Abstract
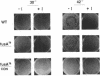
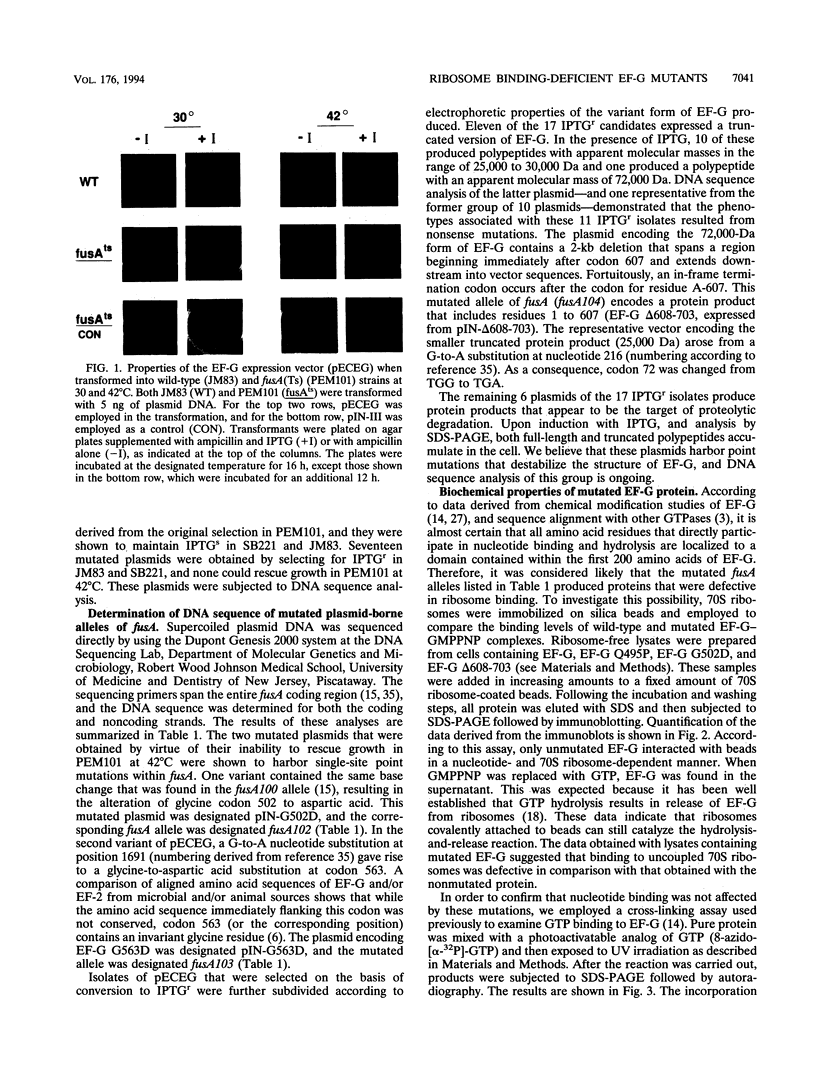
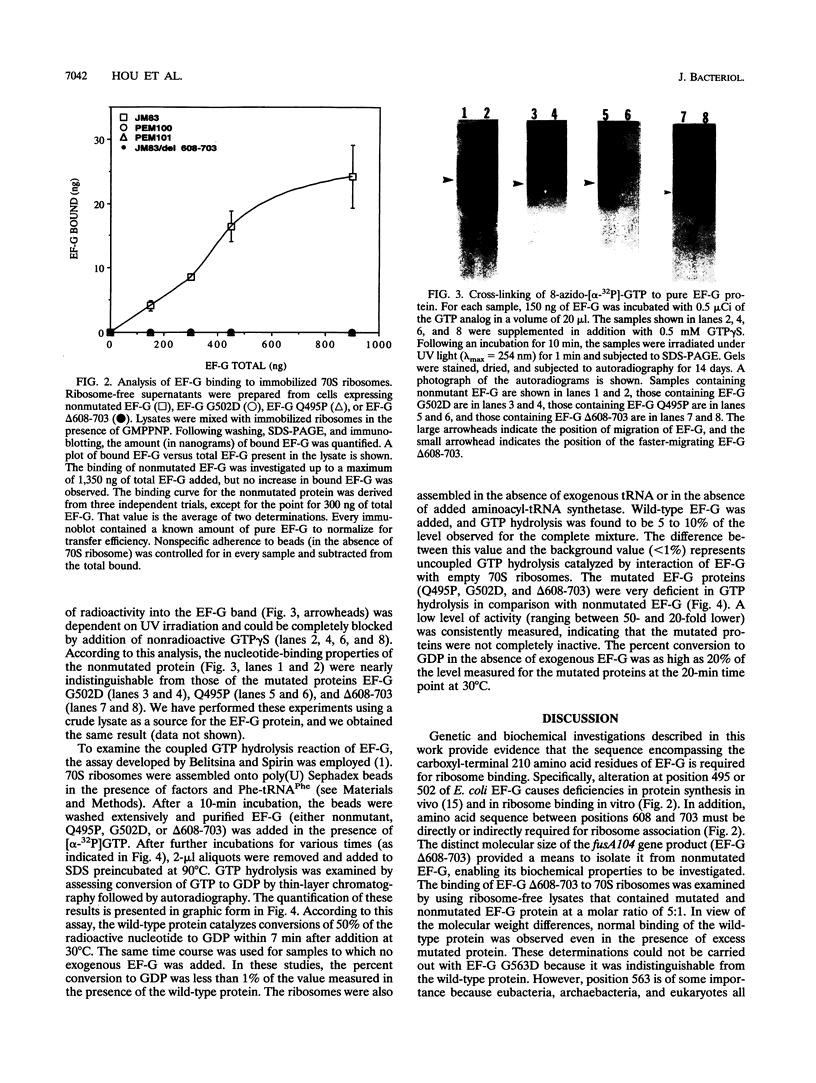
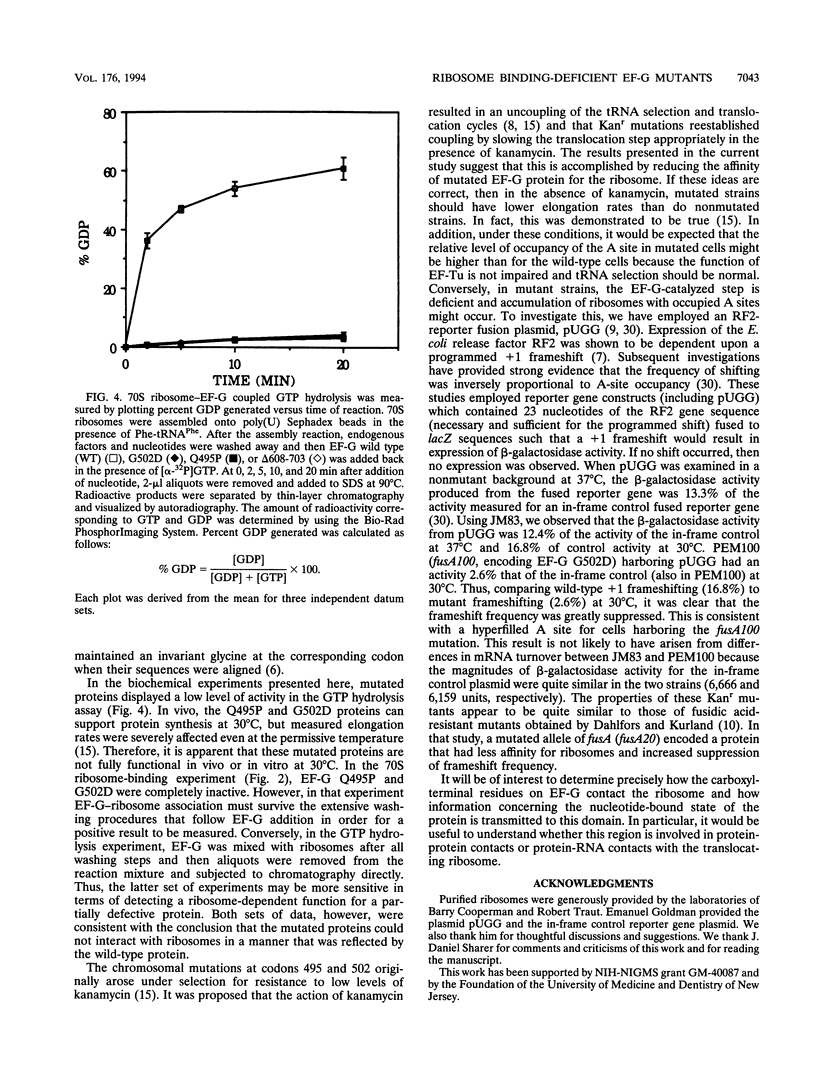
The translocation of ribosomes on mRNA is carried out by cellular machinery that has been extremely well conserved across the entire spectrum of living species. This process requires elongation factor G (EF-G, or EF-2 in archaebacteria and eukaryotes), which is a member of the GTPase superfamily. Using genetic techniques, we have identified a series of mutated alleles of fusA (the Escherichia coli gene that encodes EF-G) that were unable to support protein synthesis in vivo. These alleles encode proteins with point mutations at codons 495 (a variant with a Q-to-P change at codon 495 [Q495P]), 502 (G502D), and 563 (G563D) and a nonsense mutation at codon 608. Biochemical analyses demonstrated that EF-G Q495P, G502D, and delta 608-703 were not disrupted in guanine nucleotide binding but were deficient in ribosome-dependent GTP hydrolysis and guanine nucleotide-dependent ribosome association. We propose that all of these mutations are present in a domain that is essential for ribosome association and that GTP hydrolysis was deficient as a secondary consequence of impaired binding to 70S ribosomes.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Baca O. G., Rohrbach M. S., Bodley J. W. Equilibrium measurements of the interactions of guanine nucleotides with Escherichia coli elongation factor G and the ribosome. Biochemistry. 1976 Oct 19;15(21):4570–4574. doi: 10.1021/bi00666a004. [DOI] [PubMed] [Google Scholar]
- Belitsina N. V., Spirin A. S. Translation of matrix-bound polyuridylic acid by Escherichia coli ribosomes (solid-phase translation system). Methods Enzymol. 1979;60:745–760. doi: 10.1016/s0076-6879(79)60069-1. [DOI] [PubMed] [Google Scholar]
- Boguski M. S., McCormick F. Proteins regulating Ras and its relatives. Nature. 1993 Dec 16;366(6456):643–654. doi: 10.1038/366643a0. [DOI] [PubMed] [Google Scholar]
- Bourne H. R., Sanders D. A., McCormick F. The GTPase superfamily: a conserved switch for diverse cell functions. Nature. 1990 Nov 8;348(6297):125–132. doi: 10.1038/348125a0. [DOI] [PubMed] [Google Scholar]
