Abstract
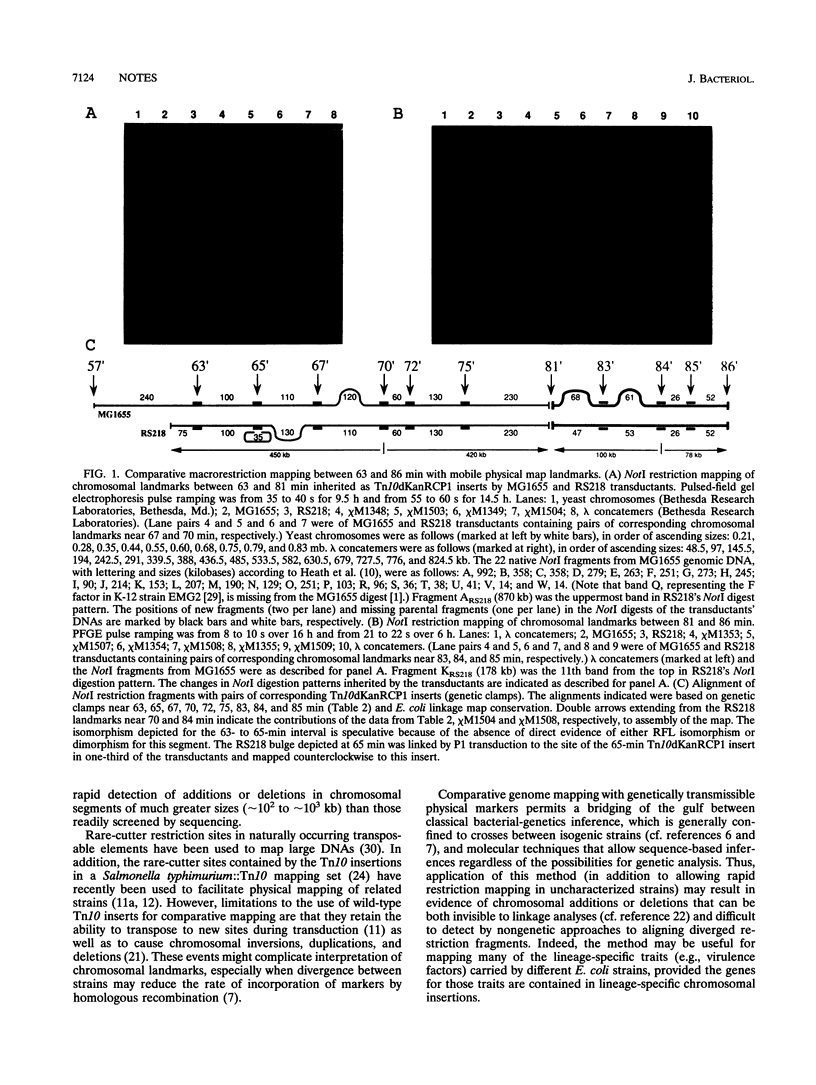
We describe a method for comparative macrorestriction mapping of the chromosomes of Escherichia coli strains. In this method, a series of physically tagged E. coli K-12 alleles serve as mobile landmarks for mapping DNAs from other strains. This technique revealed evidence of strain-specific chromosomal additions or deletions in a pathogenic isolate and can be applied to most strains, yielding information on genealogy as well as virulence. In theory, the same strategy can be used to map and compare genomic DNAs from a wide variety of species.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bachmann B. J. Linkage map of Escherichia coli K-12, edition 8. Microbiol Rev. 1990 Jun;54(2):130–197. doi: 10.1128/mr.54.2.130-197.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bender J., Kleckner N. IS10 transposase mutations that specifically alter target site recognition. EMBO J. 1992 Feb;11(2):741–750. doi: 10.1002/j.1460-2075.1992.tb05107.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloch C. A., Thorne G. M., Ausubel F. M. General method for site-directed mutagenesis in Escherichia coli O18ac:K1:H7: deletion of the inducible superoxide dismutase gene, sodA, does not diminish bacteremia in neonatal rats. Infect Immun. 1989 Jul;57(7):2141–2148. doi: 10.1128/iai.57.7.2141-2148.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DEMEREC M., OHTA N. GENETIC ANALYSES OF SALMONELLA TYPHIMURIUM X ESCHERICHIA COLI HYBRIDS. Proc Natl Acad Sci U S A. 1964 Aug;52:317–323. doi: 10.1073/pnas.52.2.317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demerec M., Adelberg E. A., Clark A. J., Hartman P. E. A proposal for a uniform nomenclature in bacterial genetics. Genetics. 1966 Jul;54(1):61–76. doi: 10.1093/genetics/54.1.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dower W. J., Miller J. F., Ragsdale C. W. High efficiency transformation of E. coli by high voltage electroporation. Nucleic Acids Res. 1988 Jul 11;16(13):6127–6145. doi: 10.1093/nar/16.13.6127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guyer M. S., Reed R. R., Steitz J. A., Low K. B. Identification of a sex-factor-affinity site in E. coli as gamma delta. Cold Spring Harb Symp Quant Biol. 1981;45(Pt 1):135–140. doi: 10.1101/sqb.1981.045.01.022. [DOI] [PubMed] [Google Scholar]
- Heath J. D., Perkins J. D., Sharma B., Weinstock G. M. NotI genomic cleavage map of Escherichia coli K-12 strain MG1655. J Bacteriol. 1992 Jan;174(2):558–567. doi: 10.1128/jb.174.2.558-567.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleckner N., Barker D. F., Ross D. G., Botstein D. Properties of the translocatable tetracycline-resistance element Tn10 in Escherichia coli and bacteriophage lambda. Genetics. 1978 Nov;90(3):427–461. doi: 10.1093/genetics/90.3.427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S. L., Hessel A., Cheng H. Y., Sanderson K. E. The XbaI-BlnI-CeuI genomic cleavage map of Salmonella paratyphi B. J Bacteriol. 1994 Feb;176(4):1014–1024. doi: 10.1128/jb.176.4.1014-1024.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S. L., Hessel A., Sanderson K. E. The XbaI-BlnI-CeuI genomic cleavage map of Salmonella enteritidis shows an inversion relative to Salmonella typhimurium LT2. Mol Microbiol. 1993 Nov;10(3):655–664. doi: 10.1111/j.1365-2958.1993.tb00937.x. [DOI] [PubMed] [Google Scholar]
- Mahillon J., Kleckner N. New IS10 transposition vectors based on a gram-positive replication origin. Gene. 1992 Jul 1;116(1):69–74. doi: 10.1016/0378-1119(92)90630-8. [DOI] [PubMed] [Google Scholar]
