L., Cairns J. A new pathway for DNA repair in Escherichia coli. Nature. 1977 May 19;267(5608):281–283. doi: 10.1038/267281a0. [DOI] [PubMed] [Google Scholar]
- Sedgwick B., Robins P. Isolation of mutants of Escherichia coli with increased resistance to alkylating agents: mutants deficient in thiols and mutants constitutive for the adaptive response. Mol Gen Genet. 1980;180(1):85–90. doi: 10.1007/BF00267355. [DOI] [PubMed] [Google Scholar]
- Smirnova G. V., Oktyabrsky O. N., Moshonkina E. V., Zakirova N. V. Induction of the alkylation-inducible aidB gene of Escherichia coli by cytoplasmic acidification and N-ethylmaleimide. Mutat Res. 1994 Jan;314(1):51–56. doi: 10.1016/0921-8777(94)90060-4. [DOI] [PubMed] [Google Scholar]
- Tanaka K., Takayanagi Y., Fujita N., Ishihama A., Takahashi H. Heterogeneity of the principal sigma factor in Escherichia coli: the rpoS gene product, sigma 38, is a second principal sigma factor of RNA polymerase in stationary-phase Escherichia coli. Proc Natl Acad Sci U S A. 1993 Apr 15;90(8):3511–3515. doi: 10.1073/pnas.90.8.3511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teo I., Sedgwick B., Kilpatrick M. W., McCarthy T. V., Lindahl T. The intracellular signal for induction of resistance to alkylating agents in E. coli. Cell. 1986 Apr 25;45(2):315–324. doi: 10.1016/0092-8674(86)90396-x. [DOI] [PubMed] [Google Scholar]
- Volkert M. R. Altered induction of the adaptive response to alkylation damage in Escherichia coli recF mutants. J Bacteriol. 1989 Jan;171(1):99–103. doi: 10.1128/jb.171.1.99-103.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volkert M. R., Hajec L. I., Nguyen D. C. Induction of the alkylation-inducible aidB gene of Escherichia coli by anaerobiosis. J Bacteriol. 1989 Feb;171(2):1196–1198. doi: 10.1128/jb.171.2.1196-1198.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volkert M. R., Nguyen D. C., Beard K. C. Escherichia coli gene induction by alkylation treatment. Genetics. 1986 Jan;112(1):11–26. doi: 10.1093/genetics/112.1.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wanner B. L. Molecular cloning of Mu d(bla lacZ) transcriptional and translational fusions. J Bacteriol. 1987 May;169(5):2026–2030. doi: 10.1128/jb.169.5.2026-2030.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]