- Bourne H. R., Sanders D. A., McCormick F. The GTPase superfamily: conserved structure and molecular mechanism. Nature. 1991 Jan 10;349(6305):117–127. doi: 10.1038/349117a0. [DOI] [PubMed] [Google Scholar]
- Bubunenko M. G., Gudkov A. T. Elongation factors Tu and G change their conformation on interaction with ribosomes. Biomed Sci. 1990 Feb;1(2):127–132. [PubMed] [Google Scholar]
- Cammarano P., Palm P., Creti R., Ceccarelli E., Sanangelantoni A. M., Tiboni O. Early evolutionary relationships among known life forms inferred from elongation factor EF-2/EF-G sequences: phylogenetic coherence and structure of the archaeal domain. J Mol Evol. 1992 May;34(5):396–405. doi: 10.1007/BF00162996. [DOI] [PubMed] [Google Scholar]
- Craigen W. J., Caskey C. T. Expression of peptide chain release factor 2 requires high-efficiency frameshift. Nature. 1986 Jul 17;322(6076):273–275. doi: 10.1038/322273a0. [DOI] [PubMed] [Google Scholar]
- Curran J. F., Yarus M. Rates of aminoacyl-tRNA selection at 29 sense codons in vivo. J Mol Biol. 1989 Sep 5;209(1):65–77. doi: 10.1016/0022-2836(89)90170-8. [DOI] [PubMed] [Google Scholar]
- De Vendittis E., Masullo M., Bocchini V. The elongation factor G carries a catalytic site for GTP hydrolysis, which is revealed by using 2-propanol in the absence of ribosomes. J Biol Chem. 1986 Apr 5;261(10):4445–4450. [PubMed] [Google Scholar]
- Duffaud G. D., March P. E., Inouye M. Expression and secretion of foreign proteins in Escherichia coli. Methods Enzymol. 1987;153:492–507. doi: 10.1016/0076-6879(87)53074-9. [DOI] [PubMed] [Google Scholar]
- Girshovich A. S., Bochkareva E. S., Kurtskhalia T. V., Pozdnyakov V. A., Ovchinnikov Y. A. Binding of GTP to elongation factor G by photoaffinity labeling. Methods Enzymol. 1979;60:726–745. doi: 10.1016/s0076-6879(79)60068-x. [DOI] [PubMed] [Google Scholar]
- Hou Y., Lin Y. P., Sharer J. D., March P. E. In vivo selection of conditional-lethal mutations in the gene encoding elongation factor G of Escherichia coli. J Bacteriol. 1994 Jan;176(1):123–129. doi: 10.1128/jb.176.1.123-129.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humphreys G. O., Willshaw G. A., Smith H. R., Anderson E. S. Mutagenesis of plasmid DNA with hydroxylamine: isolation of mutants of multi-copy plasmids. Mol Gen Genet. 1976 Apr 23;145(1):101–108. doi: 10.1007/BF00331564. [DOI] [PubMed] [Google Scholar]