- Milkman R., Bridges M. M. Molecular evolution of the Escherichia coli chromosome. IV. Sequence comparisons. Genetics. 1993 Mar;133(3):455–468. doi: 10.1093/genetics/133.3.455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson K., Selander R. K. Evolutionary genetics of the proline permease gene (putP) and the control region of the proline utilization operon in populations of Salmonella and Escherichia coli. J Bacteriol. 1992 Nov;174(21):6886–6895. doi: 10.1128/jb.174.21.6886-6895.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada N., Sasakawa C., Tobe T., Talukder K. A., Komatsu K., Yoshikawa M. Construction of a physical map of the chromosome of Shigella flexneri 2a and the direct assignment of nine virulence-associated loci identified by Tn5 insertions. Mol Microbiol. 1991 Sep;5(9):2171–2180. doi: 10.1111/j.1365-2958.1991.tb02147.x. [DOI] [PubMed] [Google Scholar]
- Perkins J. D., Heath J. D., Sharma B. R., Weinstock G. M. SfiI genomic cleavage map of Escherichia coli K-12 strain MG1655. Nucleic Acids Res. 1992 Mar 11;20(5):1129–1137. doi: 10.1093/nar/20.5.1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perkins J. D., Heath J. D., Sharma B. R., Weinstock G. M. XbaI and BlnI genomic cleavage maps of Escherichia coli K-12 strain MG1655 and comparative analysis of other strains. J Mol Biol. 1993 Jul 20;232(2):419–445. doi: 10.1006/jmbi.1993.1401. [DOI] [PubMed] [Google Scholar]
- Ross D. G., Swan J., Kleckner N. Physical structures of Tn10-promoted deletions and inversions: role of 1400 bp inverted repetitions. Cell. 1979 Apr;16(4):721–731. doi: 10.1016/0092-8674(79)90088-6. [DOI] [PubMed] [Google Scholar]
- Rothman J. L. Transduction studies on the relation between prophage and host chromosome. J Mol Biol. 1965 Jul;12(3):892–912. doi: 10.1016/s0022-2836(65)80336-9. [DOI] [PubMed] [Google Scholar]
- Sanderson K. E., Roth J. R. Linkage map of Salmonella typhimurium, edition VII. Microbiol Rev. 1988 Dec;52(4):485–532. doi: 10.1128/mr.52.4.485-532.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segall A., Mahan M. J., Roth J. R. Rearrangement of the bacterial chromosome: forbidden inversions. Science. 1988 Sep 9;241(4871):1314–1318. doi: 10.1126/science.3045970. [DOI] [PubMed] [Google Scholar]
- Silver R. P., Aaronson W., Sutton A., Schneerson R. Comparative analysis of plasmids and some metabolic characteristics of Escherichia coli K1 from diseased and healthy individuals. Infect Immun. 1980 Jul;29(1):200–206. doi: 10.1128/iai.29.1.200-206.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singer M., Baker T. A., Schnitzler G., Deischel S. M., Goel M., Dove W., Jaacks K. J., Grossman A. D., Erickson J. W., Gross C. A. A collection of strains containing genetically linked alternating antibiotic resistance elements for genetic mapping of Escherichia coli. Microbiol Rev. 1989 Mar;53(1):1–24. doi: 10.1128/mr.53.1.1-24.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith C. L., Econome J. G., Schutt A., Klco S., Cantor C. R. A physical map of the Escherichia coli K12 genome. Science. 1987 Jun 12;236(4807):1448–1453. doi: 10.1126/science.3296194. [DOI] [PubMed] [Google Scholar]
- Smith C. L., Kolodner R. D. Mapping of Escherichia coli chromosomal Tn5 and F insertions by pulsed field gel electrophoresis. Genetics. 1988 Jun;119(2):227–236. doi: 10.1093/genetics/119.2.227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sternberg N. L., Maurer R. Bacteriophage-mediated generalized transduction in Escherichia coli and Salmonella typhimurium. Methods Enzymol. 1991;204:18–43. doi: 10.1016/0076-6879(91)04004-8. [DOI] [PubMed] [Google Scholar]
- Westmoreland B. C., Szybalski W., Ris H. Mapping of deletions and substitutions in heteroduplex DNA molecules of bacteriophage lambda by electron microscopy. Science. 1969 Mar 21;163(3873):1343–1348. doi: 10.1126/science.163.3873.1343. [DOI] [PubMed] [Google Scholar]