- Jurnak F. Structure of the GDP domain of EF-Tu and location of the amino acids homologous to ras oncogene proteins. Science. 1985 Oct 4;230(4721):32–36. doi: 10.1126/science.3898365. [DOI] [PubMed] [Google Scholar]
- Kaziro Y., Inoue N., Kuriki Y., Mizumoto K., Tanaka M., Kawakita M. Purification and properties of factor G. Cold Spring Harb Symp Quant Biol. 1969;34:385–393. doi: 10.1101/sqb.1969.034.01.045. [DOI] [PubMed] [Google Scholar]
- Kaziro Y. The role of guanosine 5'-triphosphate in polypeptide chain elongation. Biochim Biophys Acta. 1978 Sep 21;505(1):95–127. doi: 10.1016/0304-4173(78)90009-5. [DOI] [PubMed] [Google Scholar]
- March P. E., Inouye M. Characterization of the lep operon of Escherichia coli. Identification of the promoter and the gene upstream of the signal peptidase I gene. J Biol Chem. 1985 Jun 25;260(12):7206–7213. [PubMed] [Google Scholar]
- March P. E., Lerner C. G., Ahnn J., Cui X., Inouye M. The Escherichia coli Ras-like protein (Era) has GTPase activity and is essential for cell growth. Oncogene. 1988 Jun;2(6):539–544. [PubMed] [Google Scholar]
- Milburn M. V., Tong L., deVos A. M., Brünger A., Yamaizumi Z., Nishimura S., Kim S. H. Molecular switch for signal transduction: structural differences between active and inactive forms of protooncogenic ras proteins. Science. 1990 Feb 23;247(4945):939–945. doi: 10.1126/science.2406906. [DOI] [PubMed] [Google Scholar]
- Parmeggiani A., Gottschalk E. M. Isolation and some properties of the amino acid polymerization factors from Escherichia coli. Cold Spring Harb Symp Quant Biol. 1969;34:377–384. doi: 10.1101/sqb.1969.034.01.044. [DOI] [PubMed] [Google Scholar]
- Richter Dahlfors A. A., Kurland C. G. Novel mutants of elongation factor G. J Mol Biol. 1990 Oct 20;215(4):549–557. doi: 10.1016/s0022-2836(05)80167-6. [DOI] [PubMed] [Google Scholar]
- Rohrbach M. S., Bodley J. W. Isolation of physically and enzymically homogeneous Escherichia coli elongation factor G. Methods Enzymol. 1979;60:606–614. doi: 10.1016/s0076-6879(79)60057-5. [DOI] [PubMed] [Google Scholar]
- Rohrbach M. S., Bodley J. W. Selective chemical modification of Escherichia coli elongation factor G. N-Ethylmaleimide modification of a cysteine essential for nucleotide binding. J Biol Chem. 1976 Feb 25;251(4):930–933. [PubMed] [Google Scholar]
- Schilling-Bartetzko S., Bartetzko A., Nierhaus K. H. Kinetic and thermodynamic parameters for tRNA binding to the ribosome and for the translocation reaction. J Biol Chem. 1992 Mar 5;267(7):4703–4712. [PubMed] [Google Scholar]
- Sipley J., Goldman E. Increased ribosomal accuracy increases a programmed translational frameshift in Escherichia coli. Proc Natl Acad Sci U S A. 1993 Mar 15;90(6):2315–2319. doi: 10.1073/pnas.90.6.2315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weijland A., Harmark K., Cool R. H., Anborgh P. H., Parmeggiani A. Elongation factor Tu: a molecular switch in protein biosynthesis. Mol Microbiol. 1992 Mar;6(6):683–688. doi: 10.1111/j.1365-2958.1992.tb01516.x. [DOI] [PubMed] [Google Scholar]
- Weissbach H., Brot N., Miller D., Rosman M., Ertel R. Interaction of guanosine triphosphate with E. coli soluble transfer factors. Cold Spring Harb Symp Quant Biol. 1969;34:419–431. doi: 10.1101/sqb.1969.034.01.048. [DOI] [PubMed] [Google Scholar]
- Wittinghofer A., Pai E. F. The structure of Ras protein: a model for a universal molecular switch. Trends Biochem Sci. 1991 Oct;16(10):382–387. doi: 10.1016/0968-0004(91)90156-p. [DOI] [PubMed] [Google Scholar]
- Zengel J. M., Archer R. H., Lindahl L. The nucleotide sequence of the Escherichia coli fus gene, coding for elongation factor G. Nucleic Acids Res. 1984 Feb 24;12(4):2181–2192. doi: 10.1093/nar/12.4.2181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Vos A. M., Tong L., Milburn M. V., Matias P. M., Jancarik J., Noguchi S., Nishimura S., Miura K., Ohtsuka E., Kim S. H. Three-dimensional structure of an oncogene protein: catalytic domain of human c-H-ras p21. Science. 1988 Feb 19;239(4842):888–893. doi: 10.1126/science.2448879. [DOI] [PubMed] [Google Scholar]